Abstract
BamA interacts with the BamBCDE lipoproteins, and together they constitute the essential β-barrel assembly machine (BAM) of Escherichia coli. The simultaneous absence of BamB and BamE confers a conditional lethal phenotype and a severe β-barrel outer membrane protein (OMP) biogenesis defect. Without BamB and BamE, wild-type BamA levels are significantly reduced, and the folding of the BamA β-barrel, as assessed by the heat-modifiability assay, is drastically compromised. Single-amino-acid substitutions in the β-barrel domain of BamA improve both bacterial growth and OMP biogenesis in a bamB bamE mutant and restore BamA levels close to the BamB+ BamE+ level. The substitutions alter BamA β-barrel folding, and folding in the mutants becomes independent of BamB and BamE. Remarkably, BamA β-barrel alterations also improve OMP biogenesis in cells lacking the major periplasmic chaperone, SurA, which, together with BamB, is thought to facilitate the transfer of partially folded OMPs to the soluble POTRA (polypeptide-transport-associated) domain of BamA. Unlike the bamB bamE mutant background, the absence of BamB or SurA does not affect BamA β-barrel folding. Thus, substitutions in the outer membrane-embedded BamA β-barrel domain overcome OMP biogenesis defects that occur at the POTRA domain of BamA in the periplasm. Based on the structure of FhaC, the altered BamA residues are predicted to lie on a highly conserved loop that folds inside the β-barrel and in regions pointing outside the β-barrel, suggesting that they influence BamA function by both direct and indirect mechanisms.
INTRODUCTION
BamA (Omp85) is a conserved component of a complex that mediates β-barrel outer membrane protein (OMP) assembly in Gram-negative bacteria, mitochondria, and chloroplasts (7, 34, 38). In Escherichia coli, the β-barrel assembly machine (BAM) complex is comprised of a β-barrel OMP, BamA, and four outer membrane lipoproteins, BamBCDE (31, 41). BamA folds into two distinct domains, an outer membrane-embedded β-barrel and five consecutive polypeptide-transport-associated (POTRA) domains (28) exposed to the soluble periplasmic compartment, which separates the outer and inner membranes of the Gram-negative envelope. The β-barrel domain of BamA is thought to catalyze the final steps of assembly of substrate β-barrel OMPs and insertion into the outer membrane (27). The POTRA domains have been shown to make direct contact with OMP polypeptides in vitro (19), presumably via β-augmentation (16).
The essential BamD lipoprotein is thought to stabilize the BamA complex through its interaction with BamC and BamE independently of BamB (21, 31). The recently solved structure of BamD suggests that its C- and N-terminal domains interact with BamA and substrate OMPs, respectively (29). Immunoprecipitation assays have indicated that BamB interacts with the POTRA domain of BamA (16). Furthermore, mutational and biochemical analyses have pinpointed BamB residues that are important for this interaction (39). Recent high-resolution structures of BamB revealed that the BamB residues previously shown to interact with BamA all are clustered on a continuous solvent-exposed surface of the protein (12, 15, 22). Interestingly, these structures also revealed a possible substrate-interacting interface. It is proposed that BamB, through its interaction with BamA's POTRA domain, assists to offload the periplasmic chaperone-bound OMPs to BamA for subsequent assembly and insertion into the outer membrane (39). The high-resolution structure of BamE has been solved recently (18). Mutational analysis found BamE-BamD and BamE-phosphatidylglycerol interaction sites (18), with the latter interactions suggesting a possible role for BamE in anchoring the BAM complex to phosphatidylglycerol-rich regions of the outer membrane.
The presentation of nascent OMPs to the soluble POTRA domains likely is mediated via periplasmic chaperones, such as SurA (1). However, it is not known how POTRA-bound OMPs interact with the BamA β-barrel for final assembly and insertion into the outer membrane (for recent reviews, see references 11 and 20). High-resolution structures of the Bam lipoproteins suggest that the Bam lipoproteins also facilitate the final assembly and insertion of OMPs, in addition to stabilizing or correctly localizing the BAM complex. Hagan et al. (10) reconstituted a functional BAM complex in vitro. While this has been an important advancement in the field, the precise roles of the individual BAM components in OMP assembly remain unclear. Defects in OMP assembly pathways trigger envelope stress response systems that lower stress by two complementary actions, one by increasing the cellular capacity to fold and assemble OMPs and the other by reducing OMP levels via increased degradation and decreased OMP synthesis (8, 9, 25).
In this study, we sought to dissect the roles of two nonessential Bam lipoproteins, BamB and BamE, in OMP biogenesis by exploiting a conditional lethal phenotype of an E. coli strain in which both BamB and BamE are absent. The analysis of BamA from a ΔbamB ΔbamE strain under permissive growth conditions revealed folding and stability defects, suggesting a role for BamB and BamE in the assembly and folding of the BamA β-barrel domain. The compensatory alterations in the β-barrel domain of BamA stabilized BamA, and its folding became independent of BamB and BamE. Based on the crystal structure of FhaC (4), the BamA suppressor residues were mapped to either a conserved loop that folds inside the β-barrel or to the external part of the β-barrel. This suggests that suppressor alterations influence BamA's function by both direct and indirect mechanisms.
MATERIALS AND METHODS
Bacterial strains, genetic methods, and growth media.
All bacterial strains used in this study were derived from RAM1292 (MC4100 Δara714) and are listed in Table 1. Null alleles of bamB, bamE, and surA were constructed by the method of Datsenko and Wanner (5). P1 transduction was carried out as described previously (30). Minimal medium (M63) and Luria-Bertani (LB) broth were prepared as described previously (30). Minimal medium was supplemented with 0.4% glycerol and 0.1% Casamino Acids. When necessary, growth media were supplemented with l-arabinose (0.1%), chloramphenicol (12.5 μg/ml), and kanamycin (25 μg/ml). All other chemicals were of analytical grade.
Table 1.
Bacterial strains used in this study
| Strain | Relevant characteristic(s) | Source |
|---|---|---|
| RAM1292 | MC4100 Δara714 | 40 |
| RAM1826 | RAM1292 BamAF474L | This study |
| RAM1827 | RAM1292 BamAN631T | This study |
| RAM1828 | RAM1292 BamAG635A | This study |
| RAM1829 | RAM1292 BamAG649A | This study |
| RAM1830 | RAM1292 BamAA652V | This study |
| RAM1831 | RAM1292 BamAE780D | This study |
| RAM1832 | RAM1292 BamA-WT | This study |
| RAM1833 | RAM1826 ΔbamB::Kmr | This study |
| RAM1834 | RAM1827 ΔbamB::Kmr | This study |
| RAM1835 | RAM1828 ΔbamB::Kmr | This study |
| RAM1836 | RAM1829 ΔbamB::Kmr | This study |
| RAM1837 | RAM1830 ΔbamB::Kmr | This study |
| RAM1838 | RAM1831 ΔbamB::Kmr | This study |
| RAM1839 | RAM1832 ΔbamB::Kmr | This study |
| RAM1840 | RAM1826 ΔbamE::Cmr | This study |
| RAM1841 | RAM1827 ΔbamE::Cmr | This study |
| RAM1842 | RAM1828 ΔbamE::Cmr | This study |
| RAM1843 | RAM1829 ΔbamE::Cmr | This study |
| RAM1844 | RAM1830 ΔbamE::Cmr | This study |
| RAM1845 | RAM1831 ΔbamE::Cmr | This study |
| RAM1846 | RAM1832 ΔbamE::Cmr | This study |
| RAM1847 | RAM1826 ΔsurA::Kmr | This study |
| RAM1848 | RAM1827 ΔsurA::Kmr | This study |
| RAM1849 | RAM1828 ΔsurA::Kmr | This study |
| RAM1850 | RAM1829 ΔsurA::Kmr | This study |
| RAM1851 | RAM1830 ΔsurA::Kmr | This study |
| RAM1852 | RAM1831 ΔsurA::Kmr | This study |
| RAM1853 | RAM1832 ΔsurA::Kmr | This study |
| RAM1854 | RAM1840 ΔbamB::Kmr | This study |
| RAM1855 | RAM1841 ΔbamB::Kmr | This study |
| RAM1856 | RAM1842 ΔbamB::Kmr | This study |
| RAM1857 | RAM1843 ΔbamB::Kmr | This study |
| RAM1858 | RAM1844 ΔbamB::Kmr | This study |
| RAM1859 | RAM1845 ΔbamB::Kmr | This study |
| RAM1860 | RAM1846 ΔbamB::Kmr | This study |
Protein and DNA methods.
Cell envelope isolation, sodium carbonate extraction, and Western blot analysis were carried out essentially as described previously (1), with two main differences. Western blot analysis and cell envelope isolation were carried out from cultures grown overnight. The following antibody dilutions were used for protein immune blotting: AcrA and OmpA/C/F, 1:17,000; BamA, 1:5,000; GroEL, 1:25,000; and LamB, 1:10,000. Protein bands were visualized with a Molecular Imager ChemiDoc-XRS system from Bio-Rad. Protein bands were quantified using Quantity One software from Bio-Rad.
The heat-modifiability test to assess BamA's folding status was carried out using purified whole-cell envelopes. Envelope samples were solubilized in 1× SDS sample buffer (5% glycerol, 5% β-mercaptoethanol, 1% SDS, and 62.5 mM Tris-HCl, pH 6.8). Prior to SDS-PAGE analysis, SDS buffer-solubilized envelope samples were either heated in a boiling water bath for 5 min or left at room temperature. BamA was detected by Western blot analysis as described above. The crystal structure of FhaC (Protein Data Bank [PDB] no. 2QDZ) was visualized using PyMol software.
Standard PCR and DNA sequencing protocols were followed to amplify and sequence the entire bamA gene and determine its nucleotide sequence. The following primers were used to sequence bamA: yaeT2rev (5′-CGGGGTCACGACAGCTTTTAC-3′), yaeT1fwd (5′-CCATTGCCAGCATTACTTTCTCCG-3′), yaeT3fwd (5′-CCAGGTCAGTCTGACGCCAG-3′), yaeT5fwd (5′-GTTATGGTACAGACGTGACGTTGG-3′), and yaeT6fwd (5′-AGATGCCGTTCTACGAGAACTTC-3′).
Bioinformatic methods.
Homologs of Escherichia coli BamA, BamB, BamC, BamD, and BamE in other bacterial species were determined using xBASE2 at http://xbase.bham.ac.uk (3).
RESULTS
Isolation of revertants that overcome the conditional lethal phenotype of a ΔbamB ΔbamE mutant.
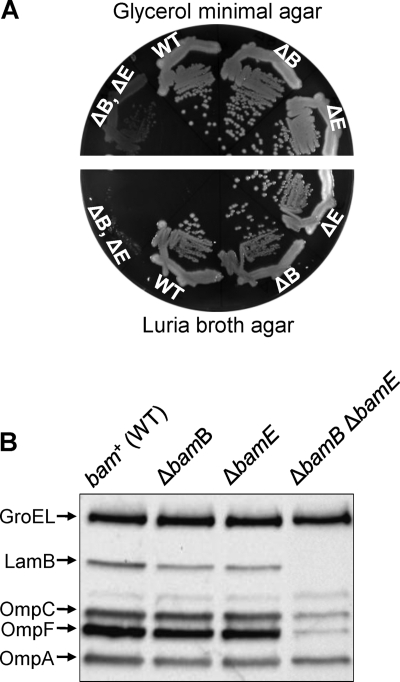
Only BamA and BamD are essential components of the BAM machinery, reflecting their indispensable roles in OMP assembly and bacterial viability (6, 21, 23, 40, 41). However, the simultaneous loss of BamB and BamE was found to be synthetically lethal (31), indicating that these two nonessential lipoproteins play an overlapping role that is critical for the BAM complex assembly and/or function. We discovered that a strain simultaneously lacking BamB and BamE can grow on glycerol minimal agar medium at 30°C, albeit poorly, with a severe OMP biogenesis defect (Fig. 1A and B). However, at 30°C on LB agar, the ΔbamB ΔbamE double mutant fails to form single colonies (Fig. 1A). This conditional lethal phenotype of the ΔbamB ΔbamE strain provided us an opportunity to isolate compensatory mutations, the characterization of which is expected to lead to a better understanding of the roles BamB and BamE play in the assembly and/or activity of the BAM complex.
Fig 1.
Growth and OMP phenotypes of various bam mutants. (A) Growth of wild type (WT), ΔbamB, ΔbamE, and ΔbamB ΔbamE strains on minimal and rich media was assessed by the single-colony isolation method. Plates were incubated for 24 (rich medium) or 36 h (minimal medium) at 30°C. (B) OMPs were detected from whole-cell lysates prepared from overnight cultures grown in glycerol minimal liquid medium at 30°C by Western blot analysis. Each lane contains protein samples from equal number of cells (based on the optical density at 600 nm). Membrane blots were probed with antibodies specific to OmpA, OmpC, OmpF, LamB, and GroEL. GroEL served as a gel loading control.
Aliquots of overnight cultures of a ΔbamB ΔbamE strain, grown in glycerol minimal liquid medium at 30°C, were spread on Luria broth agar plates and incubated overnight at 30°C. A total of 128 revertants from 8 independent cultures were purified on Luria broth agar and glycerol minimal agar plates at 30°C. Only 46 isolates formed stable colonies on Luria broth agar and/or glycerol minimal agar plates. The parental ΔbamB ΔbamE strain is resistant to the OmpF- and LamB-specific phages K20 and λ, respectively, due to dramatically reduced levels of these OMPs (Fig. 1B). If OMP assembly improves in the revertants, then they are expected to become K20- and λ-sensitive again. Indeed, the majority (42 out of 46) showed increased sensitivity to the two phages. Western blot analysis confirmed elevated OmpF and LamB levels in all phage-sensitive revertants compared to that of the ΔbamB ΔbamE parental strain. Ultimately, we picked 20 isolates for further characterization, representing all 8 independent cultures and various growth and phage phenotypes.
Identification of the reversion mutations.
We focused on bamA as the possible site of suppressor mutations, since BamA is the central component of the BAM complex and alterations in it could compensate for the absence of BamB and BamE. DNA sequence analysis of the entire bamA gene from the 20 revertants revealed that 15, all of which were incidentally the most sensitive to both K20 and λ phages, carried a point mutation in bamA. In all, six different point mutations were identified, resulting in a F474L, N631T, G635A, G649A, A652V, or E780D substitution in the mature BamA protein. F474L, G649A, and A652V substitutions occurred in 7, 2, and 3 isolates, respectively, while the other three substitutions were found once in each of the remaining three isolates. All substitutions were located in the β-barrel domain of BamA. The remaining five isolates did not have mutations in bamA and were not analyzed further.
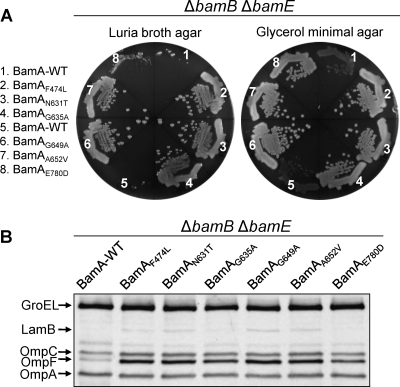
To ensure that the improved OMP biogenesis observed in the phage-sensitive revertants of the ΔbamB ΔbamE strain was due solely to alterations in bamA, the bamA locus from these revertants was moved by P1 transduction into a fresh Bam+ genetic background using a drug-resistant marker closely linked to bamA. The transfer of the suppressor bamA alleles into the fresh genetic background was confirmed by DNA sequence analysis. The null alleles of bamB and bamE then were sequentially transduced into the freshly built strains carrying either the wild-type or suppressor bamA alleles. The restoration of the growth (Fig. 2A) and OMP (Fig. 2B) phenotypes at 30°C to levels similar to those seen in the original revertant backgrounds demonstrated that the improved OMP biogenesis observed in the revertants is due solely to the presence of compensatory alterations in bamA.
Fig 2.
Growth and OMP phenotypes of ΔbamB ΔbamE in the wild-type and various mutant BamA backgrounds. (A) Growth of ΔbamB ΔbamE strains carrying wild type (WT; numbers 1 and 5) or mutant BamA proteins (numbers 2 to 4 and 6 to 8) on Luria broth agar or glycerol minimal agar plates. Plates were incubated for 24 (rich medium) or 36 h (minimal medium) at 30°C. (B) OMP levels were determined from whole-cell lysates prepared from cultures grown overnight in glycerol minimal liquid medium at 30°C by Western blot analysis. Each lane contains protein samples from equal numbers of cells (based on the optical density at 600 nm). The membrane blot was probed with antibodies specific to each protein. GroEL served as a gel loading control.
Suppressor alterations affect BamA levels and folding.
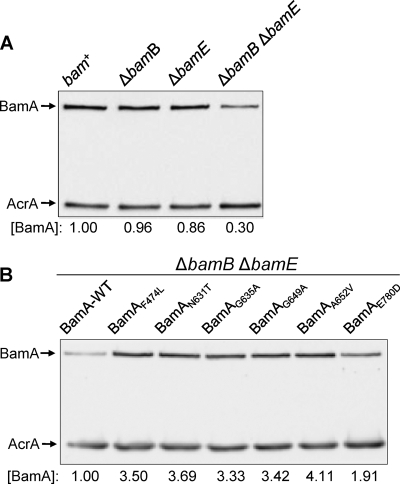
Since the single-amino-acid substitutions in the β-barrel domain of BamA overcome the synthetic lethal phenotype of the ΔbamB ΔbamE double mutant, we suspected that the BamA β-barrel folding and stability were adversely affected by the simultaneous absence of these two nonessential lipoproteins. In view of this, the suppressor alterations must improve the folding of the β-barrel domain so as to stabilize BamA and partially restore its activity without BamB and BamE. To test this possibility, we first examined BamA levels in the ΔbamB ΔbamE background, since lower levels could indicate a defect in BamA's folding/stability. Note that being a member of the σE regulon (25), bamA expression is expected to be activated in the ΔbamB ΔbamE background (31). The wild-type BamA level in the ΔbamB ΔbamE background was reduced to 30% of the level in the parental strains (Fig. 3A), indicating a defect in BamA biogenesis. In contrast to wild-type BamA, the levels of all six mutant BamA proteins were significantly elevated in the ΔbamB ΔbamE background, ranging from around 2-fold (BamAE780D) to 4-fold (BamAA652V) (Fig. 3B). The BamAE780D mutant that produced the weakest growth and OMP phenotypes (Fig. 2) had the smallest effect on the BamA level (Fig. 3B). These data indicated that the simultaneous absence of BamB and BamE destabilizes BamA and reduces its activity. Compensatory substitutions within the BamA β-barrel domain stabilize BamA and partially restore its activity in the ΔbamB ΔbamE background level.
Fig 3.
Determination of BamA levels from different genetic backgrounds. BamA levels were determined by Western blotting of whole-cell lysates prepared from cultures grown overnight in glycerol minimal liquid medium at 30°C. Membrane blots containing protein samples from control strains (A) or BamA mutants (B) were probed with antibodies specific to BamA and AcrA, with the latter serving as a gel loading control. BamA levels were determined relative to the gel loading control, AcrA, and then normalized to the wild-type value of 1.
We next assessed the folding properties of wild-type and mutant BamA proteins by examining their electrophoretic migration behavior on SDS-polyacrylamide gels. The β-barrel (but not the soluble POTRA domains of BamA) has been shown to display OmpA-like heat-modifiability behavior (16, 32). It was first reported that OmpA, a monomeric β-barrel OMP, displays a unique heat-modifiability behavior on SDS-polyacrylamide gels depending on whether the protein samples in SDS buffer are boiled or not prior to electrophoresis (13, 24). Folded OmpA from unheated samples in an SDS buffer migrates faster than the fully denatured form obtained from boiling the sample. This useful property of OmpA has since been exploited to study its folding status and kinetics (17, 33).
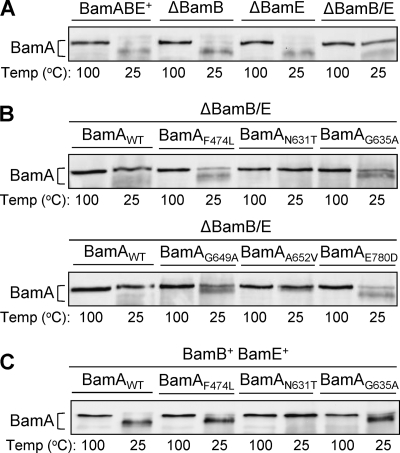
We examined the heat-modifiability behavior of wild-type BamA from envelopes obtained from wild-type, ΔbamB, ΔbamE, and ΔbamB ΔbamE strains. As expected, BamA from the wild-type strain displayed a heat-modifiable property (Fig. 4A). The same also was found to be the case in ΔbamB and ΔbamE single mutants (Fig. 4A). However, in the ΔbamB ΔbamE double mutant, a significant amount of BamA from the unheated samples migrated at the heat-denatured position, indicating an altered state of BamA folding (Fig. 4A). Presumably this altered state of BamA folding compromises its stability and activity, thus contributing to the observed synthetic lethal phenotype of the ΔbamB ΔbamE double mutant.
Fig 4.
Assessment of BamA's heat-modifiability property in different genetic backgrounds. Envelopes obtained from strains grown overnight in glycerol minimal liquid medium at 30°C were extracted with 0.1 M sodium carbonate at 4°C for 1 h. Sodium carbonate-insoluble membrane pellets were solubilized in an SDS buffer. Prior to SDS-PAGE analysis, SDS-solubilized samples either were left at room temperature (25°C) or heated (100°C). The detection of BamA was carried out by Western blot analysis using BamA-specific antibodies. Relevant genetic backgrounds of strains are labeled. BamA in four control strains (A), BamA mutants in a ΔbamB ΔbamE background (B), and three representative BamA mutants in a BamB+ BamE+ background (C) are shown.
We tested whether the suppressors affected BamA's altered heat-modifiability behavior in a ΔbamB ΔbamE background. The data showed that all six suppressor alterations changed the heat-modifiability property of BamA (Fig. 4B) and, hence, its folding. Curiously, however, in no case does the BamA folding pattern return to that seen in the wild-type, ΔbamB, or ΔbamE strain. Based on BamA electrophoretic mobility from unheated envelope samples, three different patterns emerged (Fig. 4B). First, unheated BamAF474L and BamAE780D migrated slightly slower than unheated wild-type BamA. The same also was true for BamAG635A and BamAG649A, except that they migrated even slower than BamAF474L and BamAE780D and the heat-modifiable BamA species were clearly visible (Fig. 4B). In the case of BamAN631T and BamAA652V, BamA from unheated envelope samples migrated at or very close to the denatured position.
Despite the substantial increase in the BamAN631T level (Fig. 3B) and improved OMP biogenesis in the BamAN631T BamB− BamE− background (Fig. 2B), the heat-modifiability data indicate that BamAN631T's folding was severely compromised (Fig. 4B). We tested to see if the anomalous gel mobility shifts of BamAN631T and two other variants, BamAF474L and BamAG635A, together representing the three distinct mobility patterns, depend on the simultaneous absence of BamB and BamE. Results showed that, unlike wild-type BamA, the three BamA mutants in the BamB+ BamE+ background displayed the same heat-modifiability behavior as that in the BamB− BamE− background (Fig. 4B and C), indicating that the altered folding and heat-modifiability behavior are intrinsic folding properties of the BamA mutants independent of the two lipoproteins. Moreover, as will be shown below, these BamA mutants (and the other three) did not exhibit compromised activities in a BamB+ BamE+ background, demonstrating that the in vivo folding of the BamA mutants is not severely compromised, as one might conclude by looking at the in vitro mobility shift.
Effects of BamA β-barrel alterations on OMP biogenesis in ΔbamB and ΔbamE single-mutant backgrounds.
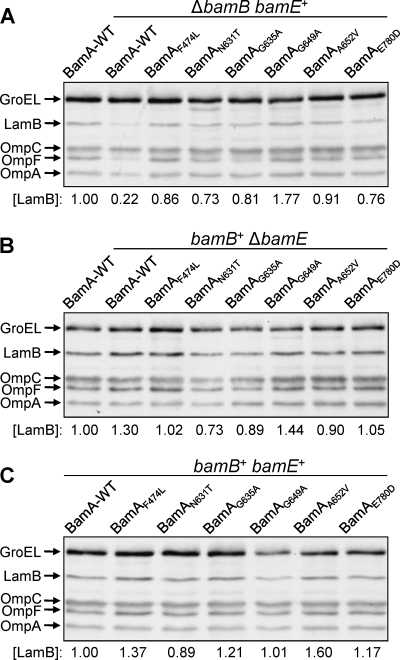
The ΔbamB and ΔbamE single mutants showed no obvious OMP biogenesis defects when steady-state OMP levels were analyzed from cultures grown on minimal medium at 30°C (Fig. 1B). However, the absence of BamB, but not BamE, impairs OMP biogenesis in cells grown on Luria broth at 30°C (Fig. 5A and B). We asked whether BamA β-barrel alterations can reverse OMP biogenesis defects in single-mutant backgrounds just as they can in the bamB bamE double mutant. The presence of BamA β-barrel alterations increased LamB levels at least 3-fold in the ΔbamB background. A similar positive effect of BamA suppressor alterations was observed for OmpF and OmpA and, to a lesser extent, for OmpC. (Fig. 5A). It is worth noting that the gamut of effects of ΔbamB on the four β-barrel OMPs observed here (Fig. 5A, lane 2) are in good agreement with a previous report (2). The fact that BamA alterations largely reverse the OMP biogenesis defect of ΔbamB, which does not influence BamA levels or folding (Fig. 3 and 4), suggests that BamA alterations directly or indirectly influence a step of OMP assembly where BamB normally acts, i.e., at the step of OMP-BamA POTRA interactions. The presence of BamA β-barrel alterations produced no significant change in the steady-state OMP levels in wild-type and ΔbamE backgrounds (Fig. 5B and C).
Fig 5.
Effects of BamA β-barrel alterations on OMP levels in different genetic backgrounds. Whole-cell extracts were obtained from cultures grown overnight in Luria broth at 30°C. OMPs were detected by Western blotting as described in the legend to Fig. 1. OMPs from ΔbamB (A), ΔbamE (B), and wild-type (BamB+ BamE+) (C) backgrounds are shown. LamB levels were determined relative to the gel loading control, GroEL, and then normalized to the wild-type value of 1.
Effects of BamA β-barrel alterations on OMP biogenesis in a ΔsurA background.
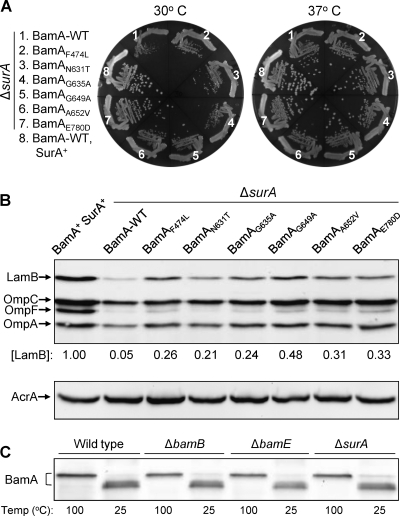
SurA is a major chaperone involved in the folding/assembly of most β-barrel OMPs (1, 36). SurA interacts directly with the POTRA 1 domain of BamA (1), presumably to offload partially folded OMPs to the BamA POTRA domain to complete their folding and assembly. Even though both BamB and SurA interact with the BamA POTRA domain, they do so independently of each other (39). If BamA β-barrel alterations influence OMP assembly at a step involving OMP-BamA POTRA interactions, as we have suggested above, then it is possible that BamA β-barrel alterations also improve OMP biogenesis in a ΔsurA background. The surA null allele confers a cold-sensitive growth phenotype on Luria broth agar, so it was introduced by P1 transduction into BamA wild-type and mutant backgrounds at 37°C. The purified transductants then were tested for growth at 30 and 37°C. All strains produced similar-sized colonies at 37°C, but at 30°C, surA strains carrying a mutant bamA allele grew somewhat or significantly better than the surA mutant expressing wild-type bamA (Fig. 6A). We tested whether improved growth at 30°C is due to improved OMP biogenesis. LamB levels in the surA strains carrying the mutant bamA alleles increased 4- to 10-fold above that of strains expressing wild-type bamA (Fig. 6B). Moreover, surA strains bearing the mutant bamA alleles displayed increased sensitivity to the λ phage, indicating the improved functional assembly of LamB. Levels of the other three major OMPs also increased in surA strains expressing the mutant bamA alleles (Fig. 6B).
Fig 6.
Effects of BamA β-barrel alterations on growth and OMP phenotypes in a ΔsurA background. (A) Growth of various strains on Luria broth agar after 24 h of incubation at 30 and 37°C. The relevant genotypes of the strains are shown. (B) Determination of OMP and AcrA levels by Western blot analysis, as described in the legend to Fig. 1. LamB levels were calculated relative to AcrA, a gel loading control, and normalized to the wild-type value of 1. (C) Examination of the BamA's heat-modifiable property in various genetic backgrounds.
The folding status of the BamA β-barrel was tested from envelopes obtained from ΔsurA cultures grown on rich medium at 30°C. The data in Fig. 6C show that the majority of BamA from unheated samples migrated at the heat-modifiable position, indicating no significant effect of ΔsurA on BamA folding. Therefore, BamA β-barrel alterations in a ΔsurA background partially overcome the OMP biogenesis defect at the step of OMP-BamA POTRA domain interactions.
DISCUSSION
Growth medium and temperature greatly influence bacterial growth rate, with faster-dividing cells requiring efficient cellular biosynthesis and assembly machinery, such as the BAM complex, to cope with proliferating cell mass. Up to a certain point, some defects in the BAM complex and/or other OMP assembly factors can be tolerated when the bacterial growth rate is slowed by lowering the incubation temperature and/or culturing on minimal medium (1, 2, 8, 23, 26, 35, 41). The same defects, however, can lead to cell death when the growth rate is increased by culturing cells at an optimum growth temperature and/or on rich medium. This basic principle of bacterial physiology allowed us to discover the conditional lethal phenotype of a mutant E. coli strain expressing a defective BAM complex due to the simultaneous absence of two nonessential lipoproteins, BamB and BamE. The conditional lethality of the ΔbamB ΔbamE double mutant provided us with the opportunity to investigate the potential role the two lipoproteins play in OMP assembly and to isolate suppressor mutations that can compensate for the simultaneous requirement of BamB and BamE.
The data presented here showed that without BamB and BamE, BamA's folding and stability were significantly impaired. This was deduced from the observation that BamA from the double mutant background displayed altered heat-modifiability behavior and was present at much lower levels than in the wild-type, ΔbamB, or ΔbamE background. Normally, BamA, like some other β-barrel OMPs, such as OmpA, migrates faster than its heat-denatured form on an SDS-polyacrylamide gel. Since this property of BamA has been attributed to its β-barrel domain (16, 32), we surmise that without BamB and BamE, BamA's β-barrel fails to fold correctly and is destabilized. We believe it is this loss of proper BamA β-barrel folding and stability, combined with the loss of additional functions of BamB and BamE, that leads to a severe OMP biogenesis defect and a conditional lethal phenotype.
From coimmunoprecipitation studies, BamB and BamE have been shown to interact with BamA independently of each other, as in BamA-BamB and BamA-BamCDE (21, 31). It follows that the absence of BamB or BamE does not affect BamA-BamCDE and BamA-BamB interactions, respectively. Furthermore, the absence of BamE has been shown to weaken the interactions with BamA and the essential BamD lipoprotein (31). Consequently, in the ΔbamB ΔbamE double mutant, BamA-BamD interactions also must be compromised, therefore it is conceivable that it is this aberrant interaction that influences BamA β-barrel's folding property and stability. However, we do not favor this possibility, because in a ΔbamE background, where BamA-BamD interactions are already weakened (31), BamA folds correctly (Fig. 4). Moreover, it is difficult to envisage a further deterioration of BamA-BamD interactions when BamB, in addition to BamE, is absent, since BamB does not influence BamA-BamCDE interactions (21, 31). Instead, we favor the possibility that individually BamB and BamE, directly or via BamD, play overlapping roles in BamA β-barrel folding, and when these two nonessential lipoproteins are simultaneously absent, BamA is partly misfolded.
Consistent with the observation of an altered folding behavior of the BamA β-barrel without BamB and BamE, suppressor alterations exclusively mapped within the β-barrel domain of BamA. These β-barrel alterations produced unique heat-modifiability characteristics, ranging from a fully heat-denatured state even without heat application to that resembling wild-type BamA in a wild-type background. Notably, these heat-modifiable properties of the BamA mutants were independent of BamB and BamE, indicating a novel folded state of the BamA β-barrel. However, despite the new and altered heat-modifiable properties, the suppressor alterations stabilized the mutant BamA proteins in a ΔbamB ΔbamE background and produced no apparent growth or OMP biogenesis defects in a BamB+ BamE+ background. Thus, the unique folded conformation of BamA affords functional benefit in a ΔbamB ΔbamE background without compromising BamA's activity in the wild-type background.
The BamA suppressor alterations also were able to improve the OMP phenotype in a ΔbamB single-mutant background. Knowing that the absence of BamB alone shows no BamA folding defect, and that BamB has been shown to interact with the BamA POTRA domain to facilitate the early events of OMP assembly (16, 39), it is possible that the BamA β-barrel alterations influence a step in OMP assembly where BamB normally acts. Likewise, we observed a marked improvement in the OMP phenotype of a surA mutation in the presence of the BamA β-barrel alterations. Because both BamB and SurA are thought to act at a step of OMP-BamA POTRA interaction (1, 16), it stands to reason that substitutions in the BamA β-barrel domain somehow influence OMP assembly events at the BamA POTRA domain. However, we must also consider the possibility that BamA β-barrel substitutions solely affect BamA β-barrel's activity, e.g., kinetically influencing OMP assembly/membrane insertion, which indirectly compensates for defects in OMP assembly at the BamA POTRA level.
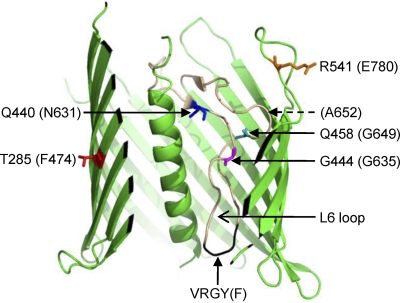
Although the six BamA β-barrel single-amino-acid substitutions we reported here significantly improved growth and/or OMP phenotypes in ΔbamB ΔbamE, ΔbamB, and ΔsurA backgrounds, the amino acid alterations themselves had modest differences in their side-chain substitutions: F474 to L, N631 to T, G635 to A, G649 to A, A652 to V, and E780 to D. It is possible that more drastic changes in the side-chain properties would more adversely affect the structure and/or function of the BamA β-barrel, and they would be eliminated from our genetic selection. Four of the six suppressor sites, N631, G635, G649, and A652, are clustered around the single most conserved region of the β-barrel among Omp85 homologues that includes a highly conserved VRGF/Y motif (in E. coli BamA it is V640R641G642F643) (38), thus emphasizing the significance of this region of BamA/Omp85 in its structure and/or function. Based on the crystal structure of FhaC (4), a member of the Omp85-TpsB superfamily, the four suppressor residues are predicted to lie in the L6 loop that folds inside the barrel (Fig. 7). A deletion of FhaC's L6 loop abolishes its secretion activity (4), indicating a key role for this loop in FhaC's function. The mapping of four BamA suppressor alterations in L6 suggests a role for this loop in BamA's function. It is important to emphasize that unlike FhaC, BamA is not involved in secreting substrates across the outer membrane. Therefore, it is unclear whether the putative L6 loop of BamA is entirely dislodged from the barrel during substrate assembly, as proposed for FhaC (4), or whether it remains within the barrel throughout the assembly process. Unlike the four aforementioned residues, F474 and E780 are predicted to lie on one of the transmembrane β-strands and on a surface-exposed loop, respectively (Fig. 7). Accordingly, alterations at these locations would be expected to influence BamA's function indirectly.
Fig 7.
Cutaway side view of the FhaC β-barrel structure (PDB no. 2QDZ). Shown are FhaC residues that correspond to BamA residues (in parentheses) affected by the suppressor alterations. The dashed arrow points to the BamA residue A652, which is missing from FhaC. The analogous FhaC and BamA residues were taken from the ClustalW analysis carried out by Jacob-Dubuisson et al. (14). Positions of the L6 loop and the conserved VRGF/Y motif in the L6 loop are shown in beige and black colors, respectively. The image was drawn using PyMol.
Although our work in E. coli has shown that the simultaneous absence of BamB and BamE confers a conditional lethal phenotype, many bacteria of the proteobacterial phylum naturally lack BamB or BamE, thus reflecting a mechanistic diversity of the BAM complex. A computer search for the BAM complex components from 12 bacterial species belonging to five different classes of proteobacteria shows a range of variability (Table 2). BamA and BamD are the most conserved components, with the exception of Helicobacter pylori, a member of the epsilon class, which appears to lack BamBCDE orthologs. BamC is the most frequently absent accessory protein, lacking in seven bacterial species, followed by BamB (five) and BamE (three). Four bacteria, Brucella melitensis, Rhodobacter sphaeroides, Rhizobium leguminosarum, and Myxococcus xanthus, lack two accessory lipoproteins of the BAM complex, although in no case does this involve the simultaneous absence of BamB and BamE. It is not known whether the remaining BamB or BamE protein in these four bacterial species performs an essential or critical role. In agreement with this view, in Neisseria meningitidis, which naturally lacks BamB, the deletion of bamE shows a much stronger OMP defect (37) than that reported for E. coli (31 and this work). Given that simple, single-amino-acid substitutions in the BamA β-barrel domain can restore the OMP phenotype of ΔbamB and ΔbamB ΔbamE mutants, it is tempting to speculate that some bacterial species that naturally lack these accessory proteins have acquired compensatory alterations in their BamA β-barrel domain. Consistent with this notion, the BamA sequence from Helicobacter pylori, which appears to lack BamBCDE orthologs, also deviates the most from other species' BamA amino acid composition (Table 2). In addition to protein sequence alterations, there must be other genetic and physiological factors that drive changes in the BAM complex composition to optimally serve the need for OMP biogenesis in different bacteria growing at various rates in different environments.
Table 2.
Composition and sequence homology of Bam proteins in various bacterial speciesa
| Proteobacteria (classb) | GenBank accession no. | Homology no. (ORF) |
||||
|---|---|---|---|---|---|---|
| BamA | BamD | BamE | BamB | BamC | ||
| Escherichia coli K-12 MG1655 (γ) | U00096 | 0.0 (b0177) | 0.0 (b2595) | 0.0 (b2512) | 0.0 (b2617) | 0.0 (b2477) |
| Vibrio cholerae O1 biovar El Tor strain N16961 (γ) | AE003852 | 0.0 (VC_2252) | 1e−68 (VC_0708) | 8e−19 (VC_0851) | 3e−88 (VC_0762) | 4e−32 (VC_2156) |
| Haemophilus influenzae 10810 (γ) | FQ312006 | 0.0 (HIB_10490) | 4e−55 (HIB_02280) | 2e−23 (HIB_09720) | 1e−07 (HIB_03150) | |
| Pseudomonas aeruginosa PA01 (γ) | AE004091 | e−166 (PA3648) | 1e−44 (PA4545) | 7e−13 (PA4765) | 3e−45 (PA3800) | |
| Burkholderia pseudomallei 1106a (β) | CP000572 | e−137 (BURPS1106A_2484) | 3e−38 (BURPS1106A_2206) | 5e−09 (BURPS1106A_3454) | 5e−38 (BURPS1106A_2225) | 0.43d (BURPS1106A_2618) |
| Neisseria meningitidis MC58 (β) | AE002098 | e−123 (NMB0182) | 1e−44 (NMB0703) | 5e−08 (NMB0204) | 0.17c (NMB0928) | |
| Brucella melitensis ATCC 23457 (α) | CP001488 | 8e−82 (Bmea_A1198) | 9e−21 (Bmea_A1470) | 2e−04 (Bmea_A0811) | ||
| Caulobacter crescentus CB15 (α) | AE005673 | 6e−77 (CC_1915) | 2e−21 (CC_1984) | 1.2c (CC_1365) | 1e−17 (CC_1653) | |
| Rhodobacter sphaeroides ATCC 17025 (α) | CP000661 | 1e−68 (Rsph17025_2146) | 1e−15 (Rsph17025_0702) | 5e−21 (Rsph17025_2161) | ||
| Rhizobium leguminosarum bv. trifolii WSM1325 (α) | CP001622 | 6e−68 (Rleg_1783) | 2e−18 (Rleg_2843) | 0.67d (Rleg_1282) | ||
| Myxococcus xanthus DK 1622 (δ) | CP000113 | 1e−62 (Mxan_4728) | 7e−20 (Mxan_1998) | 7e−21 (Mxan_3768) | ||
| Helicobacter pylori 26695 (ε) | AE000511 | 3e−24; 2e−05e (HP0655) | ||||
Homology numbers are displayed as expect values, with smaller numbers reflecting higher matches. Names of the open reading frames corresponding to the BamA-BamE proteins in different bacterial species are shown in parentheses.
Greek letters in parentheses denote various classes of bacterial species.
Presence confirmed by experimental evidence.
Presence based on conserved genetic location of bamC (in the case of Burkholderia pseudomallei) or the conserved domain of bamE (in the case of Rhizobium leguminosarum).
Two distinct homologous regions in the same open reading frame exist, corresponding to residues 26 to 458 and 646 to 807 of E. coli BamA.
ACKNOWLEDGMENTS
We are indebted to Phu Vuong for productive discussion and critical reading of the manuscript and Jon Weeks for an illustration drawn using PyMol. We thank Drew Bennion for training R.T. and his involvement in the initial stages of the project. We acknowledge Tom Silhavy of Princeton University, NJ, for BamA antibodies.
This work was supported by a grant (GM048167) from the National Institutes of Health to R.M. R.T. was supported in part by funds from the School of Life Sciences Undergraduate Research Program.
Footnotes
Published ahead of print 28 October 2011
REFERENCES
- 1. Bennion D, Charlson ES, Coon E, Misra R. 2010. Dissection of β-barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol. Microbiol. 77: 1153–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Charlson ES, Werner JN, Misra R. 2006. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 188: 7186–7194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chaudhuri RR, et al. 2008. xBASE2: a comprehensive resource for comparative bacterial genomics. Nucleic Acids Res. 36: 543–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Clantin B, et al. 2007. Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317: 957–961 [DOI] [PubMed] [Google Scholar]
- 5. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doerrler WT, Raetz CR. 2005. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem. 280: 27679–27687 [DOI] [PubMed] [Google Scholar]
- 7. Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. 2004. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol. 164: 19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gerken H, Leiser OP, Bennion D, Misra R. 2010. Involvement and necessity of the Cpx regulon in the event of aberrant β-barrel outer membrane protein assembly. Mol. Microbiol. 75: 1033–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guillier M, Gottesman S, Storz G. 2006. Modulating the outer membrane with small RNAs. Genes Dev. 20: 2338–2348 [DOI] [PubMed] [Google Scholar]
- 10. Hagan CL, Kim S, Kahne D. 2010. Reconstitution of outer membrane protein assembly from purified components. Science 328: 890–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hagan CL, Silhavy TJ, Kahne D. 2011. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem. 80: 189–210 [DOI] [PubMed] [Google Scholar]
- 12. Heuck A, Schleiffer A, Clausen T. 2011. Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J. Mol. Biol. 406: 659–666 [DOI] [PubMed] [Google Scholar]
- 13. Hindennach I, Henning U. 1975. The major proteins of the Escherichia coli outer cell envelope membrane. Eur. J. Biochem. 59: 207–213 [DOI] [PubMed] [Google Scholar]
- 14. Jacob-Dubuisson F, Villeret V, Clantin B, Delattre A-S, Saint N. 2009. First structural insights into the TpsB/Omp85 superfamily. Biol. Chem. 390: 675–684 [DOI] [PubMed] [Google Scholar]
- 15. Kim KH, Paetzel M. 2011. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J. Mol. Biol. 406: 667–678 [DOI] [PubMed] [Google Scholar]
- 16. Kim S, et al. 2007. Structure and function of an essential component of the outer membrane protein assembly machine. Science 317: 961–964 [DOI] [PubMed] [Google Scholar]
- 17. Kleinschmidt JH, Tamm LK. 1996. Folding intermediates of a β-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism. Biochemistry 35: 12993–13000 [DOI] [PubMed] [Google Scholar]
- 18. Knowles TJ, et al. 2011. Structure and function of BamE within the outer membrane and the β-barrel assembly machine. EMBO J. 12: 123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Knowles TJ, et al. 2008. Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 68: 1216–1227 [DOI] [PubMed] [Google Scholar]
- 20. Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7: 206–214 [DOI] [PubMed] [Google Scholar]
- 21. Malinverni JC, et al. 2006. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol. 61: 151–164 [DOI] [PubMed] [Google Scholar]
- 22. Noinaj N, Fairman JW, Buchanan SK. 2011. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J. Mol. Biol. 407: 248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Onufryk C, Crouch ML, Fang FC, Gross CA. 2005. Characterization of six lipoproteins in the sigma E regulon. J. Bacteriol. 187: 4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Reithmeier RA, Bragg PD. 1974. Purification and characterization of heat-modifiable protein from the outer membrane of Escherichia coli. FEBS Lett. 41: 195–198 [DOI] [PubMed] [Google Scholar]
- 25. Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the sigma E stress response in related genomes. PLoS Biol. 4: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rizzitello AE, Harper JR, Silhavy TJ. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794–6800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robert V, et al. 2006. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 4: e377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sánchez-Pulido L, Devos D, Genevrois S, Vicente M, Valencia A. 2003. POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem. Sci. 28:523–526 [DOI] [PubMed] [Google Scholar]
- 29. Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. 2011. Crystal structure of BamD: an essential component of the β-barrel assembly machinery of gram-negative bacteria. J. Mol. Biol. 409: 348–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Silhavy TJ, Berman ML, Enquist LW. 1984. Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Sklar JG, et al. 2007. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 104: 6400–6405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stegmeier JF, Andersen C. 2006. Characterization of pores formed by YaeT (Omp85) from Escherichia coli. J. Biochem. 140: 275–283 [DOI] [PubMed] [Google Scholar]
- 33. Surrey T, Jahnig F. 1992. Refolding and oriented insertion of a membrane protein into a lipid bilayer. Proc. Natl. Acad. Sci. U. S. A. 89: 7457–7461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tu SL, et al. 2004. Import pathways of chloroplast interior proteins and the outer-membrane protein OEP14 converge at Toc75. Plant Cell 16: 2078–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Typas A, et al. 2008. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat. Methods 5: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vertommen D, Ruiz N, Leverrier P, Silhavy TJ, Collet JF. 2009. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9: 2432–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Volokhina EB, Beckers F, Tommassen J, Bos MP. 2009. The β-barrel outer membrane protein assembly complex of Neisseria meningitidis. J. Bacteriol. 191: 7074–7085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. 2003. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 299: 262–265 [DOI] [PubMed] [Google Scholar]
- 39. Vuong P, Bennion D, Mantei J, Frost D, Misra R. 2008. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J. Bacteriol. 190: 1507–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Werner J, Misra R. 2005. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 57: 1450–1459 [DOI] [PubMed] [Google Scholar]
- 41. Wu T, et al. 2005. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121: 235–245 [DOI] [PubMed] [Google Scholar]