Abstract
A-type carrier (ATC) proteins of the Isc (iron-sulfur cluster) and Suf (sulfur mobilization) iron-sulfur ([Fe-S]) cluster biogenesis pathways are proposed to traffic preformed [Fe-S] clusters to apoprotein targets. In this study, we analyzed the roles of the ATC proteins ErpA, IscA, and SufA in the maturation of the nitrate-inducible, multisubunit anaerobic respiratory enzymes formate dehydrogenase N (Fdh-N) and nitrate reductase (Nar). Mutants lacking SufA had enhanced activities of both enzymes. While both Fdh-N and Nar activities were strongly reduced in an iscA mutant, both enzymes were inactive in an erpA mutant and in a mutant unable to synthesize the [Fe-S] cluster scaffold protein IscU. It could be shown for both Fdh-N and Nar that loss of enzyme activity correlated with absence of the [Fe-S] cluster-containing small subunit. Moreover, a slowly migrating form of the catalytic subunit FdnG of Fdh-N was observed, consistent with impeded twin arginine translocation (TAT)-dependent transport. The highly related Fdh-O enzyme was also inactive in the erpA mutant. Although the Nar enzyme has its catalytic subunit NarG localized in the cytoplasm, it also exhibited aberrant migration in an erpA iscA mutant, suggesting that these modular enzymes lack catalytic integrity due to impaired cofactor biosynthesis. Cross-complementation experiments demonstrated that multicopy IscA could partially compensate for lack of ErpA with respect to Fdh-N activity but not Nar activity. These findings suggest that ErpA and IscA have overlapping roles in assembly of these anaerobic respiratory enzymes but demonstrate that ErpA is essential for the production of active enzymes.
INTRODUCTION
Iron-sulfur ([Fe-S]) clusters are ubiquitous prosthetic groups of many metalloenzymes in almost all life-forms and have a variety of functions in diverse cellular processes. They play a particularly important role in electron transfer in the diverse respiratory oxidoreductases found in microorganisms. Generation of [Fe-S] clusters does not occur spontaneously but requires dedicated machineries that orchestrate their assembly and subsequent transfer to the apoprotein substrates (for reviews, see references 3, 20, and 36). There are at least three different [Fe-S] biosynthetic systems known in microbes, and they are referred to as Nif (nitrogen fixation associated), Isc (iron-sulfur cluster), and Suf (sulfur mobilization). The initial discovery of the specialized NifUS proteins for the generation of [Fe-S] clusters in the nitrogenase enzyme of the nitrogen-fixing bacterium Azotobacter vinelandii (19) made it immediately clear that further generalized [Fe-S] machineries in bacteria must exist and these are represented by the Isc and Suf systems in many microbes (45, 53).
The protein components of the Isc and Suf biogenesis systems can be roughly divided into those proteins dedicated to [Fe-S] cluster assembly and those involved in the subsequent trafficking of the preformed cluster to the ultimate apoprotein acceptor (36). The proteins proposed to be involved in transfer or trafficking of the [Fe-S] are referred to as A-type carrier (ATC) proteins, and the bacterium Escherichia coli has three of these, termed IscA, SufA, and ErpA (26), which are phylogenetically related (50). There is, however, some debate as to whether these proteins might actually be iron chaperones delivering iron to the respective assembly machineries (52), but current evidence is consistent with a role in direct cluster transfer (33, 36).
An erpA mutation severely affects the ability of E. coli to respire both in the presence of oxygen or in the presence of alternative electron acceptors such as nitrate (26). This appears to contrast with iscA or sufA mutants, which can grow anaerobically (50). E. coli must be, however, at least partially dependent on the Isc system for nitrate respiration, because a mutant lacking the isc operon exhibited a 60% reduction in activity of the global regulator FNR (31). FNR has an oxygen-sensitive [4Fe-4S] cluster, and the protein controls expression of genes required for nitrate respiration (15, 25, 51) (Fig. 1). The extent to which iscA and erpA mutations affect the maturation and activities of the key enzymes of nitrate respiration has not been examined.
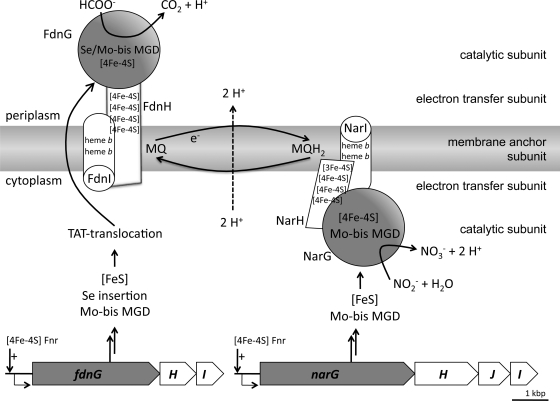
Fig 1.
Schematic representations of the organization of the formate dehydrogenase N and nitrate reductase enzymes in the cytoplasmic membrane of Escherichia coli. Shown are the FNR-regulated structural gene operons for Fdh-N (fdnGHI) and Nar (narGHJI) and the steps required from protein synthesis via cofactor insertion ([Fe-S] cluster, selenocysteine [Se], molybdo-bis-molybdopterin guanine dinucleotide [Mo-bis-MGD]) to Tat-dependent membrane transport for Fdh-N. The membrane anchor subunits FdnI and NarI also contain heme b cofactors, the electron transfer subunits FdnH and NarH have several [FeS] clusters, and the catalytic subunits FdnG and NarG have Mo-bis MGD, [4Fe-4S] and Se (only FdnG). The reaction catalyzed by each enzyme and the connecting menaquinone (MQ)-menaquinol (MQH2)-based redox loop is shown. e−, electron.
Growing E. coli anaerobically in the presence of nitrate induces the synthesis of two large membrane-associated multienzyme complexes, formate dehydrogenase N (Fdh-N) and nitrate reductase (Nar) (40). A further aerobic enzyme, Fdh-O, highly similar to Fdh-N with regard to physiological function, is also synthesized in the presence of nitrate (1, 44). Making use of endogenously generated formate as an electron donor and coupling this to nitrate reduction allow the bacterium to generate a proton gradient using a classical redox-loop chemiosmotic mechanism (8, 21–23, 32, 37) (Fig. 1). Fdh-N, Fdh-O, and Nar have similar modular architectures; they consist of a large membrane extrinsic catalytic subunit, a small electron-transferring subunit, and a membrane-anchoring subunit with a menaquinone/menaquinol-binding site (Fig. 1). The enzymes are arranged on opposite sides of the cytoplasmic membrane, which affords vectorial electron transport but subsequent to biosynthesis requires transport of the FdnGH dimer across the membrane by the twin arginine translocation (TAT) translocon where it subsequently interacts with the integral FdnI subunit to form the final active Fdh-N enzyme (34, 39). Fdh-N/O and Nar have an array of metal-containing prosthetic groups that facilitate electron transfer through each enzyme complex. All three enzymes have a molybdo-bis(molybdopterin guanine dinucleotide) (Mo-bis-MGD) cofactor and a [4Fe-4S] (iron sulfur) cluster in the catalytic subunit, four [4Fe-4S] clusters (or three [4Fe-4S] clusters and one [3Fe-4S] cluster in the case of Nar) in the small subunit and two low-potential heme groups in the membrane anchor subunit. Additionally, Fdh-N (and presumably also Fdh-O) has a selenocysteinyl residue coordinated to the Mo-bis-MGD cofactor, and this is required for enzyme activity (1, 7, 21, 22). While a considerable amount is known regarding the biosynthesis and incorporation of selenocysteine and Mo-bis-MGD into these enzymes (40, 41, 43), much less is known about the requirements for biosynthesis of the [4Fe-4S] clusters. In this study, we analyzed the biosynthesis and activities of Fdh-N and Nar during nitrate respiration of E. coli mutants devoid of the three ATC paralogues IscA, SufA, and ErpA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in this study are listed in Table 1. Strain CP1223 (MC4100 ΔsufA::cat) was constructed by introducing a 327-bp deletion within the sufA gene in strain BW25113 carrying plasmid pKD46 as described previously (13). PCR with Phusion DNA polymerase (Finnzymes, Germany) was carried out using the chloramphenicol resistance cassette from pKD3 as the template and the primers 5′_sufA (5′-CGA TGA AGT GAG GTA AAT CGA TGG ACA TGC ATT CAG GAA CCC ATG GTC CAT ATG AAT ATC CTC C-3′) and 3′_sufA (5′-TAC GAG ACA TAG TAC CGC CTA TAC CCC AAA GCT TTC GCC AGC GAT TGT GTA GGC TGG AGC T-3′) (Metabion, Germany). The mutant sufA allele was transduced by phage P1kc into strain MC4100, resulting in strain CP1223, and the replacement with the 1,028-bp pKD3 fragment was verified by PCR using the sufA cloning primers.
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant genotype or phenotypea | Reference(s) or sourceb |
|---|---|---|
| E. coli strains | ||
| MC4100 | F−araD139 Δ(argF-lac)U169 ptsF25 deoC1 relA1 flbB5301 rspL150 | 10 |
| MG1655 | F− λ−ilvGrfb-50rph-1 | 9 |
| BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ−rph-1 Δ(rhaD-rhaB)568 hsdR514 | 4, 13 |
| CP477 | As MC4100 but ΔiscA::Kanr; ECK2525 | This study |
| CP1221 | As BW25113 but ΔsufA::cat | This study |
| CP1223 | As MC4100 but ΔsufA::cat | This study |
| PB1000 | As MC4100 but Δfnr | 35 |
| LL401 | MG1655 catRExBADerpA, erpA placed under the control of the pBAD promoter | 26 |
| LL402 | MG1655 ΔerpA::Cmr; ECK0155 | 26 |
| DV1151 | MG1655 ΔerpA::Cmr ΔiscA | 49 |
| JW2512 | BW25113 ΔiscA::Kanr; ECK2525 | NBP |
| JW2513 | BW25113 ΔiscU::Kanr; ECK2526 | NBP |
| JW1470 | BW25113 ΔfdnG::Kanr; ECK1468 | NBP |
| JW1215 | BW25113 ΔnarG::Kanr; ECK1218 | NBP |
| JW3815 | BW25113 ΔtatC::Kanr; ECK3832 | NBP |
| Plasmids | ||
| pCP20 | FLP+ λcI857+ λpR Repts Ampr Cmr | 12 |
| perpA | pBluescript SK(+) containing erpA in BamHI and EcoRI site; Ampr | This study |
| pLUE-A | pUC18 expressing erpA; Ampr | 26 |
| piscA | pBluescript SK(+) containing iscA in BamHI and EcoRI site; Ampr | This study |
| psufA | pBluescript SK(+) containing sufA in BamHI and EcoRI site; Ampr | This study |
Allele numbers are given for single gene mutants and refer to E. coli K-12 nomenclature.
NBP, National BioResources Project (NIG, Japan): E. coli.
E. coli was grown aerobically in Erlenmeyer flasks filled to maximally 10% of their volume with TGYEP medium (6) on a rotary shaker (250 rpm) and incubated at 37°C. Anaerobic growth was performed at 37°C in sealed bottles filled with anaerobic growth medium in a nitrogen gas atmosphere. To elicit induction of Fdh-N and Nar enzyme synthesis, 100 mM potassium nitrate was added to the growth medium. When required, the growth medium was solidified with 1.5% (wt/vol) agar. All growth media were supplemented with 0.1% (vol/vol) SLA trace element solution (17). When growing strain DV1151, culture media were supplemented with 0.1 mM mevalonate (50). The antibiotics chloramphenicol, kanamycin, and ampicillin, when required, were added to the medium at the final concentrations of 12.5 μg ml−1, 50 μg ml−1, and 100 μg ml−1, respectively. Where indicated, 0.2% (wt/vol) l-arabinose was added to cultures to induce erpA expression in strain LL401 (arap::erpA) (26).
When required, the Kanr cassette of certain mutants was deleted by transforming with pCP20 encoding a Flp recombinase (12). Mutants were subsequently tested for sensitivity to kanamycin.
Recombinant DNA work was carried out according to published methods (38). Plasmids containing sufA, iscA, and erpA genes were constructed by PCR amplification of the respective DNA fragments from genomic DNA from strain MC4100 using Phusion DNA polymerase (Finnzymes, Germany) and the oligonucleotides sufA_FW_BamHI (F stands for forward) (5′-CGC GGA TCC ATG GAC ATG CAT TCA GGA AC-3′), sufA_RW_EcoRI (R stands for reverse) (5′-CGC GAA TTC CTA TAC CCC AAA GCT TTC GC-3′), iscA_FW_BamHI (5′-CGC GGA TCC ATG TCG ATT ACA CTG AGC GAC-3′), iscA_RW_EcoRI (5′-CGC GAA TTC TCA AAC GTG GAA GCT TTC GC-3′), erpA_FW_BamHI (5′-GCG GGA TCC ATG AGT GAT GAC GTA GCA CTG-3′), and erpA_RW_EcoRI (5′-GCG GAA TTC TTA GAT ACT AAA GGA AGA ACC-3′). The resulting 388-bp (sufA), 337-bp (iscA), and 358-bp (erpA) PCR fragments and the pBluescript SK(+) vector (Stratagene) were digested with BamHI and EcoRI and ligated, resulting in plasmids psufA, piscA, and perpA, respectively. The DNA sequence of each insert was verified (Seqlab, Germany).
Preparation of cell extracts and determination of enzyme activities.
Anaerobic cultures were harvested at an optical density at 600 nm (OD600) of approximately 0.8. Cells from cultures were harvested by centrifugation at 4,000 × g for 10 min at 4°C, resuspended in 2 to 3 ml of morpholinepropanesulfonic acid (MOPS) (pH 7.0) buffer, and lysed on ice by sonication (30-W power for 5 min with 0.5-s pulses). Unbroken cells and cell debris were removed by centrifugation for 15 min at 10,000 × g and at 4°C, and the supernatant was used as the crude cell extract. Formate dehydrogenase N and nitrate reductase enzyme activities were determined as described in reference 14 using MOPS (pH 7.0) buffer. Protein concentration of crude extracts was determined (27) with bovine serum albumin as the standard.
PAGE, activity staining, and immunoblotting.
Aliquots of 25 to 50 μg of protein from the indicated fractions were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using 10% (wt/vol) polyacrylamide (24) and transferred to nitrocellulose membranes as described previously (48). Antibodies raised against Fdh-N (1:5,000) (a kind gift from F. Sargent), and Nar (1:3,000) (a kind gift from A. Magalon) were used. Secondary antibody conjugated to horseradish peroxidase was obtained from Bio-Rad. Visualization was done by the enhanced chemiluminescence reaction (Stratagene).
Nondenaturing PAGE was performed using 5% (wt/vol) polyacrylamide gels (pH 8.5) and included 0.1% (wt/vol) Triton X-100 in the gels (5). Samples (25 μg of protein) were incubated with 5% (wt/vol) Triton X-100 prior to application to the gels. Activity staining to reveal Fdh-O using phenazinemethosulfate and nitroblue tetrazolium (PMS-NBT) and formate was done as described in reference 14, and activity staining to reveal Nar was done as described in reference 18 using nitrate, dithionite, and benzyl viologen (BV) in 50 mM MOPS (pH 7.0) as buffer.
RESULTS
The iron-sulfur cluster trafficking protein SufA is dispensable for formate dehydrogenase and nitrate reductase maturation.
The Fdh-N/O and Nar enzymes each have five [Fe-S] clusters, and we wanted to determine whether the Isc or Suf systems were required during maturation of either enzyme. Initially, we examined the effects of sufA and iscA mutations individually on Fdh-N and Nar enzyme activities in anaerobically respiring E. coli. Under the assay conditions, Fdh-O contributes only a small percentage to the overall activity, and therefore activity will be referred to as Fdh-N only (44). The sufA mutation (strain CP1223) caused an approximately 5-fold increase in both Nar and Fdh-N activity compared with the activity measured in extracts from strains MC4100 and MG1655 (Table 2). These data suggest that wild-type SufA has a negative influence on either activity or synthesis of Fdh-N and Nar, possibly due to competition for [Fe-S] clusters.
Table 2.
Activities of formate dehydrogenase and nitrate reductase are ErpA and IscA dependenta
| Strainb | Sp act of Fdh-N (U mg of protein−1)c | Sp act of Nar (U mg of protein−1)c |
|---|---|---|
| MC4100 | 0.15 ± 0.08 | 0.40 ± 0.09 |
| MG1655 | 0.15 ± 0.03 | 0.55 ± 0.40 |
| BW25113 | 0.13 ± 0.04 | 0.55 ± 0.16 |
| JW1215 (ΔnarG) | 0.21 ± 0.12 | <0.01 |
| JW1470 (ΔfdnG) | 0.03 ± 0.03 | 0.43 ± 0.30 |
| CP477 (ΔiscA) | 0.08 ± 0.03 | 0.08 ± 0.04 |
| CP477 (ΔiscA)(piscA) | 0.37 ± 0.29 | 0.32 ± 0.12 |
| CP477 (ΔiscA)(perpA) | 0.08 ± 0.01 | 0.02 ± 0.01 |
| LL402 (ΔerpA) | <0.01 | 0.02 ± 0.02 |
| LL401 (erpA) | 0.01 ± 0.01 | 0.02 ± 0.02 |
| LL401 + l-arabinose | 0.10 ± 0.06 | 0.3 ± 0.08 |
| LL401(piscA) | 0.01 ± 0.01 | 0.11 ± 0.04 |
| LL401(piscA) + l-Ara | 0.07 ± 0.01 | 0.27 ± 0.05 |
| LL401(perpA)d | 0.52 ± 0.14 | 0.24 ± 0.03 |
| CP1223 (sufA) | 0.71 ± 0.29 | 1.19 ± 0.25 |
| DV1151 (ΔiscA ΔerpA) | <0.01 | 0.02 ± 0.03 |
| JW2513 (iscU) | <0.01 | 0.01 ± 0.04 |
Cells were grown anaerobically in TGYEP medium (pH 6.5) supplemented with 100 mM KNO3.
The strain, relevant genotype, whether it carries a plasmid (e.g., plasmid piscA or perpA), and whether the strain was grown with l-arabinose is shown.
The means ± standard errors of the means of at least three independent experiments are shown.
Performed with two different plasmids encoding ErpA (perpA and pLUE-A) with both delivering similar results.
Effects of mutations in iscU and iscA on nitrate-dependent enzyme activities.
Determination of enzyme activities in a crude extract derived from a mutant unable to synthesize the ATC protein IscA after anaerobic growth in the presence of glucose and nitrate showed a reduction in Fdh-N activity of approximately 50% and in Nar activity of 80% (Table 2). Clearly, Nar activity was more strongly affected by the defect in IscA synthesis than Fdh-N activity. The fact that the activities of both enzymes were absolutely dependent on the Isc machinery was, however, demonstrated by examining enzyme activities in extracts of a mutant unable to synthesize the [Fe-S] scaffold protein IscU (2, 16), which completely lacked activity of either enzyme (Table 2). These data indicate that in the absence of IscA, another ATC protein, possibly ErpA, can accept and traffic [Fe-S] to the Fdh-N and Nar enzymes.
ErpA is essential for Fdh-N and Nar enzyme activities in nitrate-respiring E. coli.
Extracts of two different erpA mutants were totally devoid of Nar and Fdh-N activities (Table 2). As anticipated, activities of both enzymes were also absent in extracts derived from strain DV1151 (50), which carries mutations in both iscA and erpA. This result corroborates the findings described above for the iscU mutant and indicates that the Isc machinery is necessary for [Fe-S] cluster biosynthesis and delivery to the key enzymes of nitrate respiration.
Expression of the erpA gene in strain LL401 is conditional because it is under the control of the ara promoter (arap) (26). Addition of l-arabinose to the anaerobic culture medium restored Fdh-N activity to approximately 60% of the wild-type activity, while Nar activity was restored to 55% of that of strain MG1655 (Table 2). Complementation experiments with plasmids encoding IscA and ErpA revealed that multicopy iscA complemented the iscA mutation and multicopy erpA complemented the erpA mutation, restoring Fdh-N activity to levels that were 2-fold and 3-fold that of the wild type, respectively (Table 2). ErpA and IscA also restored Nar activity to the respective mutants, but only to levels between 50 and 80% of the wild-type activity (Table 2). While multicopy erpA could not restore either Fdh-N or Nar activity in the iscA mutant CP477, multicopy iscA could restore Nar activity to approximately 20% of wild-type activity in the erpA mutant (Table 2). Introduction of sufA on plasmid psufA had no significant influence on the activity of either Fdh-N or Nar in the respective mutant (data not shown). These findings imply a partial functional substitution of iscA for erpA when it is provided in excess, but only for Nar activity. Interestingly, introduction of plasmid piscA into the erpA mutant LL401 failed to result in restoration of any Fdh-N activity (Table 2).
Neither multicopy ErpA nor multicopy SufA could complement the iscA erpA double mutations in strain DV1151 (data not shown).
Taken together, these findings demonstrate that the Isc machinery but not the Suf system is required for [Fe-S] cluster insertion into the Fdh-N and Nar enzymes.
It is important to note that extracts derived from the K-12 derivatives MC4100, BW25113, and MG1655 all showed similar Nar and Fdh-N activities (Table 2). They also showed similar regulation patterns in all respects that we have tested, and thus, the findings presented allow direct comparison between the strains. Furthermore, the absence of either activity did not influence the activity of the other enzyme, e.g., deletion of fdnG did not affect Nar. This indicates that the erpA and iscA mutations affected both enzyme activities separately.
Enzyme-specific staining after native PAGE reveals that ErpA and IscA are essential for Nar and Fdh activity.
Nar enzyme activity can be visualized after nondenaturing PAGE (29). Analysis of crude extracts derived from various strains grown anaerobically with nitrate revealed a single nitrate-dependent active enzyme complex (Fig. 2A) that was absent in extracts of mutants lacking either the narG gene or fnr, which encodes the oxygen-responsive transcriptional regulator FNR of the narGHJI operon (15, 51). Extracts from strains MC4100, JW3815 (ΔtatC), and CP1223 (ΔsufA) had similarly intense activity bands like that observed in a fdnG mutant (Fig. 2A), confirming that Nar maturation does not depend on the Suf machinery. In contrast, the iscA mutant CP477 had a significantly weaker activity, while the erpA (LL402) and iscU (JW2513) mutants had a barely detectable activity band. No active Nar enzyme could be detected in an extract of the erpA iscA double null mutant (DV1151) (Fig. 2A). Complementation studies with plasmids piscA and perpA revealed that only plasmid piscA could restore Nar activity to strain CP477 (iscA) and only plasmid perpA could restore activity to detectable levels in the conditional erpA mutant LL401 (Fig. 2C). In contrast to the weak restoration of Nar activity measured in extracts (Table 2), plasmid piscA did not result in a clearly visible, active Nar enzyme. The reason for this discrepancy might be due to the activity being below the threshold that can be detected by this method. Introduction of sufA on plasmid psufA had no significant influence on Nar activity in the respective mutants (data not shown).
Fig 2.
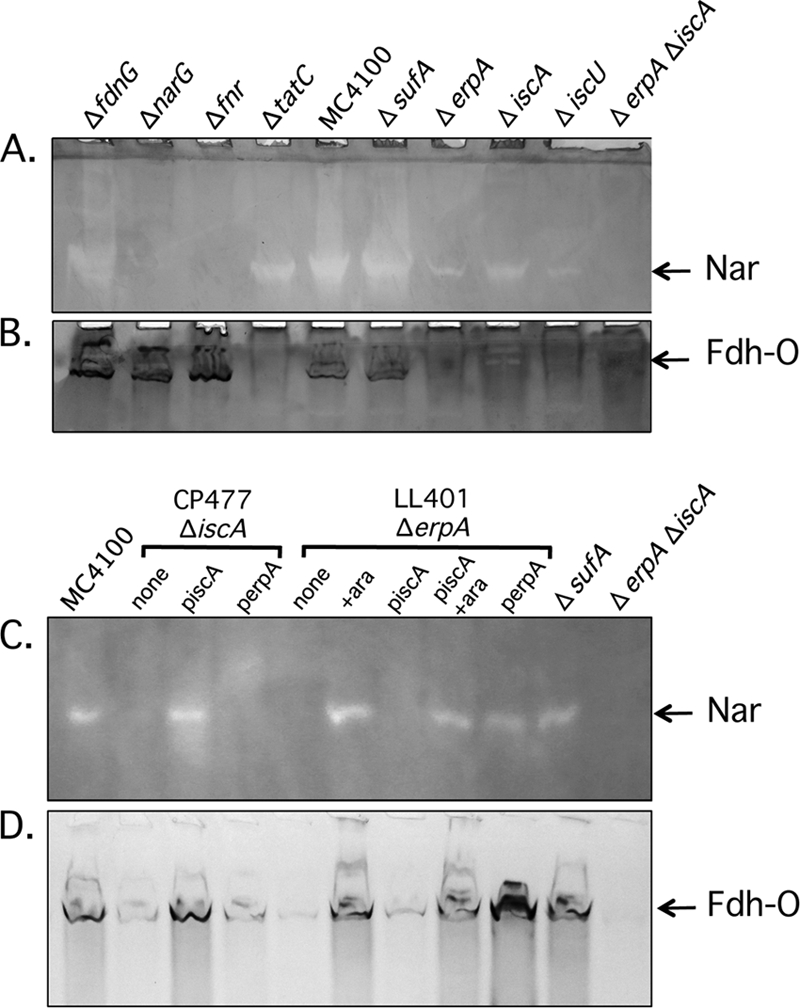
Staining for Nar and Fdh-O activities. In the interest of clarity, only the relevant genotypes of the strains analyzed are shown. Samples (50 μg protein) of crude extracts from strains JW1470 (ΔfdnG), JW1215 (ΔnarG), PB1000 (Δfnr), JW3815 (ΔtatC), MC4100 (wild type), CP1223 (ΔsufA), LL401 (conditional erpA), LL402 (ΔerpA), CP477 (ΔiscA), JW2513 (ΔiscU), and DV1151 (ΔerpA ΔiscA) after anaerobic growth in TGYEP medium (pH 6.5) with 100 mM nitrate were separated in SDS-polyacrylamide gels (7.5% [wt/vol] polyacrylamide) and stained for Nar (A and C) or Fdh-O (B and D) activities as described in Materials and Methods. In a similar manner, samples (25 μg protein) of crude extract from strains CP477 (ΔiscA) and LL401 (araP::erpA) transformed with plasmid piscA (iscA+) or perpA (erpA+) as indicated were stained for Nar (C) or Fdh-O (D) activity. Conditional recovery of erpA gene expression in strain LL401 was induced by supplementing the growth medium with 0.2% (wt/vol) l-arabinose (26).
The activities of the highly similar Fdh-N and Fdh-O enzymes cannot be readily distinguished using standard enzyme assays; however, by specific staining using formate and PMS-NBT after nondenaturing PAGE, the Fdh-O enzyme complex can be readily visualized (44). A single active enzyme complex present in extracts from fdnG, narG, sufA, and fnr mutants could be clearly distinguished and had a similar intensity to that seen in extracts from strain MC4100 (Fig. 2B). Expression of the fdoGHI operon is independent of the FNR regulator (1, 44).
In contrast to Nar, however, but like Fdh-N, maturation of Fdh-O is completely dependent on a functional TAT translocon (49), as no enzyme activity could be detected in extracts derived from the tatC mutant JW3815 (Fig. 2B). No Fdh-O enzyme activity could be visualized in extracts derived from iscA, erpA, iscU, or erpA iscA mutants, which demonstrates that Fdh-O activity was strongly dependent on both ErpA and IscA.
Extracts derived from the single-knockout mutants CP477 (ΔiscA) and LL401 (conditional erpA mutant) complemented with plasmid perpA or piscA were analyzed using the same approach as described above. Each plasmid could restore Fdh-O enzyme activity to the respective mutant (Fig. 2D). Although this assay determines activity using a cumulative approach, and is therefore only semiquantitative, some weak cross-complementation was observed for each plasmid. The findings of this experiment indicate that with respect to Fdh-O enzyme activity, ErpA and IscA exhibit a degree of redundancy.
Mutants lacking ErpA or IscA are devoid of the small electron-transferring subunit and exhibit aberrant Fdh-N and Nar polypeptide patterns.
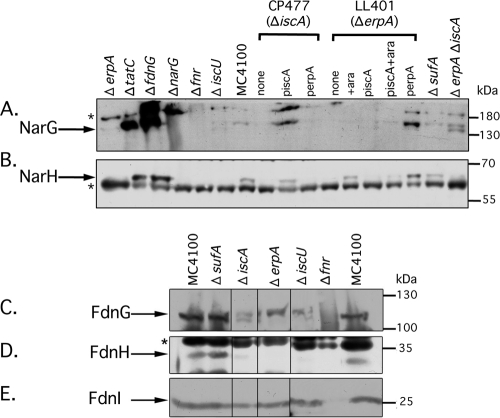
In an attempt to understand the reason for the lack of Fdh-N and Nar enzyme activity in the [Fe-S] cluster trafficking mutants, extracts derived from the various mutants grown under nitrate-respiring conditions were prepared and subjected to Western blot analysis with antiserum raised against either Fdh-N or Nar (Fig. 3). The Fdh-N enzyme comprises the 113-kDa FdnG catalytic subunit, the 32.2-kDa FdnH electron transfer subunit, and the 25.3-kDa FdnI membrane anchor subunit (22), while the Nar enzyme comprises the 140.5-kDa NarG catalytic subunit, the 58.6-kDa NarH electron transfer subunit, and the 25.5-kDa NarI membrane anchor (8). Like the catalytic subunit of Fdh-N, the catalytic subunit of Nar has a [4Fe-4S] cluster (8, 23); however, in contrast to FdnG, NarG lacks a TAT signal peptide. Western blot analysis of extracts from strains MC4100 and PB1000 (Δfnr) allowed unequivocal identification of the NarG subunit in a MC4100 crude extract (Fig. 3A). A mutant lacking either Fdh-N (ΔfdnG) or TatC exhibited very strong signals in Western blots for the NarG polypeptide (Fig. 3A). This might suggest that in the absence of active Fdh-N, Nar is more efficiently assembled.
Fig 3.
Western blot analysis of Nar and Fdh-N proteins. In the interest of clarity, only the relevant genotypes of the strains analyzed are shown. Crude extracts (25 μg protein) derived from strains MC4100 (wild type), CP1223 (ΔsufA), CP477 (ΔiscA), LL401 (conditional erpA), LL402 (ΔerpA), DV1151 (ΔerpA ΔiscA), JW2513 (ΔiscU), JW1470 (ΔfdnG), JW1215 (ΔnarG), PB1000 (Δfnr), and JW3815 (ΔtatC) and where indicated transformed with plasmid piscA (iscA+) or perpA (erpA+), grown in TGYEP medium (pH 6.5) with 100 mM nitrate were separated in SDS–10% (wt/vol) polyacrylamide gels (12.5% [wt/vol] polyacrylamide in panel B) and transferred to nitrocellulose membranes. Samples were probed with antiserum raised against NarG (A), NarH (B), FdnG (C), FdnH (D), or FdnI (E). The asterisks signify unidentified polypeptide species that show cross-reaction with the antiserum and were used as loading controls in the experiments. The migration positions of molecular mass standards (PageRuler Prestained; Fermentas) (in kilodaltons) are shown to the right of the blots. The polypeptides corresponding to the respective Nar and Fdh-N subunits are indicated to the left of the blots.
In extracts derived from mutants that had significantly reduced, or that lacked, Nar enzyme activity, a second, faster migrating NarG polypeptide species could occasionally be observed. This is best exemplified in an extract derived from the erpA iscA double null mutant DV1151 (see the rightmost lane in Fig. 3A). Extracts derived from strains CP477 and LL401 phenotypically or genotypically complemented revealed that multicopy iscA restored high levels of the more slowly migrating NarG polypeptide species to the iscA mutant, and similarly, multicopy erpA restored NarG to LL401 (Fig. 3C) in correlation with the restoration of Nar enzyme activity in the extract (Table 2).
The antiserum raised against Nar also recognized the 58.6-kDa electron-transferring subunit of Nar, NarH (Fig. 3B). Analysis of crude extracts from the various strains revealed a direct correlation between loss of Nar enzyme activity (Table 2) and the absence of detectable NarH polypeptide (Fig. 3B). Extracts of the fnr mutant PB1000 acted as a negative control, and as anticipated (51), no NarH polypeptide could be detected. Similarly, no NarH could be detected in a mutant lacking the gene encoding the large subunit, suggesting that the NarH polypeptide was unstable in the absence of the catalytic subunit. Importantly, extracts derived from mutants devoid of ErpA, IscA, or IscU also lacked NarH, and upon reintroduction of plasmid piscA or perpA, NarH could be restored to the corresponding mutants CP477 (iscA) and LL401, respectively (Fig. 3B). These results suggest that one reason for the lack of Nar activity in mutants devoid of ATC proteins is due to the absence of the iron-sulfur-containing, electron transfer subunit.
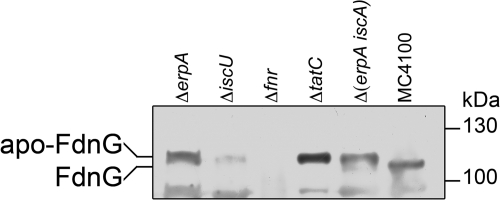
In the Western blots of Fdh-N shown in Fig. 3, FdnG, FdnH, and FdnI could all be identified to migrate as single polypeptides in crude extracts from strain MC4100, each with the approximate size predicted from the deduced molecular mass (Fig. 3C, D, and E). The identities of all three subunits were confirmed by analysis of an extract derived from the fnr mutant PB1000, which lacked all three polypeptides. No transcription of the fdnGHI operon occurs in a fnr mutant (25, 51). An extract from a sufA mutant revealed a polypeptide pattern like that seen for strain MC4100 (Fig. 3C, D, and E), which was anticipated because the mutant showed increased rather then decreased Fdh-N enzyme activity (Table 2). In an extract derived from the iscA mutant CP477, the FdnG polypeptide migrated as two forms, the lower form corresponded to that in strain MC4100, while the second form of the polypeptide migrated marginally more slowly with a molecular mass that would be expected if the TAT signal peptide were not cleaved (49). The two mutants (erpA and iscU) devoid of any measureable Fdh-N enzyme activity (Table 2) showed only the more slowly migrating polypeptide, suggesting that in these strains FdnG could not pass the TAT translocon (Fig. 3A). Analysis of the migration pattern of FdnG in the tatC mutant JW3815 (Fig. 4) revealed that only the more slowly migrating form of the FdnG polypeptide could be identified, thus strongly supporting the contention that in the erpA and iscU mutants, FdnG was not transported across the cytoplasmic membrane.
Fig 4.
Correlation between the slower migrating FdnG polypeptide species identified in certain ATC trafficking mutants with that observed in a ΔtatC strain. In the interest of clarity, only the relevant genotypes of the strains analyzed are shown. Samples (50 μg protein) of crude extracts derived from strains LL402 (ΔerpA), JW2513 (ΔiscU), PB1000 (Δfnr) JW3815 (ΔtatC), DV1151 (ΔerpA ΔiscA), and MC4100 (wild type) after anaerobic growth in TGYEP medium (pH 6.5) with 100 mM nitrate were separated in SDS–8% (wt/vol) polyacrylamide gels, transferred to nitrocellulose membranes, and probed with antiserum raised against FdnG. The migration positions of the apoprotein form (apo-FdnG) and the processed species of FdnG are indicated to the left of the gel, and the sizes (in kilodaltons) of the molecular mass standards (PageRuler Prestained; Fermentas) are given to the right of the gel.
Analysis of the electron-transferring subunit FdnH of the Fdh-N enzyme revealed that in the crude extracts of strains CP477 (ΔiscA), LL402 (ΔerpA), and JW2513 (ΔiscU), no, or barely detectable, FdnH polypeptide could be visualized (Fig. 3B).
With the exception of the fnr mutant PB1000, all of the strains analyzed had essentially comparable levels of the membrane anchor subunit FdnI (Fig. 3B). This indicates that the reason for the smaller amount, or absence, of FdnH was more rapid degradation.
DISCUSSION
In this study, we have provided a plausible explanation for the observation originally made by Loiseau et al. (26) that an E. coli erpA mutant is unable to grow by nitrate respiration. In the absence of this proposed [Fe-S] cluster trafficking protein, maturation through metal cofactor insertion of the key respiratory enzymes Fdh-N and Nar cannot be completed and the enzymes are essentially inactive. Because transcription of both the fdnGHI and narGHJI operons is absolutely dependent on the [Fe-S] protein FNR (15, 25, 51), it was conceivable that this could have been the explanation for the respiratory defect in the erpA mutant. However, identification of both the FdnG and FdnI polypeptides in the erpA single mutant and erpA iscA double mutant indicated that FNR function was not completely compromised and implies that insertion of the [4Fe-4S] cluster into FNR is not totally dependent on the Isc system or ErpA (31). Western blot analysis confirmed the presence of essentially wild-type levels of FNR in extracts of the ATC protein mutants (data not shown). These findings support the results of Mettert et al. (31) who demonstrated that Isc− mutants retain 40% functionality of FNR. As the Suf machinery does not appear to be involved in [Fe-S] cluster assembly into FNR, this might suggest the existence of another route of delivery for particular clusters also under anaerobic conditions. FNR as a key [Fe-S] protein controlling expression of a comparatively large regulon is clearly not reliant on only one pathway for cluster assembly and delivery, whereas for cluster insertion into the electron-transfer subunits of the anaerobic oxidoreductases, the Isc system appears to be the main route.
We could demonstrate for both Fdh-N and Nar that the likely reason for the lack of activity and incomplete maturation of the enzymes is presumably because the electron-transferring small subunit, and possibly also the large catalytic subunit, fails to receive its complement of [4Fe-4S] clusters. This conclusion is based on the apparent enhanced turnover of the FdnH and NarH subunits and the appearance of a slowly migrating form of the FdnG catalytic subunit that correlates in its migration characteristics with the species observed in a tatC mutant. Isolation and spectroscopic characterization of the FdnG polypeptide from an erpA mutant or an iscU mutant, which shared the same phenotype with respect to nitrate respiration, will be required to demonstrate whether the catalytic subunit indeed lacks its [4Fe-4S] cluster or whether the reason for the apparent lack of TAT-dependent transport is because the small subunit was absent. The identification of the intact FdnI anchor polypeptide in the various mutants confirmed that the mature FdnGH dimer is first transported by the TAT machinery and then forms the final active Fdh-N complex after membrane translocation (39).
Supporting evidence that the [4Fe-4S] cluster might indeed be missing or aberrantly inserted into the large subunit of these enzymes was provided by the observation of a faster migrating form of the NarG polypeptide detectable in the iscU single mutant and in the erpA iscA double mutant. In this instance, however, because Nar is not a TAT substrate (39), the faster migrating form of the protein cannot result from processing. Rather, this suggests that either the [4Fe-4S] cluster or the Mo-bis-MGD cofactor (or both) was not properly inserted into the protein in the mutant, thus making the protein more susceptible to endoproteolytic cleavage. Why only maximally half of the population of NarG polypeptides migrated more rapidly in the gel, despite the enzyme being completely inactive, is currently unclear.
It should be noted that in the assembly of Fdh-N, Fdh-O, and Nar, [Fe-S] cluster-containing proteins also play a role in Mo-bis-MGD cofactor biosynthesis (43), while in the case of Fdh-N and Fdh-O maturation, the [Fe-S] protein FdhE has a chaperone function (28), and mutants unable to synthesize FdhE have no Fdh-N enzyme activity (30, 42). Thus, there are several possible steps along the maturation pathway of these enzymes that could be affected in mutants deficient in [Fe-S] cluster trafficking function (Fig. 1).
The stronger dependence on ErpA than on IscA for Fdh and Nar maturation, together with the similar phenotypes of erpA and iscU mutants, indicates that ErpA is central to [Fe-S] cluster insertion by the Isc machinery in nitrate-respiring cells (36, 50). The fact that ErpA and IscA have different but overlapping apoprotein targets is suggested by the observation that, although the approximately 110-amino-acid ErpA and IscA proteins are phylogenetically related, they share only 40% amino acid sequence identity. Increasing the amount of IscA in an erpA mutant by introducing the iscA gene on a plasmid demonstrated that IscA can, to a certain extent, functionally substitute for ErpA and confirms that the proteins have overlapping functions. This has been previously suggested (36). Interestingly, a sufA mutation increased both Nar and Fdh enzyme activities, which suggests that although SufA is not required for iron-sulfur cluster insertion into Nar or Fdh-N polypeptides, it might compete with the IscA and ErpA ATC proteins for iron-sulfur cluster substrate. Notably, SufA shares 32% amino acid identity with ErpA and 48% amino acid identity with IscA. Taken together, these findings suggest that the absolute level of each ATC protein in the cell, together with variations in primary sequence, govern apoprotein substrate specificity.
With regard to anaerobic respiration, IscU sits above IscA, and most probably also ErpA, in the hierarchy of [Fe-S] cluster biogenesis. It has been shown that IscU can transfer a [Fe-S] cluster to IscA but not vice versa (33). The transfer does not occur directly, as IscA and IscU do not interact (47), but occurs possibly via the chaperone HscA, which is encoded at the isc locus on the chromosome. IscU has been proposed to transfer [Fe-S] clusters indirectly to ErpA via IscA in aerobically growing cells (50). The findings presented in this study strongly suggest that IscU can transfer [Fe-S] clusters to ErpA independently of IscA; however, whether this occurs via an intermediate protein, e.g., HscA, remains to be established. In analogy with IscU, the SufBCD scaffold complex donates [Fe-S] clusters to SufA (11) under oxidative stress and during iron limitation (36). SufA appeared only to impede transfer of [Fe-S] clusters to the Fdh-N, Fdh-O, and Nar apoproteins via IscA and ErpA, based on enzyme activity data, indicating a high degree of specificity of the ErpA/IscA for the respective anaerobic apoprotein targets. It will be interesting to determine what controls this specificity and whether apoprotein target-specific chaperone proteins might recruit specific ATC proteins.
Finally, the question arises whether any of the ATC proteins might transfer [3Fe-4S] clusters preferentially compared to [4Fe-4S] clusters into the electron transfer subunits of the enzymes or whether ErpA and IscA do not distinguish between these cluster types. For example, recent studies looking at the roles of IscA and SufA in aerobic metabolism have indicated that a distinct and as yet undefined [Fe-S] cluster assembly pathway is responsible for introduction of [2Fe-2S] clusters into FhuF and the SoxR regulator (46). The overlapping, yet differential dependence on both IscA and ErpA also might be consistent with the suggestion that IscA and ErpA assemble different [Fe-S] clusters into certain apoprotein substrates with different efficiencies. While Fdh-N has only [4Fe-4S] clusters in the holoenzyme, Nar has a single [3Fe-4S] cluster, as well as [4Fe-4S] clusters. Whether ErpA and IscA exhibit differential cluster transfer to different apoprotein targets will be one of the questions addressed in future studies.
ACKNOWLEDGMENTS
We thank Frédéric Barras, Marseille, France, for supplying strains and plasmids and for helpful discussions. We are also indebted to Axel Magalon, Marseille, France, for supplying antibodies against E. coli nitrate reductase and Frank Sargent and Tracy Palmer, both at the University of Dundee, for supplying antibodies against Fdh-N. The “National BioResources Project (NIG, Japan): E. coli” is thanked for providing E. coli mutants.
This work was supported by grant Sa 494/3-1 from the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print 11 November 2011
REFERENCES
- 1. Abaibou H, Pommier J, Benoit S, Giordano G, Mandrand-Berthelot MA. 1995. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J. Bacteriol. 177: 7141–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agar JN, et al. 2000. IscU as a scaffold for iron-sulfur cluster biosynthesis: sequential assembly of [2Fe-2S] and [4Fe-4S] clusters in IscU. Biochemistry 39: 7856–7862 [DOI] [PubMed] [Google Scholar]
- 3. Ayala-Castro C, Saini A, Outten FW. 2008. Fe-S cluster assembly pathways in bacteria. Microbiol. Mol. Biol. Rev. 72: 110–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baba T, et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ballantine S, Boxer D. 1985. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J. Bacteriol. 163: 454–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Begg Y, Whyte J, Haddock B. 1977. The identification of mutants of Escherichia coli deficient in formate dehydrogenase and nitrate reductase activities using dye indicator plates. FEMS Microbiol. Lett. 2: 47–50 [Google Scholar]
- 7. Berg BL, Li J, Heider J, Stewart V. 1991. Nitrate-inducible formate dehydrogenase in Escherichia coli K-12. I. Nucleotide sequence of the fdnGHI operon and evidence that opal (UGA) encodes selenocysteine. J. Biol. Chem. 266: 22380–22385 [PubMed] [Google Scholar]
- 8. Bertero MG, et al. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat. Struct. Biol. 10: 681–687 [DOI] [PubMed] [Google Scholar]
- 9. Blattner FR, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- 10. Casadaban MJ. 1976. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J. Mol. Biol. 104: 541–555 [DOI] [PubMed] [Google Scholar]
- 11. Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW. 2009. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry 48: 10644–10653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cherepanov P, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14 [DOI] [PubMed] [Google Scholar]
- 13. Datsenko K, Wanner B. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Enoch HG, Lester RL. 1975. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J. Biol. Chem. 250: 6693–6705 [PubMed] [Google Scholar]
- 15. Green J, Paget MS. 2004. Bacterial redox sensors. Nat. Rev. Microbiol. 2: 954–966 [DOI] [PubMed] [Google Scholar]
- 16. Hoff KG, Silberg JJ, Vickery LE. 2000. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97: 7790–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hormann K, Andreesen J. 1989. Reductive cleavage of sarcosine and betaine by Eubacterium acidaminophilum via enzyme systems different from glycine reductase. Arch. Microbiol. 153: 50–59 [Google Scholar]
- 18. Hucklesby DP, Hageman RH. 1973. A staining method for nitrite reductase on polyacrylamide gels after electrophoresis. Anal. Biochem. 56: 591–592 [DOI] [PubMed] [Google Scholar]
- 19. Jacobson MR, et al. 1989. Biochemical and genetic analysis of the nifUSVWZM cluster from Azotobacter vinelandii. Mol. Gen. Genet. 219: 49–57 [DOI] [PubMed] [Google Scholar]
- 20. Johnson DC, Dean DR, Smith AD, Johnson MK. 2005. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 74: 247–281 [DOI] [PubMed] [Google Scholar]
- 21. Jormakka M, Törnroth S, Byrne B, Iwata S. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 295: 1863–1868 [DOI] [PubMed] [Google Scholar]
- 22. Jormakka M, Byrne B, Iwata S. 2003. Formate dehydrogenase–a versatile enzyme in changing environments. Curr. Opin. Struct. Biol. 13: 418–423 [DOI] [PubMed] [Google Scholar]
- 23. Jormakka M, Richardson D, Byrne B, Iwata S. 2004. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure 12: 95–104 [DOI] [PubMed] [Google Scholar]
- 24. Laemmli U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- 25. Li J, Stewart V. 1992. Localization of upstream sequence elements required for nitrate and anaerobic induction of fdn (formate dehydrogenase-N) operon expression in Escherichia coli K-12. J. Bacteriol. 174: 4935–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loiseau L, et al. 2007. ErpA, an iron-sulfur (Fe-S) protein of the A-type essential for respiratory metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104: 13626–13631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lowry O, Rosebrough N, Farr A, Randall R. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193: 265–275 [PubMed] [Google Scholar]
- 28. Lüke I, et al. 2008. Biosynthesis of the respiratory formate dehydrogenases from Escherichia coli: characterization of the FdhE protein. Arch. Microbiol. 190: 685–696 [DOI] [PubMed] [Google Scholar]
- 29. Lund K, DeMoss JA. 1976. Association-dissociation behavior and subunit structure of heat-released nitrate reductase from Escherichia coli. J. Biol. Chem. 251: 2207–2216 [PubMed] [Google Scholar]
- 30. Mandrand-Berthelot MA, Couchoux-Luthaud G, Santini CL, Giordano G. 1988. Mutants of Escherichia coli specifically deficient in respiratory formate dehydrogenase activity. J. Gen. Microbiol. 134: 3129–3139 [DOI] [PubMed] [Google Scholar]
- 31. Mettert EL, Outten FW, Wanta B, Kiley PJ. 2008. The impact of O2 on the Fe-S cluster biogenesis requirements of Escherichia coli FNR. J. Mol. Biol. 384: 798–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitchell P. 1979. Keilin's respiratory chain concept and its chemiosmotic consequences. Science 206: 1148–1159 [DOI] [PubMed] [Google Scholar]
- 33. Ollagnier-de-Choudens S, Sanakis Y, Fontecave M. 2004. SufA/IscA: reactivity studies of a class of scaffold proteins involved in [Fe-S] cluster assembly. J. Biol. Inorg. Chem. 9: 828–838 [DOI] [PubMed] [Google Scholar]
- 34. Palmer T, Sargent F, Berks B. 2005. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 13: 175–180 [DOI] [PubMed] [Google Scholar]
- 35. Pinske C, Bönn M, Krüger S, Lindenstrauß U, Sawers RG. 2011. Metabolic deficiencies revealed in the biotechnologically important model bacterium Escherichia coli BL21(DE3). PLoS One 6: e22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Py B, Barras F. 2010. Building Fe-S proteins: bacterial strategies. Nat. Rev. Microbiol. 8: 436–446 [DOI] [PubMed] [Google Scholar]
- 37. Richardson D, Sawers RG. 2002. Structural biology. PMF through the redox loop. Science 295: 1842–1843 [DOI] [PubMed] [Google Scholar]
- 38. Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39. Sargent F. 2007. Constructing the wonders of the bacterial world: biosynthesis of complex enzymes. Microbiology 153: 633–651 [DOI] [PubMed] [Google Scholar]
- 40. Sawers RG. 1994. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Van Leeuwenhoek 66: 57–88 [DOI] [PubMed] [Google Scholar]
- 41. Sawers RG, Clark DP. July 2004, posting date Module 3.5.3, Fermentative pyruvate and acetyl-coenzyme A metabolism. In Curtiss R, III, et al. (ed), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC: http://www.ecosal.org [DOI] [PubMed] [Google Scholar]
- 42. Schlindwein C, Giordano G, Santini CL, Mandrand MA. 1990. Identification and expression of the Escherichia coli fdhD and fdhE genes, which are involved in the formation of respiratory formate dehydrogenase. J. Bacteriol. 172: 6112–6121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwarz G, Mendel RR, Ribbe MW. 2009. Molybdenum cofactors, enzymes and pathways. Nature 460: 839–847 [DOI] [PubMed] [Google Scholar]
- 44. Soboh B, et al. 2011. The respiratory molybdo-selenoprotein formate dehydrogenases of Escherichia coli have hydrogen: benzyl viologen oxidoreductase activity. BMC Microbiol. 11: 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takahashi Y, Tokumoto U. 2002. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J. Biol. Chem. 277: 28380–28383 [DOI] [PubMed] [Google Scholar]
- 46. Tan G, Lu J, Bitoun JP, Huang H, Ding H. 2009. IscA/SufA paralogues are required for the [4Fe-4S] cluster assembly in enzymes of multiple physiological pathways in Escherichia coli under aerobic growth conditions. Biochem. J. 420: 463–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tokumoto U, et al. 2002. Network of protein-protein interactions among iron-sulfur cluster assembly proteins in Escherichia coli. J. Biochem. 131: 713–719 [DOI] [PubMed] [Google Scholar]
- 48. Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U. S. A. 76: 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tullman-Ercek D, et al. 2007. Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J. Biol. Chem. 282: 8309–8316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vinella D, Brochier-Armanet C, Loiseau L, Talla E, Barras F. 2009. Iron-sulfur (Fe/S) protein biogenesis: phylogenomic and genetic studies of A-type carriers. PLoS Genet. 5: e1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walker MS, DeMoss JA. 1991. Promoter sequence requirements for Fnr-dependent activation of transcription of the narGHJI operon. Mol. Microbiol. 5: 353–360 [DOI] [PubMed] [Google Scholar]
- 52. Yang J, Bitoun JP, Ding H. 2006. Interplay of IscA and IscU in biogenesis of iron-sulfur clusters. J. Biol. Chem. 281: 27956–27963 [DOI] [PubMed] [Google Scholar]
- 53. Zheng L, Cash VL, Flint DH, Dean DR. 1998. Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273: 13264–13272 [DOI] [PubMed] [Google Scholar]