Abstract
“Candidatus Methylomirabilis oxyfera” is a newly discovered denitrifying methanotroph that is unrelated to previously known methanotrophs. This bacterium is a member of the NC10 phylum and couples methane oxidation to denitrification through a newly discovered intra-aerobic pathway. In the present study, we report the first ultrastructural study of “Ca. Methylomirabilis oxyfera” using scanning electron microscopy, transmission electron microscopy, and electron tomography in combination with different sample preparation methods. We observed that “Ca. Methylomirabilis oxyfera” cells possess an atypical polygonal shape that is distinct from other bacterial shapes described so far. Also, an additional layer was observed as the outermost sheath, which might represent a (glyco)protein surface layer. Further, intracytoplasmic membranes, which are a common feature among proteobacterial methanotrophs, were never observed under the current growth conditions. Our results indicate that “Ca. Methylomirabilis oxyfera” is ultrastructurally distinct from other bacteria by its atypical cell shape and from the classical proteobacterial methanotrophs by its apparent lack of intracytoplasmic membranes.
INTRODUCTION
Methanotrophic bacteria (methanotrophs) are included in the subset of bacteria known as methylotrophs. These organisms play a critical role in the global carbon cycle and are defined by their ability to utilize methane (CH4) as their sole carbon and energy source (reviewed in references 6, 13, 24, and 34). Since their discovery more than a century ago (33), methanotrophs were found in a variety of ecosystems, including soils, sediments, wetlands, freshwater, and marine habitats.
Until recently, methanotrophs were assigned to a rather limited group of bacteria within the γ- and α-subclasses of Proteobacteria (13, 34). A major extension was made by the isolation of three Verrucomicrobia species, which were able to oxidize methane in extremely acidophilic environments (8, 16, 25), and the identification of the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera” in freshwater enrichment cultures (12, 15, 19, 20, 26). For “Ca. Methylomirabilis oxyfera,” various enrichment cultures using different inocula have been described; they show >97.5% 16S rRNA gene sequence similarity (40).
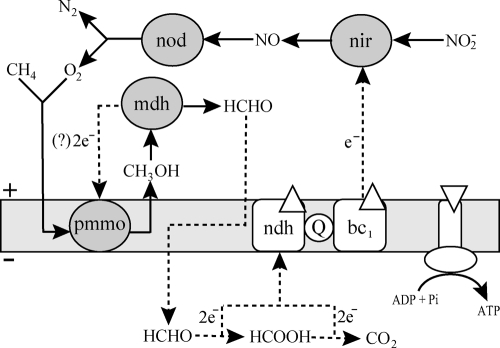
So far, at least two properties set “Ca. Methylomirabilis oxyfera” apart from the other known methanotrophs. First, its phylogenetic association with the deep-branching NC10 phylum (26), a phylum without any cultivated representatives in pure culture (27), opened a new phylogenetic branch within the otherwise well-defined methanotrophs. Second, “Ca. Methylomirabilis oxyfera” is neither an aerobic methane oxidizer (like all other known methanotrophs) nor an sensu stricto anaerobic methane oxidizer, with the only known case of anaerobic methane oxidation (AMO) represented by the consortium of methanotrophic archaea and sulfate-reducing bacteria through reverse methanogenesis (17). It seems that “Ca. Methylomirabilis oxyfera” has developed a new way of living on methane, by combining AMO coupled to denitrification with normal respiration through a newly discovered intra-aerobic pathway for the production of oxygen (Fig. 1). The oxygen is produced through an atypical denitrification pathway, which proceeds by the dismutation of nitric oxide into dinitrogen and oxygen. Part of the produced oxygen then is used for the activation and oxidation of methane (10); the remaining oxygen is proposed to be used in normal respiration by terminal respiratory oxidases (39) (Fig. 1).
Fig 1.
Postulated model for central catabolism and energy conservation in “Ca. Methylomirabilis oxyfera.” White diamonds, direction of proton flow. bc1, cytochrome bc1 complex; mdh, methanol dehydrogenase; ndh, NAD(P)H dehydrogenase complex; nir, nitrate reductase; nod, nitric oxide dismutase; pmmo, particulate methane monooxygenase; Q, coenzyme Q. Figure based on reference 40.
With respect to cell shape, methanotrophs harbor a variety of types; rods, cocci, and, occasionally, crescent- and pear-shaped forms are described (13). There is, however, one ultrastructural feature of methanotrophs that is shared by most: the intracytoplasmic membranes (ICMs). The ICMs harbor the key enzyme for the methane oxidation, the particulate form of methane monooxygenase (pMMO). Some methanotrophs also posses the soluble form of this enzyme (sMMO), which resides in the cytoplasm (13, 34). The physical arrangement of pMMO in ICMs results in an increase of the amount of this enzyme, which can reach up to 80% of total ICM content, and it might be reflected in an enhancement of metabolic speed (23, 29). With the exception of Methylocella species (7), which contains only the soluble form of the methane monooxygenase enzyme, and the three currently known verrucomicrobial species (24), all other methanotrophs possess the pMMO enzyme as well as ICM structures. The ICMs occur in two main types of arrangements: as bundles of vesicular disks in Gammaproteobacteria (type I methanotrophs) and as paired peripheral layers in Alphaproteobacteria (type II methanotrophs) (13).
Since its discovery in 2006 (26), some of the key features of “Ca. Methylomirabilis oxyfera” have been unraveled, including the genome, transcriptome, and proteome, as well as the major catabolic pathways (10, 40). However, unlike the case for many proteobacterial methanotrophs, knowledge of the ultrastructure of “Ca. Methylomirabilis oxyfera” so far is nonexistent. Being evolutionarily completely unrelated to previously known methanotrophs, “Ca. Methylomirabilis oxyfera” is very interesting from an ultrastructural point of view. Here, we investigated the ultrastructure of “Ca. Methylomirabilis oxyfera” using an array of electron microscopy techniques in combination with various sample preparation methods. We observed that “Ca. Methylomirabilis oxyfera” cells possess an atypical polygonal shape. Also, the outermost layer of the cell consisted of a putative protein surface layer (S-layer). Further, this study revealed that, at least under the growth conditions used in this study, “Ca. Methylomirabilis oxyfera” does not develop ICMs.
MATERIALS AND METHODS
“Ca. Methylomirabilis oxyfera” enrichment culture.
Samples were taken from a 15-liter sequencing batch reactor containing the “Ca. Methylomirabilis oxyfera” enrichment culture (modified from reference 26).
Fluorescence in situ hybridization (FISH).
Cells from the “Ca. Methylomirabilis oxyfera” enrichment culture were harvested, and hybridizations with a fluorescent probe were performed as described previously (11), using a stringency of 50% formamide in the hybridization buffer. The probe was purchased as a Cy3-labeled derivative from Thermo Electron Corporation (Ulm, Germany). The probe S-*-DBACT-0193-a-A-18 was used for NC10 bacteria (26). The preparation was counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) and mounted with Vectashield (Vector Laboratories, Inc., CA). The percentage of “Ca. Methylomirabilis oxyfera” cells was estimated by counting the number of cells that hybridized with the S-*-DBACT-0193-a-A-18 probe and the number of cells that showed only a DAPI signal from a total of 600 counted cells.
Sample preparation for cryo-SEM.
Cells from the “Ca. Methylomirabilis oxyfera” enrichment culture were frozen by both plunge-freezing and high-pressure-freezing methods. For plunge freezing, the cells were placed between two cryostubs, forming a sandwich, and plunge frozen in liquid nitrogen slush. For high-pressure freezing, cells were transferred into a 100-μm cavity of a planchette (3-mm diameter; 0.1 to 0.2-mm depth; Engineering Office M. Wohlwend GmbH, CH-9466 Sennwald, Switzerland) and closed with the flat side of a lecithin-coated planchette (3-mm diameter; 0.3-mm depth). The cells then were cryofixed by high-pressure freezing (Leica EMHPF; Leica Microsystems, Vienna, Austria) and transferred to cryostubs. Samples then were placed into a cryotransfer system (Gatan Alto 2500; Oxford, United Kingdom). The top cryostub from plunge-frozen samples was fractured by a razor. Subsequently, both samples were processed in a similar manner. The water layer was sublimated for 10 min at −80°C, sputter coated with a thin layer of Au-Pd (60/40 ratio) for 45 s using a Cressington 208HR sputter coater fitted with an MTM-20 thickness controller (Cressington Scientific Instruments Ltd., United Kingdom), and analyzed by cryoscanning electron microscopy (cryo-SEM).
For cryo-SEM, cells were taken from the “Ca. Methylomirabilis oxyfera” enrichment culture at six different time points. In total, 195 typical images were obtained containing different amounts of cells (ranging from 1 to ca. 30 cells).
Sample preparation for transmission electron microscopy (TEM). (i) Chemical fixation (tannic acid-mediated osmium impregnation), Epon embedding, and sectioning.
Cells from the “Ca. Methylomirabilis oxyfera” enrichment culture were immersed for 30 min at room temperature in aldehyde-based fixative (1.5% glutaraldehyde and 2% paraformaldehyde in 0.08 M sodium cacodylate trihydrate buffer, pH 7.4), postfixed with 1% osmium tetroxide (OsO4) and 1.5% K4[Fe(CN)6] for 90 min at 4°C in darkness, incubated with 1% tannic acid in 0.1 M sodium cacodylate trihydrate buffer (pH 7.4) for 30 min at room temperature, and treated with 1% OsO4 in distilled water for 30 min on ice in darkness. Finally, cells were dehydrated in a graded ethanol series (70, 80, 90, 96, and 100% ethanol), gradually infiltrated with Epon resin, sectioned (70-nm sections) using a Reichert Ultracut E Microtome (Leica Microsystems, Vienna, Austria), and collected on carbon-Formvar-coated 100-mesh hexagonal copper grids.
For chemical fixation, cells were taken from the “Ca. Methylomirabilis oxyfera” enrichment culture at one time point. This sample was chemically fixed in triplicate using the described protocol both with and without the tannic acid-mediated osmium impregnation (6 samples). For each fixation, three Epon blocks were produced, used for thin sectioning, and investigated by TEM (6 blocks). Based on contrast and ultrastructural preservation, the fixation with tannic acid-mediated osmium impregnation was used for further investigation. These blocks were extensively examined by TEM, and in total 50 typical images were obtained containing different amounts of cells (ranging from 1 to ca. 50 cells). In instances where cells were counted, cells were chosen from the images at random to the best of our ability.
(ii) Cryofixation, freeze-substitution, Epon embedding, and sectioning.
Cells from the “Ca. Methylomirabilis oxyfera” enrichment culture were cryofixed by high-pressure freezing as described above. Freeze-substitution was performed in acetone containing 2% OsO4, 0.2% uranyl acetate, and 1% H2O (37). Subsequently, samples were kept at −90°C for 47 h; brought to −60°C at 2°C per hour; kept at −60°C for 8 h; brought to −30°C at 2°C per hour; and kept at −30°C for 8 h in a freeze-substitution unit (AFS; Leica Microsystems, Vienna, Austria). Uranyl acetate was removed by washing the samples four times for 30 min in the AFS device at −30°C with acetone containing 2% OsO4 and 1% H2O. Fixation then was continued for 1 h on ice. OsO4 and H2O were removed by two washes for 30 min on ice with anhydrous acetone. Samples were gradually infiltrated with Epon resin and polymerized for 72 h at 60°C (22). Ultrathin sections (for TEM; 70 nm) and semithin sections (for electron tomography [ET]; 400 nm) were cut using a Reichert Ultracut E microtome (Leica Microsystems, Vienna, Austria) and collected on carbon-Formvar-coated 100-mesh hexagonal and 50-mesh square copper grids, respectively. The ultrathin sections were poststained with 20% uranyl acetate in 70% methanol for 4 min and Reynolds lead citrate for 2 min (28).
For cryofixation, cells were taken from “Ca. Methylomirabilis oxyfera” enrichment cultures at four different time points. All four samples were freeze-substituted in duplicate in both acetone containing 2% OsO4 and acetone containing 2% OsO4, 0.2% uranyl acetate, and 1% water (16 samples in total). For each fixation, two Epon blocks were produced and used for thin sectioning and investigation by TEM (16 blocks). Based on contrast and ultrastructural preservation, the substitution in acetone containing 2% OsO4, 0.2% uranyl acetate, and 1% water was used for further investigation. These blocks were extensively examined by TEM, and in total 131 typical images were obtained containing different amounts of cells (ranging from 1 to ca. 50 cells). In instances where cells were counted, cells were chosen from the images at random to the best of our ability.
(iii) ET.
Electron tomography (ET) was performed as described previously (35). Ten-nanometer colloidal gold particles were applied to one surface of grids bearing 200- to 400-nm semithin sections of high-pressure-frozen, freeze-substituted, and Epon-embedded “Ca. Methylomirabilis oxyfera” cells to serve as fiducial markers in the alignment of the tilt series. The sections were poststained with 2% uranyl acetate in water for 10 min. Specimens were placed in a high-tilt specimen holder, and dual-axis data sets were automatically recorded at 200 kV using a Tecnai-20 microscope (FEI Company, Eindhoven, The Netherlands) by rotating the grid 90° inside the microscope (Fischione rotation holder; Fischione Instruments, Pittsburgh, PA). The angular tilt range was from −65° to 65°, with an increment of 1°. Binned (two by two) images (1,024 by 1,024 pixels) were recorded using a charge-coupled device (CCD) camera (TemCam F214; TVIPS GmbH, Gauting, Germany). Automated data acquisition of the tilt series was carried out using Xplore 3D (FEI Company, Eindhoven, The Netherlands). Tomograms from each tilt axis were computed with the R-weighted back-projection algorithm and combined into one double-tilt tomogram using the IMOD software package (18). In total, 30 “Ca. Methylomirabilis oxyfera” cells were imaged in six double-tilt tomograms.
(iv) Freeze-etching.
Freeze-etching was performed on concentrated (by a 4-min centrifugation step at 10,000 or 12,000 × g) “Ca. Methylomirabilis oxyfera” cells from the enrichment culture, of which 1.7 μl per gold carrier was plunge frozen in liquid nitrogen by hand. The samples then were introduced into a Cressington freeze-etch machine at <−170°C and a pressure of below 10−6 bar. The samples were kept at −97°C for 7 min before being fractured. The water was left to sublimate from the samples for 4 min (freeze-etching) before the samples were shadowed with 1 nm Pt-C (angle 45°) and 10 nm C (angle 90°). The biological material was removed from the replicas by overnight incubation in 70% sulfuric acid; the replicas were washed twice on bidistilled water and picked up with 700-mesh hexagonal copper grids before investigation by TEM.
For freeze-etching, cells were taken from the “Ca. Methylomirabilis oxyfera” enrichment culture at two different time points. The replicas were extensively examined by TEM, and in total 180 typical images were obtained containing different amounts of cells (ranging from one to three cells).
TEM.
Cells, ultrathin sections, and replicas were investigated with a TEM at 60, 80, or 120 kV (CM12, Tecnai10, or Tecnai12; FEI Company, Eindhoven, The Netherlands), and images were recorded using a CCD camera (MegaView II, OSIS 0124; TVIPS, Gauting, Germany).
Cryo-SEM.
The coated samples were analyzed with a field emission SEM (JSM-6330F; JEOL, Tokyo, Japan) at a sample temperature of −170°C using an accelerating voltage of 3 kV.
RESULTS
Quantification of “Ca. Methylomirabilis oxyfera” cells in the enrichment culture.
Until now, it has not been possible to grow “Ca. Methylomirabilis oxyfera” in pure culture. In the culture used for this study, the level of enrichment was about 71% when assessed by FISH using a previously described oligonucleotide probe (26). “Ca. Methylomirabilis oxyfera” cells appeared as small rods with an intense DAPI signal in the center. They occurred as single cells or as multicellular aggregates, which occasionally included cells from other species that made up 29% of the community.
Cryo-SEM.
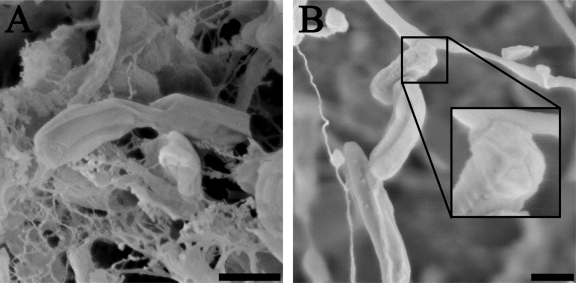
The cell shape of “Ca. Methylomirabilis oxyfera” was investigated using cryo-SEM (Fig. 2). The general appearance of “Ca. Methylomirabilis oxyfera” cells was similar when prepared with plunge-freezing or high-pressure-freezing methods. The rod-shaped cells were, on average, 1,158 ± 323 nm long and 259 ± 64 nm wide (measured from a total of 50 cells). The cell surface had a relatively smooth appearance, except for the presence of several distinct longitudinal ridges that ran along the entire cell length and joined at the cell poles in a circular, cap-like structure (Fig. 2B, inset). Different cells had different amounts of these longitudinal ridges, resulting in a polygonal cell shape for “Ca. Methylomirabilis oxyfera.” Further, “Ca. Methylomirabilis oxyfera” cells were observed to divide by binary fission in growing cultures (Fig. 2A).
Fig 2.
Cryoscanning electron micrographs of “Ca. Methylomirabilis oxyfera” cells showing the longitudinal ridges along the cell length. (A) Plunge-frozen “Ca. Methylomirabilis oxyfera” cells undergoing cell division. (B) Plunge-frozen “Ca. Methylomirabilis oxyfera” cells showing the cap-like structure (inset) at the cell poles. Scale bars, 500 nm.
Freeze-etching.
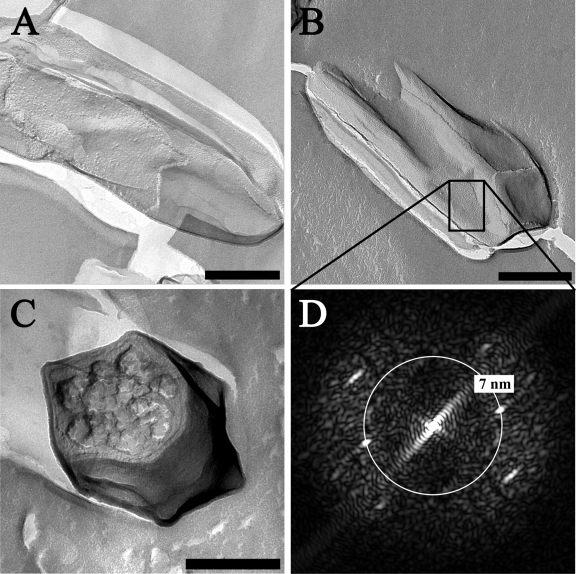
Freeze-etching also revealed the polygonal cell shape of “Ca. Methylomirabilis oxyfera” (Fig. 3), especially in cross-section (Fig. 3C). The ridges had a granular appearance compared to the rest of the cell surface (Fig. 3A and B). Further, “Ca. Methylomirabilis Ca. Methylomirabilis oxyfera” cells were observed to contain an additional layer, outside the cell wall, as the outermost sheath (Fig. 3A and B). This layer could be a (glyco)protein surface layer (S-layer) with an oblique or square lattice symmetry (p2 or p4). The power spectrum (Fig. 3D) indicated a repetitive pattern with frequencies at 7 nm−1 and, in some cases, at 5 nm−1. Those values are likely to correspond to a center-to-center spacing of the S-layer units. Compared to S-layer center-to-center spacings found in the literature, which are in the range of 3 to 35 nm(30), these are rather small values. However, small values have been found previously for p2 symmetry S-layers (32).
Fig 3.
Transmission electron micrographs of freeze-etched “Ca. Methylomirabilis oxyfera” cells. (A and B) “Ca. Methylomirabilis oxyfera” cells fractured longitudinally over the cell wall and displaying a putative S-layer. (C) “Ca. Methylomirabilis oxyfera” cell fractured transversely through the cell. (D) FFT power spectrum of the putative S-layer cut-out shown in panel B. Repetitive patterns occur after ca. 7 nm. Scale bars, 200 nm.
Ultrathin sections of cryofixed, freeze-substituted, and Epon-embedded cells.
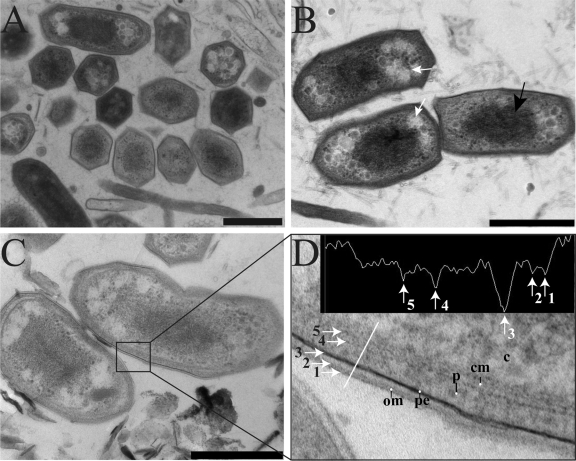
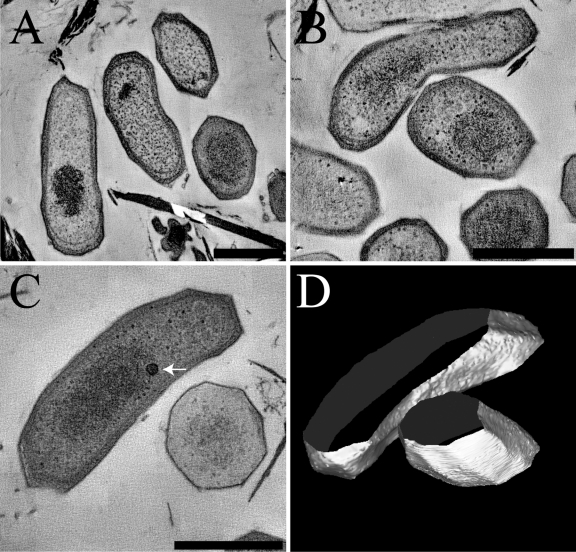
Transmission electron microscopy of ultrathin sections of cryofixed, freeze-substituted, and Epon-embedded “Ca. Methylomirabilis oxyfera” cells showed a cell envelope typical of Gram-negative bacteria (Fig. 4). The cell envelope had a total width of about 40 nm and consisted, from inside to outside, of a cytoplasmic membrane, peptidoglycan, and an outer membrane (Fig. 4D). The peptidoglycan comprised an electron-dense layer within the periplasmic space in close vicinity to the outer membrane (Fig. 4D). “Ca. Methylomirabilis oxyfera” cells differed from prototypical bacterial cell shapes. Transversely sectioned cells had a polygonal cell shape with variant numbers of sides for different cells, and longitudinally sectioned cells showed cornered cell poles (Fig. 4). Inside the cytoplasm, electron light granules were observed which possibly contain reserve material (Fig. 4B, white arrows). The nucleoid, which is visible as a densely stained area in the middle of the cell, occupied much of the cell content and appeared to be quite condensed (Fig. 4B, black arrow). Surprisingly, intracytoplasmic membranes (ICMs), commonly found in proteobacterial methanotrophs (13, 35), were never observed.
Fig 4.
Transmission electron micrographs of cryofixed, freeze-substituted, and Epon-embedded “Ca. Methylomirabilis oxyfera” cells. (A) Overview showing the dominant polygonal cell shape in the sample. (B) Longitudinally sectioned “Ca. Methylomirabilis oxyfera” cells showing electron light granules (white arrows) and condensed nucleoid (black arrow). (C) Longitudinally sectioned cells showing a Gram-negative cell envelope. (D) Cut-out and density profile of part of the cell wall shown in panel C. The density profile is measured across the white line. The numbered arrows in the density profile correspond to the numbered arrows in the cell wall. om, outer membrane; pe, peptidoglycan; p, periplasm; cm, cytoplasmic membrane; c, cytoplasm. Scale bars, 500 nm.
Ultrathin sections of chemically fixed and Epon-embedded cells.
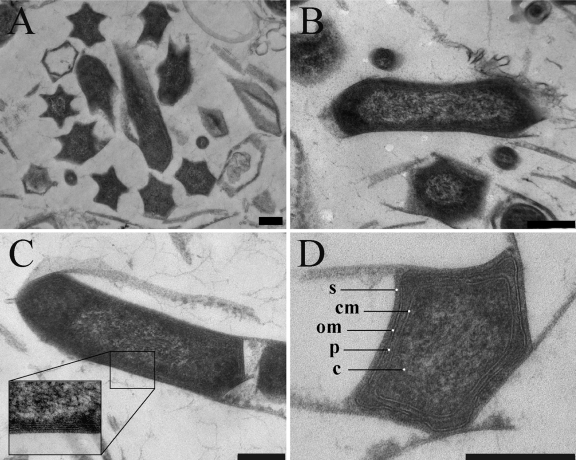
There was a significant variation in the appearance of the cell envelope in the chemically fixed cells compared to the cryofixed cells. The chemically fixed cells appeared more shrunk (Fig. 5), possibly due to dehydration, a phenomenon that is commonly reported for chemically fixed cells, particularly when glutaraldehyde is used as an initial chemical fixative in combination with dehydration at room temperature (14). In the chemically fixed cells, the cell wall often was collapsed while the ridges stayed in position, resulting in a more pronounced star-like appearance of the cells (Fig. 5). The polygonal cell shape was the dominant morphotype in the sample from the enrichment culture; from 980 cells counted, cells with a polygonal shape made up 69%.
Fig 5.
Transmission electron micrographs of chemically fixed and Epon-embedded “Ca. Methylomirabilis oxyfera” cells. (A) Overview showing the star-like cell shape caused by dehydration and cell wall collapse. Longitudinal (B and C) and transverse (D) sections show the Gram-negative cell envelope and the presence of a putative S-layer on the top of the outer membrane. om, outer membrane; p, periplasm; cm, cytoplasmic membrane; c, cytoplasm; s, putative S-layer. Scale bars, 200 nm.
In general, when comparing the chemically fixed cells to cryofixed cells, the bilayer of the outer membrane appeared thicker and denser, and the membranes appeared more distorted and wavy. In some instances, an additional layer with a thickness of about 8 nm was observed on top of the outer membrane (Fig. 5C, inset, and D). This could correspond to the putative S-layer observed in the freeze-etching preparations.
ET.
Three-dimensional imaging using ET confirmed the polygonal shape of “Ca. Methylomirabilis oxyfera” cells after high-pressure freezing and freeze-substitution (Fig. 6; also see Movies S1 to S3 in the supplemental material). Occasionally, an electron-dense, vesicular body with a diameter of about 55 nm was observed within the cytoplasm (Fig. 6C, arrow). These structures contained a rather rough and irregular boundary, and the absence of any membrane connections suggests that they are separate entities and possibly storage vesicles.
Fig 6.
(A, B, and C) Tomographic slices of “Ca. Methylomirabilis oxyfera” cells showing the polygonal cell shape. See also Movies S1, S2, and S3 in the supplemental material, respectively. (D) Model of the tomogram shown in panel C. Arrow, electron-dense vesicular body. Scale bars, 500 nm.
DISCUSSION
In the present paper, we performed a detailed ultrastructural study of the newly discovered denitrifying methanotroph “Ca. Methylomirabilis oxyfera.” This bacterium is a member of the deep-branching ‘NC10’ phylum, thus it is evolutionary unrelated to the previously known methanotrophs (10, 26). To avoid misinterpretation due to artifacts inherent to one single technique or sample preparation method, we used scanning electron microscopy (SEM), transmission electron microscopy (TEM), and electron tomography (ET) in combination with various sample preparation methods, including plunge freezing, high-pressure freezing, chemical fixation, cryofixation, and freeze-etching, to investigate the ultrastructure of this bacterium.
At a first glance, “Ca. Methylomirabilis oxyfera” cells appeared as typical rod-shaped Gram-negative bacteria. However, careful inspection revealed that “Ca. Methylomirabilis oxyfera” possesses a unique and not yet described ultrastructure. The cell wall contained multiple longitudinal ridges, which conferred a distinctive polygonal shape to the cells (Fig. 2 to 6). This atypical cell shape was observed in all the independent methods and sample preparations employed. The percentage of cells that depicted the polygonal shape (69%) was in the same range as assessed by FISH using specific probes for “Ca. Methylomirabilis oxyfera” (71%). Taken together, this strongly suggests that this polygonal shape is a real feature of “Ca. Methylomirabilis oxyfera” and is not artificial.
The mechanism by which a certain cell shape is acquired and maintained is not always clearly understood, but it often involves exo- or endoskeletal elements. The exoskeleton-like (glyco)protein surface layer (S-layer) and peptidoglycan components are known to play a role in osmotic and mechanical cell stabilization, as well as in shape maintenance (4, 9, 31). The endoskeletal-like elements, such as the actin-like protein MreB and the tubulin-like protein FtsZ, are known to act as internal scaffolds that influence the cell shape (21, 41). Another endoskeletal protein is the intermediate filament crescentin (CreS). The presence of crescentin is essential for the formation of the vibrioid and helical cell shape of Caulobacter crescentus, and the absence of it leads to cells with a straight, rod cell shape (2). The genome of “Ca. Methylomirabilis oxyfera” contains both mreB (open reading frame [ORF] identifier DAMO_3131) and ftsZ (ORF identifier DAMO_2292) genes, but no homologue of creS, the gene encoding crescentin, is found. However, these known shape-determining elements alone cannot explain how complex cell shapes, like the polygonal cell shape of “Ca. Methylomirabilis oxyfera,” are formed and maintained. The square shape of Haloquadratum walsbyi (36) is an example of another complex and unusual cell shape. In this archaeon, it is hypothesized that the square shape is derived from the presence of a cross-linked matrix of poly-gamma-glutamate that forms a capsule outside the cell (3). It is possible that the polygonal cell shape in “Ca. Methylomirabilis oxyfera” also is derived from the presence of a capsular matrix, as has been suggested for H. walsbyi. If so, the composition of this matrix most likely is different from the one in H. walsbyi, since “Ca. Methylomirabilis oxyfera” lacks the genes encoding the poly-gamma-glutamate biosynthesis protein complex CapBCA (1). However, in contrast to the prototypical Gram-negative bacteria, where the outer membrane gives the bacteria a rough appearance when examined by SEM, the cell surface of “Ca. Methylomirabilis oxyfera” was relatively smooth (Fig. 2). This smoothness was consistent with the presence of an additional layer as the outermost sheath, a putative S-layer (Fig. 3 and 5C, inset, and D). This layer might play a role in cell shape maintenance and/or determination. Nevertheless, this hypothesis requires further investigation, and the possibility that the polygonal cell shape is derived from an endoskeleton-like element cannot be ruled out.
To our knowledge, the presence of a star-like cell shape was reported only once in the literature (38). It was found in a branching, filamentous bacterium from a deep-surface mine slime. The authors named the bacteria “star-shaped bacteria” due to their appearance as stars in transverse sections of chemically fixed cells. Unfortunately, there is no genetic information available about these star-shaped bacteria; otherwise, it would be interesting to investigate whether it is phylogenetically related to “Ca. Methylomirabilis oxyfera,” and whether both organisms share unique genes involved in cell shape determination.
Strikingly, one ultrastructural feature was not observed in “Ca. Methylomirabilis oxyfera” cells under the present growth conditions, namely, ICMs. With the exception of verrucomicrobial species, ICMs are common to all known pMMO-containing methanotrophs (13, 34). The extension and arrangement of the ICM might, however, differ from species to species and with growth conditions (5). In this study, we did not observe ICMs of any kind in “Ca. Methylomirabilis oxyfera” cells. Such negative results, however, must be interpreted cautiously because they do not necessarily imply a genetic incapability to produce such structures. “Ca. Methylomirabilis oxyfera” is an extremely slow-growing organism; the estimated doubling time is 1 to 2 weeks under laboratory conditions (11), with a metabolic rate of 1.7 nmol methane oxidized min−1 mg protein−1 (12). It was suggested that the slow metabolism was due to suboptimal growth conditions (i.e., the lack of an essential growth cofactor) (40), but this still needs further investigation. One possible explanation for the lack of ICMs is that it is energetically disadvantageous for “Ca. Methylomirabilis oxyfera” to produce ICMs, since to do so requires a considerable energy investment that does not comply with its slow metabolism. Hence, the question remains whether optimal growth conditions would trigger the development of ICMs or whether the lack of these structures is an intrinsic property of “Ca. Methylomirabilis oxyfera.”
In conclusion, we found that “Ca. Methylomirabilis oxyfera” cells possess an unusual polygonal cell shape. The mechanism of polygonal cell shape determination, however, remains a puzzle to be solved. The unique cell shape of “Ca. Methylomirabilis oxyfera” provides clear differentiation from other morphotypes and might be valuable as a morphology-based tool for identification. Other observations, such as the putative S-layer and the apparent absence of ICMs, are interesting and a challenge for future research on the formation of the atypical polygonal cell shape and the actual subcellular localization of the pMMO enzyme.
Supplementary Material
ACKNOWLEDGMENTS
We thank Katinka van de Pas-Schoonen for support in maintaining the enrichment cultures, Bruno Humbel and Rob Mesman for the operation of the high-pressure freezer, Geert-Jan Janssen for support in operating the cryoscanning electron microscope, and Katharina F. Ettwig, Francisca Luesken, Bas Dutilh, Huub Op den Camp, and Jan T. Keltjens for stimulating discussions.
L.V.N. is supported by the Netherlands Organization for Scientific Research (VENI grant 863.09.009), M.L.W. by a Horizon grant (050-71-058), and M.S.M.J. by ERC 232937.
Footnotes
Published ahead of print 21 October 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ashiuchi M. 2002. Biochemistry and molecular genetics of poly-gamma-glutamate synthesis. Appl. Microbiol. Biotechnol. 59:9–14 [DOI] [PubMed] [Google Scholar]
- 2. Ausmees N, Kuhn JR, Jacobs-Wagner C. 2003. The bacterial cytoskeleton: an intermediate filament-like function in cell shape. Cell 115:705–713 [DOI] [PubMed] [Google Scholar]
- 3. Bolhuis H, et al. 2006. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics 7:169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cabeen MT, Jacobs-Wagner C. 2005. Bacterial cell shape. Nat. Rev. Microbiol. 3:601–610 [DOI] [PubMed] [Google Scholar]
- 5. De Boer W, Hazeu W. 1972. Observations on the fine structure of a methane-oxidizing bacterium. Antonie Van Leeuwenhoek 38:33–47 [DOI] [PubMed] [Google Scholar]
- 6. Dedysh SN. 2009. Exploring methanotroph diversity in acidic northern wetlands: molecular and cultivation-based studies. Microbiology 78:655–669 [Google Scholar]
- 7. Dedysh SN, et al. 2004. Methylocella tundrae sp. nov., a novel methanotrophic bacterium from acidic tundra peatlands. Int. J. Syst. Evol. Microbiol. 54:151–156 [DOI] [PubMed] [Google Scholar]
- 8. Dunfield PF, et al. 2007. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature 450:879–882 [DOI] [PubMed] [Google Scholar]
- 9. Engelhardt H. 2007. Are S-layers exoskeletons? The basic function of protein surface layers revisited. J. Struct. Biol. 160:115–124 [DOI] [PubMed] [Google Scholar]
- 10. Ettwig KF, et al. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548 [DOI] [PubMed] [Google Scholar]
- 11. Ettwig KF, et al. 2008. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ. Microbiol. 10:3164–3173 [DOI] [PubMed] [Google Scholar]
- 12. Ettwig KF, van Alen T, van de Pas-Schoonen KT, Jetten MSM, Strous M. 2009. Enrichment and molecular detection of denitrifying methanotrophic bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75:3656–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanson R, Hanson T. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hayat MA. 1986. Glutaraldehyde: role in electron microscopy. Micron Microsc. Acta 17:115–135 [Google Scholar]
- 15. Hu S, et al. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ. Microbiol. Rep. 1:377–384 [DOI] [PubMed] [Google Scholar]
- 16. Islam T, Jensen S, Reigstad LJ, Larsen ØBirkeland NK. 2008. Methane oxidation at 55 °C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. U. S. A. 105:300–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knittel K, Boetius A. 2009. Anaerobic oxidation of methane: progress with an unknown process. Annu. Rev. Microbiol. 63:311–334 [DOI] [PubMed] [Google Scholar]
- 18. Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 19. Luesken FA, et al. 2011. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl. Microbiol. Biotechnol. doi:10.1007/s00253-011-3361-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luesken FA, et al. 2011. pmoA primers for detection of anaerobic methanotrophs. Appl. Environ. Microbiol. 77:3877–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Margolin W. 2009. Sculpting the bacterial cell. Curr. Biol. 19:R812–R822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mollenhauer HH. 1964. Plastic embedding mixtures for use in electron microscopy. Stain Technol. 39:111–114 [PubMed] [Google Scholar]
- 23. Nguyen H-HT, Elliott SJ, Yip JH-K, Chan SI. 1998. The particulate methane monooxygenase from Methylococcus capsulatus (Bath) is a novel copper-containing three-subunit enzyme. J. Biol. Chem. 273:7957–7966 [DOI] [PubMed] [Google Scholar]
- 24. Op den Camp HJM, et al. 2009. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1:293–306 [DOI] [PubMed] [Google Scholar]
- 25. Pol A, et al. 2007. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature 450:874–878 [DOI] [PubMed] [Google Scholar]
- 26. Raghoebarsing AA, et al. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921 [DOI] [PubMed] [Google Scholar]
- 27. Rappé MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369–394 [DOI] [PubMed] [Google Scholar]
- 28. Reynolds ES. 1963. Use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ribbons DW, Michalover JL. 1970. Methane oxidation by cell-free extracts of Methylococcus capsulatus. FEBS Lett. 11:41–44 [DOI] [PubMed] [Google Scholar]
- 30. Sleytr UB. 1997. I. Basic and applied S-layer research: an overview. FEMS Microbiol. Rev. 20:5–12 [Google Scholar]
- 31. Sleytr UB, Beveridge TJ. 1999. Bacterial S-layers. Trends Microbiol. 7:253–260 [DOI] [PubMed] [Google Scholar]
- 32. Smarda J, Smajs D, Komrska J, Krzyzánek V. 2002. S-layers on cell walls of cyanobacteria. Micron 33:257–277 [DOI] [PubMed] [Google Scholar]
- 33. Sohngen NL. 1906. Uber bakterien, welche methan ab knhlenstoffnahrung and energioquelle gebrauchen. Parasitendk. Infectionskr. Abt. 2:513–517 [Google Scholar]
- 34. Trotsenko YA, Murrell JC, Allen SS, Laskin I, Geoffrey MG. 2008. Metabolic aspects of aerobic obligate methanotrophy, p 183–229 In Laskin AI, Sariaslani S, Gadd GM. (ed), Advances in applied microbiology, vol 63 Academic Press, San Diego, CA: [DOI] [PubMed] [Google Scholar]
- 35. van Niftrik L, et al. 2008. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: cell plan, glycogen storage, and localization of cytochrome c proteins. J. Bacteriol. 190:708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Walsby AE. 1980. Square bacterium. Nature 283:69–71 [Google Scholar]
- 37. Walther P, Ziegler A. 2002. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 208:3–10 [DOI] [PubMed] [Google Scholar]
- 38. Wanger G, Onstott TC, Southam G. 2008. Stars of the terrestrial deep subsurface: a novel “star-shaped” bacterial morphotype from a South African platinum mine. Geobiology 6:325–330 [DOI] [PubMed] [Google Scholar]
- 39. Wu ML, et al. 2011. Physiological role of the respiratory quinol oxidase in the anaerobic nitrite-reducing methanotroph “Candidatus Methylomirabilis oxyfera.” Microbiology 157:890–898 [DOI] [PubMed] [Google Scholar]
- 40. Wu ML, et al. 2011. A new intra-aerobic metabolism in the nitrite-dependent anaerobic methane-oxidizing bacterium “Candidatus Methylomirabilis oxyfera.” Biochem. Soc. Trans. 39:243–248 [DOI] [PubMed] [Google Scholar]
- 41. Young KD. 2003. Bacterial shape. Mol. Microbiol. 49:571–580 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.