Abstract
Transcription initiation is a critical step in bacterial gene regulation and is often controlled by transcription regulators. The alternate sigma factor (σ54) is one such regulator that facilitates activator-dependent transcription initiation and thus modulates the expression of a variety of genes involved in metabolism and pathogenesis in bacteria. This study describes the role of σ54 in the nosocomial pathogen Enterococcus faecalis. Biofilm formation is one of the important pathogenic mechanisms of E. faecalis, as it elevates the organism's potential to cause surgical site and urinary tract infections. Lysis of bacterial cells within the population contributes to biofilm formation by providing extracellular DNA (eDNA) as a key component of the biofilm matrix. Deletion of rpoN rendered E. faecalis resistant to autolysis, which in turn impaired eDNA release. Despite the significant reduction in eDNA levels compared to the parental strain, the rpoN mutant formed more robust biofilms as observed using laser scanning confocal microscopy and Comstat analysis, indicating and emphasizing the presence of other matrix components. Initial adherence to a polystyrene surface was also enhanced in the mutant. Proteinase K treatment at early stages of biofilm development significantly reduced the accumulation of biofilm by the rpoN mutant. In conclusion, our data indicate that other factors in addition to eDNA might contribute to the overall composition of the enterococcal biofilm and that the regulatory role of σ54 governs the nature and composition of the biofilm matrix.
INTRODUCTION
As opportunistic pathogens, enterococci are the third leading cause of hospital-acquired or associated infections, as they are responsible for 11.2% of surgical site infections (SSI), 14.9% of urinary tract infections (UTI), and 16% of reported bloodstream infections (25). The ability to form a biofilm is an important aspect of the lifestyle of the organism, as biofilm formation is thought to be a property associated with the establishment of SSI and UTI (34), both of which can serve as foci to establish bloodstream infections. Biofilms are aggregates of bacteria that are covered in exoploymer matrix and are more resistant to antibiotics than their planktonic counterparts (15, 26). In several bacterial species, nucleic acids, polysaccharides, proteins, and lipids constitute the exopolymer matrix (19). The components of the biofilm matrix form a physical barrier that enhances the inaccessibility of the biofilm cells to antibiotics and the immune system, thereby making the infection difficult to eradicate (33). Extracellular DNA (eDNA) serves as an important biofilm matrix component in several microbial model systems, including but not limited to Neisseria meningitidis, Listeria monocytogenes, Pseudomonas aeruginosa, Enterococcus faecalis, Staphylococcus aureus, and Staphylococcus epidermidis (2, 23, 29, 32, 36, 47, 48, 54). The expression of the two secreted E. faecalis proteases, gelatinase and serine protease, is regulated in a quorum-dependent manner by the Fsr regulatory system (22, 45, 46), and these proteases direct biofilm development by modulating the eDNA matrix via regulation of the extent of autolysis (54) in a fratricidal manner (51). In an attempt to identify other factors that govern eDNA release in E. faecalis, we identified rpoN, which encodes σ54, in a preliminary transposon mutagenesis screen.
Transcription initiation is one of the important stages of gene regulation, and sigma factors play a crucial role in determining the controlled response of a subset of genes tied to a given environmental stimulus. Sigma factors reversibly bind to RNA polymerases and drive promoter-specific transcription initiation. In prokaryotes, two distinct families of sigma factors have been studied, sigma 70 (σ70) and σ54. The σ70 family also includes several related alternate sigma factors. Sigma 54 shares no structural homology with sigma 70, possesses a distinct consensus binding sequence (−24/−12; TTGGCACNNNNNTTGCT) and, unlike sigma 70, facilitates activator-dependent transcription initiation (24, 38).
Sigma 54 plays an important role in the virulence of several bacteria but does not share the same function in all pathogens (30). In Vibrio fischeri, σ54 influences biofilm formation, motility, and symbiotic colonization of squid and negatively regulates bioluminescence (58). Quorum-sensing regulation in Vibrio cholerae O1 strains is dependent on rpoN (28). Sigma 54 is required for biofilm formation by Burkholderia cenocepacia as well as its survival within macrophages (51). In the major food-borne pathogen Listeria monocytogenes, σ54 is essential for its osmotolerance potential (41) and is responsible for mesentericin sensitivity (14, 41), whereas in Pseudomonas aeruginosa, σ54 influences the activity of isocitrate lyase (21), alginate biosynthesis (6), and pilin and flagellin production, in addition to several other virulence determinants (44). Sigma 54 also regulates biofilm formation, enterocyte effacement, acid tolerance, flagellar biosynthesis, and several other processes in Escherichia coli (3, 49, 60).
In E. faecalis, σ54 is responsible for sensitivity to class IIa bacteriocins, such as mesentericin and divercin (9, 13). The basis for the class IIa bacteriocin sensitivity is due to the role of σ54 in regulating four distinct sugar phosphotransferase systems (PTSs) that are dependent on four known σ54 enhancer binding proteins (LpoR, MphR, MpoR, and MptR) (13). MptD, a component of the mannose PTS, is thought to serve as the cellular receptor for the class IIa bacteriocins (24). However, additional roles for σ54 in enterococcal biology remain to be elucidated.
In this study, we investigated the role of σ54 in eDNA release, autolysis, and biofilm formation, and we demonstrate a functional role for σ54 in regulating initial adherence of cells to substrate as well as the overall composition of the biofilm matrix.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Tables 1 and 2, respectively. E. coli Electro10 Blue (E10-Blue; Stratagene) was used for construction of plasmids and was cultured in Luria-Bertani (LB) broth supplemented with appropriate antibiotics. E. faecalis strains were cultured in either Trypticase soy broth containing 0.25% glucose (TSB) or Todd-Hewitt broth (THB; BD Biosciences) containing appropriate antibiotics whenever required. Chloramphenicol (Cm) and spectinomycin (Spec) were used for selection of E. coli at concentrations of 10 μg/ml and 150 μg/ml, respectively. For E. faecalis, Cm, Spec, and tetracycline (Tet) were used at 15 μg/ml, 500 μg/ml, and 15 μg/ml, respectively. When required, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Amresco) was used at a concentration of 80 μg/ml for both E. coli and E. faecalis.
Table 1.
E. faecalis strains used in this study
Table 2.
Plasmid constructs used in this study
In-frame markerless deletion of rpoN.
An E. coli-Enterococcus temperature-sensitive cloning vector, pLT06 (56), was used to generate an isogenic in-frame deletion of rpoN in E. faecalis V583. Upstream and downstream regions flanking rpoN (ef0782) were amplified by PCR from a V583 genomic template by using the primer pair RpoNP1/RpoNP2 and RpoNP3/RpoNP4, respectively (Table 3). The primers RpoNP1/RpoNP2 and RpoNP3/RpoNP4 were designed with EcoRI/BamHI and BamHI/PstI restriction sites, respectively. The resultant PCR products were digested with BamHI, ligated, and reamplified with primers RpoNP1 and RpoNP4. For the construction of the deletion vector, the amplified product was digested with EcoRI and PstI followed by ligation to similarly digested pLT06. The ligation was electroporated into competent E10-Blue cells for propagation, and blue colonies were selected on LB agar containing chloramphenicol and X-Gal at 30°C. Clones were screened for the appropriate insert by using the primers OriF and SeqR. A positive plasmid designated pKS70 was confirmed by restriction digestion and electroporated into E. faecalis V583 cells (12), and VI01 was subsequently generated following the protocol previously described (56) and confirmed by PCR using the primers RpoNUp and RpoNDown. Using pKS70, ∼98% of the rpoN gene was deleted, leaving seven codons at the 5′ end and two codons at the 3′ end. The next adjacent gene is ef0783, which encodes an O-acetyltransferase. This gene is located approximately 200 bp downstream of rpoN, and the strategy used to delete rpoN does not alter the expression of ef0783 (data not shown).
Table 3.
Oligonucleotides used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| RpoNP1 | GAGAGAATTCACAACGGTACAGTAAAATGG |
| RpoNP2 | CTCTGGAATCCCATTCGTTGCTCAAATTTCAT |
| RpoNP3 | GAGAGGATCCGAGTAAACAACCAAAGATTAT |
| RpoNP4 | CTCTCTGCAGGAACTAAGGCACTTAAACCA |
| RpoN UP | AGTCCAAGGAAGAGTCGTG |
| RpoN DOWN | AAGACAGTGGCTGCCAAAC |
| OriF | CAATAATCGCATCCGATTGCA |
| SeqR | CCTATTATACCATATTTTGGAC |
Markerless complementation of VI01 (ΔrpoN).
The temperature-sensitive cloning vector pLT06 (56) was used to generate markerless gene complementation of rpoN in VI01. The rpoN gene (ef0782) along with flanking regions was amplified by PCR from a V583 genomic template using primers RpoNP1 and RpoNP4 (Table 3). For the construction of rpoN markerless complementation vector pVI12, the amplified product was digested with EcoRI and PstI followed by ligation with similarly digested plasmid vector pLT06. The ligation was electroporated into competent E10-Blue cells for propagation, and blue colonies were selected on LB agar containing chloramphenicol and X-Gal at room temperature. Clones were screened for the appropriate insert by using the primers OriF and SeqR. A positive plasmid designated pVI12 was confirmed by restriction digestion and electroporated into E. faecalis VI01 cells (12), and VI40 (markerless complement) was generated following the protocol previously described (56) and was confirmed by PCR using primers RpoNUp and RpoNDown.
2DG resistance.
E. faecalis V583, VI01, and VI40 were grown on LB agar containing 0.2% fructose and 10 mM 2-deoxy-d-glucose (2DG) (24). 2DG is a toxic homologue of glucose and enters cells via the mannose PTS permease (5). In E. faecalis, mannose PTS expression is controlled by σ54. Strains resistant to 2DG do not express a functional mannose PTS permease (24). Hence, growth on medium containing 2DG was used as a marker to confirm deletion of rpoN.
Detection and precipitation of extracellular DNA.
Overnight cultures were centrifuged for 10 min at 13,000 rpm, and the resulting supernatant was filtered (0.2-μm pore size; Nalgene) to obtain cell-free supernatants. The supernatants were tested for the presence of eDNA by using 1 μM SYTOX Green (Invitrogen, Molecular Probes).
The eDNA was also precipitated from the culture filtrate with an equal volume of isopropanol. The precipitated eDNA was washed in 75% ethanol, air dried, and dissolved in TE buffer (10 mM Tris-Cl, 1 mM EDTA; pH 8.0) and visualized on 1% agarose gels after staining with ethidium bromide.
Autolysis assay.
The autolysis assay was performed as previously described (15).
Quantitative detection of eDNA in biofilms.
eDNA in biofilms was quantified using a previously described protocol with some modifications (36). Briefly, biofilms were grown in a 96-well polystyrene plate in TSB for 24 h at 37°C. After 24 h, the supernatant was discarded and the biofilm was suspended in resuspension buffer (50 mM Tris-Cl [pH 8], 10 mM EDTA, 500 mM NaCl). The resuspended biofilm was centrifuged, and eDNA was quantified in the supernatant with 1 μM SYTOX Green (Invitrogen, Molecular Probes).
CSLM.
Confocal laser scanning microscopy (CLSM) was performed on 1-day-old biofilms as described previously (54). E. faecalis strains VI01 and VI40 were transformed with pMV158GFP (39) to generate VI29 and VI41, respectively, both of which expressed green fluorescent protein (GFP) constitutively. VT09 [V583(pMV158GFP)] (54) along with VI29 and VI41 were used for confocal imaging. Briefly, biofilms were grown on sterile glass coverslips placed in six-well tissue culture plates. The coverslip was submerged in 5 ml of TSB containing tetracycline for plasmid maintenance. After 24 h of growth, the biofilm was gently washed with sterile phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.4]) and stained with 1 μM SYTOX orange (Invitrogen) for 6 to 7 min. The coverslip were inverted on a clean glass slide and sealed using clear nail polish. The biofilm was visualized using a Zeiss LSM 5 Pa laser scanning confocal microscope.
Macroscopic biofilm.
To visualize the biofilms formed by VT09, VI29, and VI41 macroscopically, biofilms were grown as described for confocal analysis, with the exception that after 24 h of growth, the biofilms were gently washed with sterile phosphate buffer, then fresh TSB was added, and the biofilms were grown for an additional 24 h, at which time the biofilm was washed and imaged with an AlphaImager system (Alpha Innotech, San Leandro, CA).
Adherence assay.
Adherence of E. faecalis strains to flat-bottom polystyrene plates (BRAND, Germany) was tested using a previously described protocol (27) with some modifications. Cultures grown overnight were diluted 1:10 in fresh TSB, and 200 μl was transferred to a flat-bottom 96-well polystyrene microtiter plate. After 2 h of incubation at 37°C, the supernatant was discarded and the wells were gently washed with sterile PBS. The adherent cells were resuspended in 200 μl PBS by vigorous pipetting, diluted, and plated on THB agar for colony counting. Also, the initial load was calculated by plating the diluted culture on THB agar for colony counting. The adherence potential of the strains was calculated as the percentage of initial load that adhered.
Proteinase K treatment of biofilm.
Biofilms were grown on 96-well round-bottom tissue culture-treated polystyrene plates (Techno Plastic Products, Switzerland) as previously described (22). At 6, 12, and 24 h, the biofilms were treated with 1 μg/ml proteinase K (Amresco), and this treatment remained for the remainder of the experiment. The 24-h treatment was allowed to stand for 1 h prior to processing the biofilm. An untreated control was included to determine the effect of treatment. After 24 h of growth, the biofilm was quantified by the crystal violet staining method (22). Each assay was performed in triplicate and repeated four times.
Statistical analysis.
Statistical analysis of quantitative detection of eDNA, adherence assay results, and Comstat analysis of biofilms were performed using GraphPad Prism 4 software (San Diego, CA). One-way analysis of variance followed by Dunn's multiple comparison tests was performed to determine statistical significance.
RESULTS
Construction of the E. faecalis V583 isogenic rpoN mutant and its complement.
The rpoN deletion mutant VI01 (ΔrpoN) was constructed using the markerless deletion vector pKS70. Initial growth curves of the wild-type V583 strain, the rpoN deletion mutant (VI01), and its complement (VI40) were assessed in TSB. No alterations in the growth of the 3 strains were observed (see Fig. S1 in the supplemental material). The 2DG-resistant phenotype was confirmed by growth on medium containing 2DG. VI01 grew to the final dilution of 10−8, while the parental strain V583 and the rpoN complement VI40 were significantly inhibited and grew only at dilutions of 10−3 and 10−4. Complementation confirmed that there were no polar effects of the gene deletion and attributed the 2DG-resistant phenotype to the targeted deletion of rpoN (Fig. 1).
Fig 1.
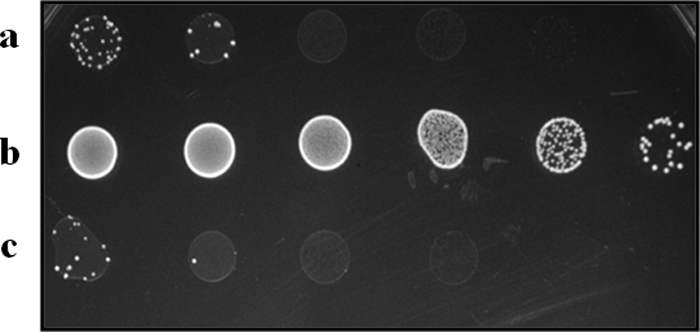
2DG resistance analysis of VI01. Wild-type strain, V583 (a) and the complement strain VI40 (c) are sensitive to 2DG because of a functional mpt operon under the control of intact rpoN. rpoN mutant VI01 (b) is resistant to 2DG. This confirms the deletion and complementation of rpoN in E. faecalis.
Sigma 54 alters eDNA in the supernatant of planktonic and biofilm cultures.
On the basis of phenotype characterization from a preliminary transposon mutagenesis screen, we tested for eDNA in the supernatants of planktonic cultures by using SYTOX Green. A smaller amount of eDNA was detected in VI01 culture supernatants than with the wild-type V583 strain, whereas the markerless complementation of the rpoN mutant restored the phenotype to wild type (Fig. 2A). eDNA in the supernatants of the various strains was also confirmed by visualization on an ethidium bromide-stained 1% agarose gel after precipitation of eDNA with isopropanol (Fig. 2B).
Fig 2.
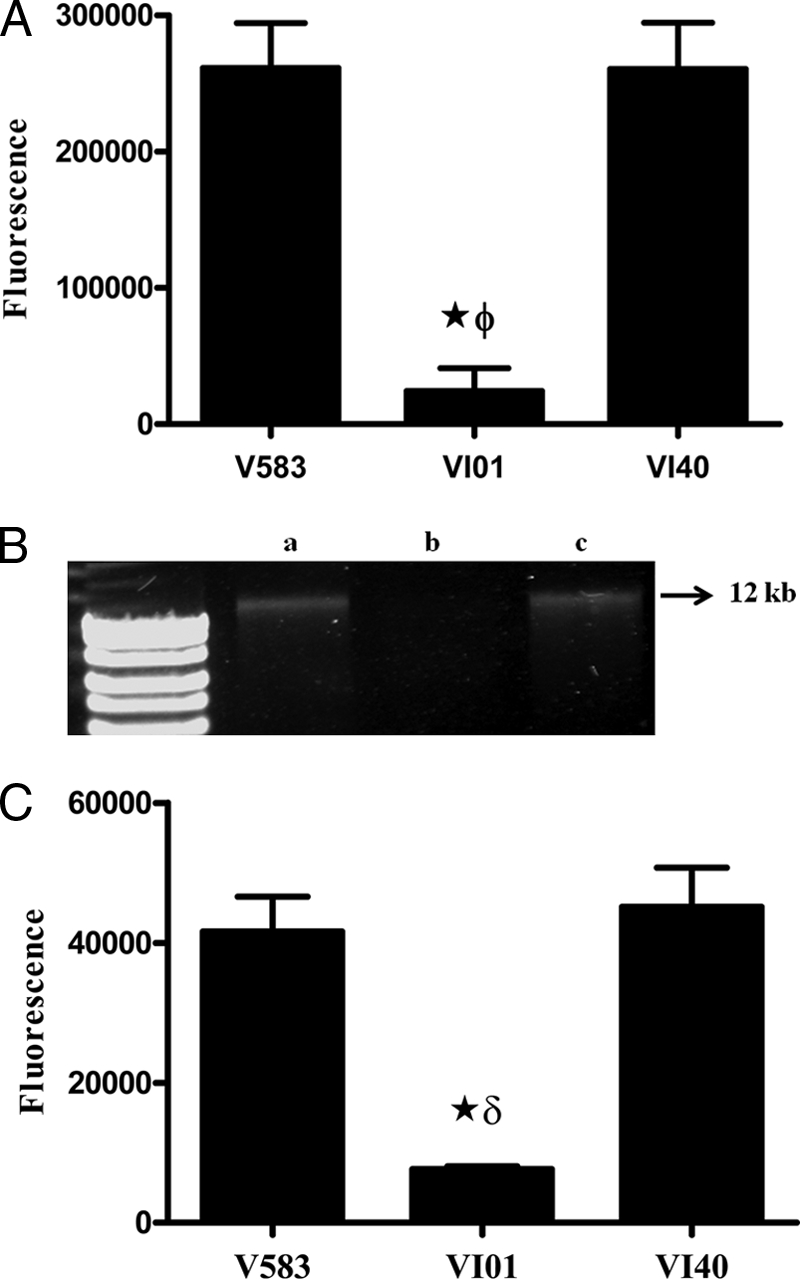
(A) Quantitative detection of eDNA in culture supernatants with SYTOX Green. eDNA was quantified in the culture supernatants of cultures grown overnight by using 1 μM SYTOX Green. Assays were performed in quadruplicate, and error bars indicate the standard errors of the means. ★, significant decrease (P < 0.001) relative to wild-type V583; ϕ, significant decrease (P < 0.001) relative to rpoN complement (VI40). (B) Qualitative detection of eDNA in culture supernatant by isopropanol precipitation. Lane a, wild type (V583); lane b, rpoN mutant (VI01); lane c, rpoN complement (VI40). (C) Detection of eDNA in biofilm by using SYTOX Green. Assays were performed in sextuplets, and error bars indicate standard errors of means. ★, significant decrease (P < 0.001) relative to wild-type V583; δ, significant decrease (P < 0.001) relative to the rpoN complement (VI40).
Given the fact that planktonic growth and biofilms are two different lifestyles of the bacterium, we tested to see the effect of rpoN deletion on eDNA during biofilm development. As observed in planktonic cultures, a lesser amount of eDNA was detected in the VI01 biofilm than in the wild type, and this was attributed to the deletion of rpoN, as the complementation restored the eDNA detected in the biofilm to wild-type levels (Fig. 2C).
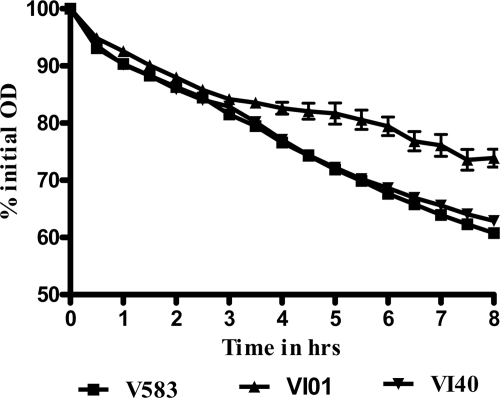
Sigma 54 alters the rate of autolysis in E. faecalis V583.
Because eDNA release in E. faecalis is dependent upon cell death by autolysis (54) and the rpoN mutant is defective in eDNA release, we hypothesized that σ54 may differentially modify the rate of autolysis in E. faecalis. In the autolysis assay, we observed that VI01 showed a significant decrease in the rate of autolysis, a phenotype readily complemented by introducing the gene in single copy to its native locus (Fig. 3).
Fig 3.
RpoN alters the rate of autolysis in E. faecalis. Differences in autolysis rates of the wild type (V583), rpoN mutant (VI01), and complemented strain (VI40) are plotted as the percentage of the initial optical density (OD) at 600 nm. Assays were performed in triplicate and repeated four times; error bars represent standard errors of means.
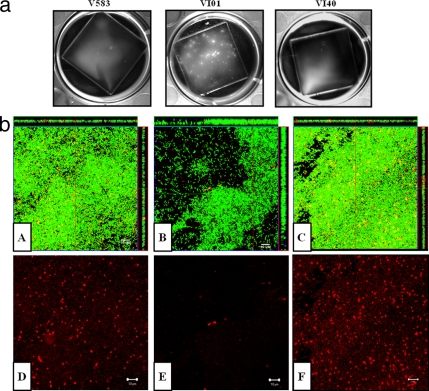
Sigma 54 alters biofilm development by E. faecalis.
eDNA has been shown to be an important matrix component in E. faecalis biofilms (54). The decreased levels of eDNA in VI01 led us to the hypothesis that VI01 may form less-dense biofilms than the wild-type V583. However, CLSM analysis of 24-h-old biofilms grown on glass coverslips showed that VI29 (ΔrpoN, GFP+) formed a thicker biofilm (as measured based on z-stack thickness, using an LSM image examiner) than those formed by the wild-type strain VT09 or the complemented strain VI41 (Fig. 4b). The appearance of the VI29 biofilm suggested early initiation of microcolony development, which was confirmed by macroscopic examination of the biofilms after 2 days of growth on coverslips (Fig. 4a). Despite the increased thickness and overall biofilm biomass of the rpoN mutant strain (VI29) compared to the parental and complemented strains (Table 4), very few random dead cells and DNA (as detected by SYTOX orange staining) were observed within the biofilm. Consistent with earlier observations on the role of cell death and of eDNA as a matrix component (54), regions within the wild-type VT09 contained concentrated foci of DNA and dead cells, which was phenocopied by the complement VI41 strain (Fig. 4b).
Fig 4.
(a) Macroscopic views of biofilms (produced by V583, VI01, and VI40) grown on glass coverslips in TSB medium. (b) Confocal analysis of 1-day-old biofilms grown on glass coverslips. The wild type, mutant, and complement constitutively expressed GFP from pMV158gfp as mentioned in Materials and Methods. Biofilms were grown on glass coverslips in TSB medium. Dead cells and eDNA were stained with SYTOX orange (1 μM). Live bacteria appear green, while dead cells and eDNA are red. Biofilm orthogonal projections are shown for VT09 (A), VI29 (B), and VI41 (C) and show merged green and red staining. Panels D, E, and F correspond to dead cell and eDNA staining of VT09, VI29, and VI41 biofilms, respectively (matched pairs to biofilms in panels A, B, and C). Bars, 10 μm.
Table 4.
Comstat analysis of 1-day-old biofilma
| Strain | Biomass (μm3/μm2) | Avg thickness (μm) |
|---|---|---|
| VT09 | 2.09 ± 0.522§ | 2.40 ± 0.589§ |
| VI29 | 4.723 ± 1.28 | 5.97 ± 2.01 |
| VI41 | 2.19 ± 0.638§ | 2.14 ± 0.624§ |
Values are means ± standard deviations.
, significantly different (P < 0.05) from VI29 result.
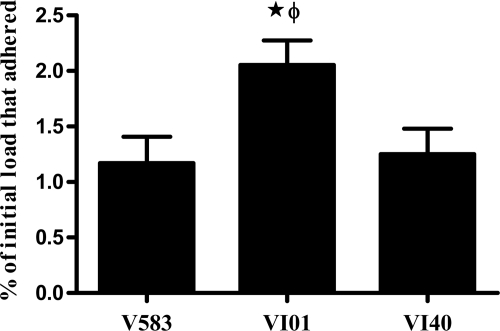
Deletion of rpoN increases adherence to polystyrene plates.
In order to determine whether increased biofilm formation by the rpoN mutant was due to its initial adherence ability, we calculated the percentage of the initial inoculum that adhered to 96-well microtiter plates after 2 h. Adherence of VI01 to a polystyrene plate was significantly enhanced in comparison to the wild type. In addition, markerless complementation of VI01 (VI40) reduced the adherence potential to wild-type levels (Fig. 5).
Fig 5.
Polystyrene plate adherence assay results. Deletion of rpoN increased adherence of E. faecalis to 96-well polystyrene plates. ★, significant increase (P < 0.05) relative to wild-type V583; ϕ, significant increase (P < 0.05) relative to the rpoN complement (VI40).
Sigma 54 modulates the composition of E. faecalis V583 biofilms.
On the basis of the macroscopic observations and CLSM of the VI29 biofilm and the relative lack of eDNA detection for this mutant, we hypothesized a role for a different polymer matrix that promotes biofilm formation in the rpoN mutant. To test the role of proteins in VI01 biofilm, we examined the effect of proteinase K treatment on biofilm development. The wild-type and complemented strains exhibited decreased biofilm when treated with proteinase K only after 24 h of biofilm growth. In contrast, reduction in VI01 biofilm was significant when treated with proteinase K after 6 h of biofilm growth and continued to respond to treatment after 12 h and 24 h of biofilm growth (Fig. 6).
Fig 6.
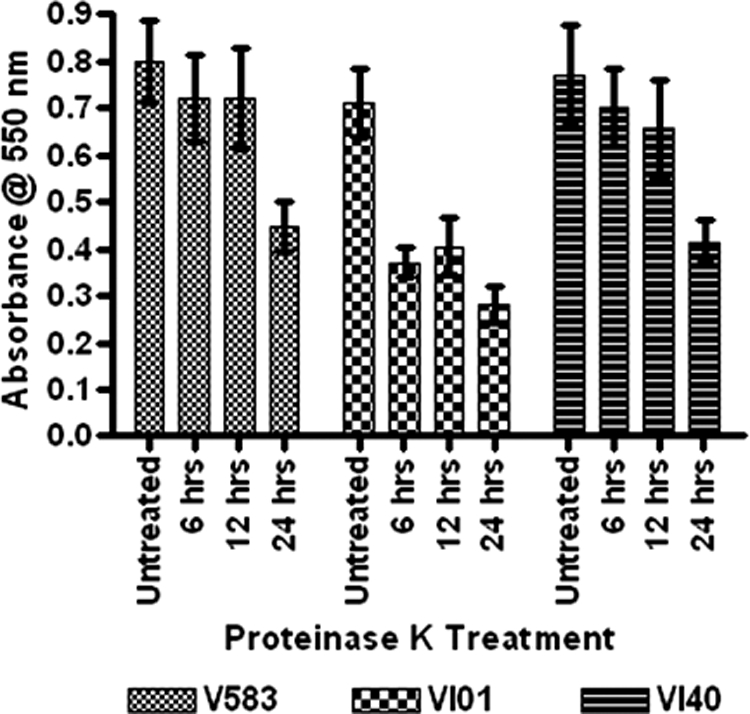
Proteinase K inhibits biofilm development of the rpoN mutant on polystyrene plates. Biofilms were seeded at time zero for V583, VI01, and VI40, and the untreated biofilms were stained 24 h later. At the indicated times after seeding the biofilm, proteinase K (1 μg/ml) was added, and the treatment was allowed to continue for the remainder of the assay. Each assay was performed in triplicate and repeated four times. Error bars indicate standard errors of the means.
DISCUSSION
The role of σ54 in regulating numerous biological properties, including those related to virulence, has been well documented for a variety of bacterial species (3, 10, 21, 42, 49, 51, 52, 55, 58, 59). However, its role in E. faecalis has been limited to observations made regarding its contribution to sensitivity to class IIa bacteriocins through the regulation of sugar PTSs (9, 13, 22). Identification of σ54 as a potential regulatory protein in the cascade of biofilm development was an interesting breakthrough, and we focused our efforts on elucidating its effect on E. faecalis V583 biofilm. The role of autolysis (54) and fratricide (53) has been well documented in enterococcal biofilm formation and has been shown to be important in providing eDNA as a key biofilm matrix component. However, the observation that biofilm formation was enhanced in the rpoN mutant despite the increased resistance to autolysis and the absence of eDNA was an unexpected finding.
One possible explanation for the increased resistance to autolysis observed in the rpoN mutant could be novel modifications of the cell wall or alteration of the modifications, such as O-acetylation (43) or d-alanylation (17) on the cell wall that protect against lysis. Deletion of rpoN did not alter the autolysin profile of E. faecalis when the micrococcal cell wall was used as a zymogram substrate (data not shown), ruling out the possibility of inactive autolysins. Also, the deletion of rpoN did not have a measurable effect on the secretion of the extracellular proteases GelE and SprE (data not shown), which have been previously shown to contribute to autolysis in E. faecalis (54, 57). A significant reduction in cell death due to impaired cell lysis occurred in E. faecalis V583 (ΔrpoN) planktonic and biofilm cultures, suggesting the requirement of a functional σ54 for regulation of susceptibility to cell lysis. In P. aeruginosa, deletion of rpoN abolishes cell death in the microcolonies during biofilm maturation and has been related to the expression of surface structures (type 4 pili and flagella) whose expression is regulated by σ54 (56). Additionally, σ54-dependent gene regulation promotes phage-induced lysis in P. aeruginosa (10). There are seven phages associated with E. faecalis V583, with one of them being a part of the core genome (37). It will be interesting to test the role of σ54-dependent transcription of phage particle proteins and host cell lysis and the contribution of σ54 to biofilm development.
Enhancement of biofilm formation in the absence of a well-characterized matrix component in the rpoN mutant indicates that a substantial knowledge gap still exists in unraveling factors associated with E. faecalis biofilm development. Cellular processes regulated by σ54 are attractive in this regard, to begin revealing the interplay between metabolism and biofilm development, as one of the few characterized roles for σ54 is the regulation of four sugar PTS pathways. It is noteworthy that deletion of the genes encoding the four known enhancer binding proteins (LpoR, MphR, MpoR, and MptR) did not reduce eDNA release, impair autolysis, or alter biofilm development (data not shown), suggesting that σ54 might act as a repressor of genes independent of enhancer protein function. The idea that σ54 levels in the cell or within the population might be regulated raises an interesting experimental question. Our observation that the wild-type and rpoN complement strains could grow on 2DG at a much lower frequency (10−4 and 10−5, respectively) relative to the rpoN mutant parallels a recent report by Flanagan et al. (18) that resistance to the E. faecalis plasmid-encoded bacteriocin MC4-1 (a class IIa bacteriocin) was dependent on point mutations within the rpoN gene that occurred at high frequency (10−3 to 10−4). This resistance was shown to be reversible to a susceptible phenotype by point mutations that also occurred within rpoN as second-site suppressors. These combined observations suggest that there are hot spots for mutation within rpoN and could be a mechanism for phase variation within the E. faecalis population.
In Vibrio vulnificus, σ54 positively regulates the gene encoding ADP-glycero-manno-heptose-6-epimerase (gmhD), which is responsible for production of lipopolysaccharide and exopolysaccharide, both of which are required for biofilm formation (31), while in B. cenocepacia σ54 controls motility, which in turn plays a role in biofilm formation (51). However, in E. coli K-12, rpoN deletion enhances biofilm formation (3). Such different effects of rpoN on the biofilm-forming potential of bacteria provide a clear example of how the knowledge regarding a gene whose function was first reported to be restricted to nitrogen assimilation has evolved to govern virulence-related functions in addition to bacterial metabolism. Our data provide additional support for the expanding role of σ54 in the world of low-GC Gram-positive bacteria.
Biofilm formation is a multistep process that begins with the attachment of bacteria to a substrate, followed by colonization via further recruitment of more bacteria or by cell division. Initial attachment of a bacterial cell to a surface is an important stage in biofilm development and determines the fate of this process. In P. aeruginosa (8, 35), Staphylococcus aureus (11), and Streptococcus spp. (40) it has been shown that a defect in initial adherence of a bacterium affects biofilm formation and subsequently influences the virulence of these pathogens. Our data indicate a similar influence of the attachment process in enterococcal biofilm development, wherein deletion of rpoN increases the adherence potential of the pathogen, which subsequently results in a more dense biofilm.
Other than DNA, other molecules, such as proteins and polysaccharides, have been suggested to be important constituents in the polymer matrix of several bacteria (16, 19). Robust biofilm formation by VI01 despite the significant reduction in eDNA led us to test for the presence of other matrix components by using compounds capable of dissolving the aforementioned components. The reduced ability of VI01 (ΔrpoN) to form a biofilm when treated with proteinase K suggests a role for protein in either adhesion or matrix composition to promote E. faecalis biofilm and is consistent with recent observations by Guiton et al. (20). Those authors observed that colonization of an implanted piece of urinary catheter as well as the bladder epithelium was dependent on a functioning sortase enzyme for the proper anchoring of proteins to the cell wall, which in turn promoted cellular adhesion. In S. aureus, a biofilm defect in mutants that overproduce extracellular protease was rectified by the addition of α2-macroglobulin, a general protease inhibitor, indicating a vital role for proteins in either cellular adhesion or the biofilm matrix (4). Similarly, in Bacillus subtilis, TasA is required for the structural integrity and development of biofilms (7). In E. faecalis biofilms (54), eDNA is known to be a crucial matrix component in the early stages of biofilm development, but by 24 h of growth in the biofilm DNase has a minimal effect on disrupting the biofilm. Here we show that in E. faecalis V583, proteins are likely to serve as important matrix components during the later stages of biofilm development, as a reduction in biomass was observed only at 24 h and not at earlier time points. This suggests the time-dependent involvement of different polymers in the overall development of the biofilm.
Complementation studies of the rpoN mutant by using a low-copy-number plasmid did not result in complete reversal of the phenotype to wild-type levels in experiments that involved stressing of cells (osmotic shock and 2DG toxicity) (data not shown). This was primarily due to plasmid loss in the absence of selection and suggested a survival advantage for E. faecalis in the absence of σ54 under certain stress conditions. The inability to fully complement an rpoN mutant has also been reported for L. monocytogenes (41). Similarly, in a V. fischeri squid colonization model (58), the level of colonization varied with the complemented strain, and only some animals exhibited wild-type levels of colonization. For this reason, we utilized a complementation strategy that restored the function of the gene by placing it at its native locus in single copy.
A literature survey for σ54 and its biological roles revealed a bias toward Gram-negative species, with P. aeruginosa, Vibrio spp., and E. coli being the most studied. In an attempt to identify the distribution of rpoN in low-GC Gram-positive organisms, we performed a BLAST search using σ54 of E. faecalis V583 as the query. Among the organisms queried, only L. monocytogenes, B. subtilis, C. difficle, and C. perfringens appeared to have homologues, whereas in S. aureus, S. pneumoniae, and S. pyogenes homologues to σ54 were absent. The basis for this distribution among enterically adapted organisms as well as the potential genes regulated by σ54 await further study.
In conclusion, the results from this study show that σ54 in E. faecalis V583 contributes to cell death and eDNA release and that in its absence, E. faecalis adapts an alternate matrix to establish biofilms. Understanding the mechanism underlying the phenotypes observed in this study is the main focus of ongoing studies in our laboratory.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Vinai Thomas for the preliminary screen of the E. faecalis transposon library and to Dan Boyle for assistance with confocal imaging.
This work was supported by Public Health Service grant AI77782 from the National Institutes of Health.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Acebo P, Nieto C, Corrales MA, Espinosa M, Lopez P. 2000. Quantitative detection of Streptococcus pneumoniae cells harbouring single or multiple copies of the gene encoding the green fluorescent protein. Microbiology 146: 1267–1273 [DOI] [PubMed] [Google Scholar]
- 2. Bass JIF, et al. 2010. Extracellular DNA: a major proinflammatory component of Pseudomonas aeruginosa biofilms. J. Immunol. 184: 6386–6395 [DOI] [PubMed] [Google Scholar]
- 3. Belik AS, Tarasova NN, Khmel IA. 2008. Regulation of biofilm formation in Escherichia coli K12: effect of mutations in the genes hns, strA, lon, and rpoN. Mol. Gen. Microbiol. Virol. 23: 159–162 [PubMed] [Google Scholar]
- 4. Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. Plos One 5: e10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bond DR, Tsai BM, Russell JB. 1999. Physiological characterization of Streptococcus bovis mutants that can resist 2-deoxyglucose-induced lysis. Microbiology 145: 2977–2985 [DOI] [PubMed] [Google Scholar]
- 6. Boucher JC, Schurr MJ, Deretic V. 2000. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol. Microbiol. 36: 341–351 [DOI] [PubMed] [Google Scholar]
- 7. Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 59: 1229–1238 [DOI] [PubMed] [Google Scholar]
- 8. Byrd MS, Pang B, Mishra M, Swords WE, Wozniak DJ. 2010. The Pseudomonas aeruginosa exopolysaccharide Psl facilitates surface adherence and NF-kappa B activation in A549 cells. Mbio 1(3): e00140–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calvez S, Rince A, Auffray Y, Prevost H, Drider D. 2007. Identification of new genes associated with intermediate resistance of Enterococcus faecalis to divercin V41, a pediocin-like bacteriocin. Microbiology 153: 1609–1618 [DOI] [PubMed] [Google Scholar]
- 10. Ceyssens PJ, et al. 2008. The genome and structural proteome of YuA, a new Pseudomonas aeruginosa phage resembling M6. J. Bacteriol. 190: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clarke SR, Foster SJ. 2006. Surface adhesins of Staphylococcus aureus. Adv. Microb. Physiol. 51: 187–224 [DOI] [PubMed] [Google Scholar]
- 12. Cruz-Rodz AL, Gilmore MS. 1990. High-efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol. Gen. Genet. 224: 152–154 [DOI] [PubMed] [Google Scholar]
- 13. Dalet K, Briand C, Cenatiempo Y, Hechard Y. 2000. The rpoN gene of Enterococcus faecalis directs sensitivity to subclass IIa bacteriocins. Curr. Microbiol. 41: 441–443 [DOI] [PubMed] [Google Scholar]
- 14. Dalet K, Cenatiempo Y, Cossart P, Hechard Y, European Listeria Genome Consortium. 2001. A σ54-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147: 3263–3269 [DOI] [PubMed] [Google Scholar]
- 15. Del Papa MF, Hancock LE, Thomas VC, Perego M. 2007. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J. Bacteriol. 189: 8835–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dunne WM. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15: 155–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fabretti F, et al. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74: 4164–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flanagan S, Clewell D, Sedgley C. 2010. Enterococcus faecalis alternates between resistance and sensitivity to the class IIa enterococcal bacteriocin MC4-1, abstr. 35A.3rd Int. ASM Conf. Enterococci. ASM Press, Washington, DC [Google Scholar]
- 19. Flemming HC, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8: 623–633 [DOI] [PubMed] [Google Scholar]
- 20. Guiton PS, Hung CS, Hancock LE, Caparon MG, Hultgren SJ. 2010. Enterococcal biofilm formation and virulence in an optimized murine model of foreign body-associated urinary tract infections. Infect. Immun. 78: 4166–4175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hagins JM, Scoffield JA, Suh SJ, Silo-Suh L. 2010. Influence of RpoN on isocitrate lyase activity in Pseudomonas aeruginosa. Microbiology 156: 1201–1210 [DOI] [PubMed] [Google Scholar]
- 22. Hancock LE, Perego M. 2004. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. J. Bacteriol. 186: 5629–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harmsen M, Lappann M, Knochel S, Molin S. 2010. Role of extracellular DNA during biofilm Formation by Listeria monocytogenes. Appl. Environ. Microbiol. 76: 2271–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hechard Y, Pelletier C, Cenatiempo Y, Frere J. 2001. Analysis of σ54-dependent genes in Enterococcus faecalis: a mannose PTS permease (EIIMan) is involved in sensitivity to a bacteriocin, mesentericin Y105. Microbiology 147: 1575–1580 [DOI] [PubMed] [Google Scholar]
- 25. Hidron AI, et al. 2008. Antimicrobial-resistant pathogens associated with healthcare-associated infections: annual summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2006-2007. Infect. Control Hosp. Epidemiol. 29: 996–1011 [DOI] [PubMed] [Google Scholar]
- 26. Hoiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. 2010. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 35: 322–332 [DOI] [PubMed] [Google Scholar]
- 27. Huelves L, et al. 2007. Adherence of Streptococcus pneumoniae to polystyrene plates, effect of serum on adhesion, and virulence in the gerbil otitis media model. Microb. Pathog. 43: 114–119 [DOI] [PubMed] [Google Scholar]
- 28. Ishikawa T, Rompikuntal PK, Lindmark B, Milton DL, Wai SN. 2009. Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. Plos One 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jefferson KK. 2004. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 236: 163–173 [DOI] [PubMed] [Google Scholar]
- 30. Kazmierczak MJ, Wiedmann M, Boor KJ. 2005. Alternative sigma factors and their roles in bacterial virulence. Microbiol. Mol. Biol. Rev. 69: 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim HS, Lee MA, Chun SJ, Park SJ, Lee KH. 2007. Role of NtrC in biofilm formation via controlling expression of the gene encoding an ADP-glycero-manno-heptose-6-epimerase in the pathogenic bacterium, Vibrio vulnificus. Mol. Microbiol. 63: 559–574 [DOI] [PubMed] [Google Scholar]
- 32. Lappann M, et al. 2010. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 75: 1355–1371 [DOI] [PubMed] [Google Scholar]
- 33. Lewis K. 2001. Riddle of biofilm resistance. Antimicrob. Agents Chemother. 45: 999–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindsay D, von Holy A. 2006. Bacterial biofilms within the clinical setting: what healthcare professionals should know. J. Hosp. Infect. 64: 313–325 [DOI] [PubMed] [Google Scholar]
- 35. Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. 2006. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 188: 8213–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mann EE, et al. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. Plos One 4: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McBride SM, Fischetti VA, LeBlanc DJ, Moellering RC, Gilmore MS. 2007. Genetic diversity among Enterococcus faecalis. Plos One 2: e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Merrick MJ. 1993. In a class of its own: the RNA polymerase sigma factor 54 (sigma N). Mol. Microbiol. 10: 903–909 [DOI] [PubMed] [Google Scholar]
- 39. Nieto C, Espinosa M. 2003. Construction of the mobilizable plasmid pMV158GFP, a derivative of pMV158 that carries the gene encoding the green fluorescent protein. Plasmid 49: 281–285 [DOI] [PubMed] [Google Scholar]
- 40. Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73: 407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okada Y, et al. 2006. The sigma factor RpoN (sigma 54) is involved in osmotolerance in Listeria monocytogenes. FEMS Microbiol. Lett. 263: 54–60 [DOI] [PubMed] [Google Scholar]
- 42. O'Toole R, Milton DL, Horstedt P, Wolf-Watz H. 1997. RpoN of the fish pathogen Vibrio (Listonella) anguillarum is essential for flagellum production and virulence by the water-borne but not intraperitoneal route of inoculation. Microbiology 143: 3849–3859 [DOI] [PubMed] [Google Scholar]
- 43. Pfeffer JM, Strating H, Weadge JT, Clarke AJ. 2006. Peptidoglycan O acetylation and autolysin profile of Enterococcus faecalis in the viable but nonculturable state. J. Bacteriol. 188: 902–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Potvin E, Sanschagrin F, Levesque RC. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 32: 38–55 [DOI] [PubMed] [Google Scholar]
- 45. Qin X, Singh KV, Weinstock GM, Murray BE. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183: 3372–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Qin X, Singh KV, Weinstock GM, Murray BE. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68: 2579–2586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qin ZQ, et al. 2007. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology 153: 2083–2092 [DOI] [PubMed] [Google Scholar]
- 48. Rice KC, et al. 2007. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 104: 8113–8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Riordan JT, Tietjen JA, Walsh CW, Gustafson JE, Whittam TS. 2010. Inactivation of alternative sigma factor 54 (RpoN) leads to increased acid resistance, and alters locus of enterocyte effacement (LEE) expression in Escherichia coli O157:H7. Microbiology 156: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sahm DF, et al. 1989. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 33: 1588–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saldias MS, Lamothe J, Wu R, Valvano MA. 2008. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infect. Immun. 76: 1059–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stevens MJA, Molenaar D, de Jong A, De Vos WM, Kleerebezem M. 2010. σ54-mediated control of the mannose phosphotransferase sytem in Lactobacillus plantarum impacts on carbohydrate metabolism. Microbiology 156: 695–707 [DOI] [PubMed] [Google Scholar]
- 53. Thomas VC, et al. 2009. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 72: 1022–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Thomas VC, Thurlow LR, Boyle D, Hancock LE. 2008. Regulation of autolysis-dependent extracellular DNA release by Enterococcus faecalis extracellular proteases influences biofilm development. J. Bacteriol. 190: 5690–5698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thompson LS, Webb JS, Rice SA, Kjelleberg S. 2003. The alternative sigma factor RpoN regulates the quorum sensing gene rhlI in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 220: 187–195 [DOI] [PubMed] [Google Scholar]
- 56. Thurlow LR, Thomas VC, Hancock LE. 2009. Capsular polysaccharide production in Enterococcus faecalis and contribution of CpsF to capsule serospecificity. J. Bacteriol. 191: 6203–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Waters CM, Antiporta MH, Murray BE, Dunny GM. 2003. Role of the Enterococcus faecalis GelE protease in determination of cellular chain length, supernatant pheromone levels, and degradation of fibrin and misfolded surface proteins. J. Bacteriol. 185: 3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wolfe AJ, Millikan DS, Campbell JM, Visick KL. 2004. Vibrio fischeri σ54 controls motility, biofilm formation, luminescence, and colonization. Appl. Environ. Microbiol. 70: 2520–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang TC, Leu YW, Chang-Chien HC, Hu RM. 2009. Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma Factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM. J. Bacteriol. 191: 2266–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhao K, Liu MZ, Burgess RR. 2010. Promoter and regulon analysis of nitrogen assimilation factor, σ54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 38: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.