Abstract
The nonsulfur purple bacteria that exhibit unusual metabolic versatility can produce hydrogen gas (H2) using the electrons derived from metabolism of organic compounds during photoheterotrophic growth. Here, based on 13C tracer experiments, we identified the network of glucose metabolism and quantified intracellular carbon fluxes in Rhodobacter sphaeroides KD131 grown under H2-producing conditions. Moreover, we investigated how the intracellular fluxes in R. sphaeroides responded to knockout mutations in hydrogenase and poly-β-hydroxybutyrate synthase genes, which led to increased H2 yield. The relative contribution of the Entner-Doudoroff pathway and Calvin-Benson-Bassham cycle to glucose metabolism differed significantly in hydrogenase-deficient mutants, and this flux change contributed to the increased formation of the redox equivalent NADH. Disruption of hydrogenase and poly-β-hydroxybutyrate synthase resulted in a significantly increased flux through the phosphoenolpyruvate carboxykinase and a reduced flux through the malic enzyme. A remarkable increase in the flux through the tricarboxylic acid cycle, a major NADH producer, was observed for the mutant strains. The in vivo regulation of the tricarboxylic acid cycle flux in photoheterotrophic R. sphaeroides was discussed based on the measurements of in vitro enzyme activities and intracellular concentrations of NADH and NAD+. Overall, our results provide quantitative insights into how photoheterotrophic cells manipulate the metabolic network and redistribute intracellular fluxes to generate more electrons for increased H2 production.
INTRODUCTION
Rhodobacter sphaeroides is a purple nonsulfur bacterium that exhibits extraordinary metabolic versatility. It can grow photoheterotrophically using a variety of organic compounds, including organic acids and sugars, as the carbon source or photoautotrophically using carbon dioxide as the sole carbon source (28). In addition, it can grow chemoheterotrophically and chemoautotrophically in the dark. It is one of the most often used models for photobiological production of hydrogen gas (H2). During photoheterotrophic growth, H2 can be produced by R. sphaeroides and other purple nonsulfur bacteria via nitrogenase, an enzyme that converts dinitrogen to ammonia with H2 as an obligatory product. In the absence of dinitrogen, nitrogenase produces H2 as the sole product using the electrons generated from carbon metabolism and the energy from photosynthesis (18, 39). The synthesis and activity of nitrogenase are repressed by the presence of ammonium (26). Thus, H2 production experiments are usually carried out in medium containing a poor nitrogen source. The highest H2 yields and production rates have been achieved by using glutamate as the nitrogen source (18).
The metabolic versatility of R. sphaeroides is largely owed to its complicated metabolic network. For example, the Calvin-Benson-Bassham (CBB) cycle, Embden-Meyerhof-Parnas (EMP) pathway, Entner-Doudoroff (ED) pathway, pentose phosphate (PP) pathway, and the tricarboxylic acid (TCA) cycle were all reported to be present in this organism (5, 28). This allows R. sphaeroides to flexibly select and operate metabolic pathways and reactions in response to genetic and environmental perturbations. However, despite the recent progress on genomics and transcriptomics (29) and stoichiometric modeling (15) of this bacterium, knowledge of how its various pathways interact and contribute to intracellular metabolism is limited. Particularly, to manipulate R. sphaeroides for efficient photobiological H2 production, it is important to gain insight into how the metabolic network in its entirety is operated to generate the reducing equivalents required for H2 production.
Based on 13C tracer experiments, metabolic flux analysis emerged as a key approach to identify the active pathways and to quantify the intracellular fluxes in a complex metabolic network (35, 43). In this approach, the 13C labeling patterns in products of metabolism, which reflect the in vivo activity of metabolic pathways and enzymes, are analyzed by nuclear magnetic resonance or mass spectrometry (MS). Direct interpretation of the detected 13C patterns using algebraic equations by so-called metabolic flux ratio analysis enables identification of active pathways in a bioreaction network and determination of the ratios of some converging fluxes (10, 33, 36). This method has demonstrated its value in providing direct evidence for novel or unexpected metabolic pathways (11, 19). By combining the 13C data with biomass composition and quantitative physiological data, the absolute intracellular fluxes may be estimated by finding the best fit to all the available data (41). Since the fluxes represent the mathematically best estimate for the given biochemical reaction network, the validity of the network itself may affect the flux result. To avoid this, the bioreaction network identified by flux ratio analysis may be used for flux quantification. The calculated fluxes provide a holistic view of cellular metabolism and can be used for quantitative analysis of redox homeostasis and energy metabolism (6, 16, 32).
In this study, we used 13C-based metabolic flux analysis to quantitatively investigate the intracellular metabolism in R. sphaeroides KD131 grown photoheterotrophically in mineral medium containing glucose and glutamate as the carbon and nitrogen sources, respectively. The network of active pathways was identified by metabolic flux ratio analysis based on gas chromatography (GC)-MS analysis of cellular amino acids, glycerol, and glucose from [U-13C]glucose and [1-13C]glucose labeling experiments. The in vivo fluxes through various pathways were then quantified by 13C-constrained flux analysis. By using this approach, the influence of knocking out H2-uptake hydrogenase or/and poly-β-hydroxybutyrate (PHB) synthase on intracellular flux distribution was investigated. The mutants have been reported to exhibit increased H2 evolution compared to that of the wild type (23). The flux analysis allowed insights into how photoheterotrophic cells manipulate the metabolic network to generate more reducing equivalents for increased H2 production.
MATERIALS AND METHODS
Strains, media, and growth conditions.
The strains used in this study were Rhodobacter sphaeroides wild-type KD131 (KCTC12085) and its knockout mutants including an PHB synthase knockout mutant (ΔphbC strain), an H2-uptake hydrogenase knockout mutant (ΔhupSL strain), and a double-knockout mutant (ΔhupSL ΔphbC strain) (25).
The strains were precultured aerobically at 30°C on Luria-Bertani medium to the mid-exponential growth phase, washed twice, and diluted to the same optical density at 660 nm ([OD660] ∼0.03) using RCVB minimal medium (38) containing (per liter) 1.0 g of sodium glutamate, 0.1 g of yeast extract, 5.0 g of NaHCO3, 0.2 g of MgSO4, 0.075g of CaCl2 · 2H2O, 0.012 g of FeSO4 · 7H2O, 0.02 g of Na2EDTA, 1 ml of trace salts solution, 1 ml of vitamin solution, and 20 mM potassium phosphate buffer (pH 7.0). The trace salts solution contained (per 100 ml) 0.21 g of MnSO4 · 4H2O, 0.28 g of H3BO3, 0.004 g of Cu(NO3)2 · 7H2O, 0.024 g of ZnSO4 · 7H2O, and 0.075 g of Na2MoO4 · 2H2O. The vitamin solution contained (per 100 ml) 0.01 g of biotin, 0.5 g of thiamine HCl, and 1.0 g of nicotinic acid. Filter-sterilized glucose was added to the medium at a final concentration of 5.4 g per liter. For 13C labeling experiments, glucose was added either entirely in the form of the 1-13C-labeled isotope isomer (99% pure; Sigma) or in the form of a mixture of 30% (wt/wt) U-13C-labeled glucose (>99%; Cambridge Isotope Laboratories, Andover, MA) and 70% (wt/wt) natural glucose. Anaerobic photoheterotrophic cultures were performed at 30°C in 125-ml glass bottles containing 110 ml of the minimal medium with illumination of 4,000 lx at the surface of bottles. The glass bottles were sealed with rubber stoppers, and 60-ml syringes were used to collect the gas samples from the headspace of cultures.
Analytical methods.
Cell growth was monitored by measuring the optical density at 660 nm. Cell dry weight (CDW) was determined from cell pellets of 100-ml culture aliquots that were centrifuged for 15 min at 4°C and 9,000 × g, washed twice with distilled water, and dried at 85°C until the weight was constant.
For analysis of extracellular metabolites, culture samples were centrifuged for 10 min at 4°C and 15,000 × g to remove the cells. Glucose and glutamate concentrations were determined with enzymatic test kits (r-Biopharm, Darmstadt, Germany). Organic acids were detected by high-pressure liquid chromatography analysis (Agilent model 1100) at a wavelength of 210 nm, using a Shodex KC-811 column (8 by 300 mm; Shodex Inc., Tokyo, Japan) and 6 mM HClO4 as the mobile phase at a flow rate of 1.0 ml min−1 at 50°C. For determination of extracellular polysaccharides, culture samples were centrifuged at 9,000 × g for 10 min, and the pellets were washed twice with 0.9% (wt/vol) NaCl. The polysaccharide concentration in the pooled supernatants was measured with the phenol-sulfuric method by using glucose for calibration. The hydrogen and carbon dioxide concentrations in the headspace of cultures were measured by GC (GC7900; Techcomp, Shanghai, China) using an instrument equipped with a thermal conductivity detector. The bicarbonate concentration was determined by adding 6 M HCl to convert bicarbonate to carbon dioxide and measuring carbon dioxide by GC. The specific uptake rate of a substrate (e.g., glucose) and specific secretion rate of a product (e.g., acetate) were determined during the exponential growth phase as the coefficient of a linear regression of the change in the substrate or product concentration versus biomass concentration, divided by the growth rate (36).
The macromolecular composition of biomass was measured using previously reported methods (4, 17). Total protein content was determined using the Lowry method, total carbohydrate content was determined by the phenol-sulfuric method, total RNA was assayed spectrophotometrically through alkali degradation and extraction by perchloric acid (2), DNA was quantified through a colorimetric procedure that involves the reaction of DNA with diphenylamine in the presence of perchloric acid, and total lipids were measured using the sulfo-phospho-vanillin method (21). Bovine serum albumin, glucose, calf thymus DNA, and thiolein were used as standards for the protein, carbohydrate, DNA, and lipid measurements, respectively. PHB was extracted by chloroform, treated with sulfuric acid, and then measured by a gas chromatograph (Agilent model 7890A) equipped with a capillary column (Alltech EC-WAX; 30 m by 0.32 mm) and a flame ionization detector (3).
Enzyme assays.
In vitro enzyme activities were determined in crude cell extracts from 10-ml culture aliquots that were centrifuged at 4°C and 9,000 × g for 10 min. The cell pellets were washed twice and resuspended in 20 mM HEPES buffer (pH 7.0) containing 100 mM NaCl and 2 mM β-mercaptoethanol. After sonication, cell debris was removed by centrifugation for 15 min at 4°C and 20,000 × g. The supernatant was used for determination of enzyme activities and protein concentration. Citrate synthase activity was assayed using a previously published colorimetric technique based on reaction of coenzyme A (CoA) with 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (30). Briefly, 1 μl of the cell extract was added to 200 μl of 50 mM Tris buffer (pH 8.0) containing 0.3 mM acetyl-CoA, 0.5 mM oxaloacetate, and 0.25 mM DTNB (Sigma). The formation of chromophoric thionitrobenzoate was monitored at 412 nm using a Beckman DU800 spectrophotometer. Malate dehydrogenase activity was determined by measuring the utilization of NADH (42). Briefly, 1 μl of the cell extract was added to 200 μl of 50 mM Tris buffer (pH 8.0) containing 0.2 mM NADH and 0.5 mM oxaloacetate. The change in NADH concentration was monitored spectrophotometrically at 340 nm. 6-Phosphofructokinase activity was measured by monitoring the decrease in NADH concentration using triose-3-P isomerase and glycerol-3-P dehydrogenase as coupling enzymes (1). 6-P-gluconate dehydrogenase activity was determined by monitoring the NADPH concentration spectrophotometrically at 340 nm (7).
Determination of intracellular NADH and NAD+ concentrations.
The intracellular NADH and NAD+ were extracted and assayed by using a fluorescent NAD/NADH detection kit (Cell Technology Inc., CA), which utilizes a nonfluorescent detection reagent that is reduced in the presence of NADH to produce its fluorescent analog. Briefly, cells were harvested at late-exponential-growth phase by centrifuging 1.5 to 2 ml of culture broth at 9,000 × g and 4°C for 10 min. Intracellular NADH and NAD+ were extracted using respective extraction buffers by following the manufacturer's instructions. NADH reacted with nonfluorescent detection reagent to form NAD+ and the fluorescent analog that was monitored at 550-nm excitation and 595-nm emission wavelengths by using a spectrofluorometer (Varioskan Flash; Thermo Scientific Co.). NAD+ is further converted to NADH via an enzyme-coupled reaction. The enzyme reaction specifically reacts with NAD+/NADH and not with NADP+/NADPH.
Sample preparation and GC-MS analysis.
Cell aliquots were harvested during late exponential growth phase (OD660 of ∼2.0) by centrifuging 3 ml of culture broth at 9,000 × g and 4°C for 10 min. The pellet was washed with 1 ml of 0.9% (wt/vol) NaCl, resuspended in 0.75 ml of 6 M HCl, and then separated into two fractions. The first fraction (0.5 ml) was hydrolyzed at 105°C for 24 h in sealed 2-ml glass vials and used for analysis of amino acids and glycerol, and the second fraction (0.25 ml) was hydrolyzed at 105°C for only 30 min and used for glucose analysis. The filtrate of hydrolysate was dried in a vacuum centrifuge at room temperature. Amino acids and glycerol were derivatized at 85°C for 1 h in 120 μl pyridine (Sigma) and 30 μl of N-methyl-N-[tert-butyldimethylsilyl] trifluoroacetamide (Sigma). Glucose was derivatized at 70°C for 3 h in 100 μl of pyridine and 50 μl of bis(trimethylsilyl)trifluoroacetamide containing 1% (vol/vol) trimethylchlorosilane (Sigma). After filtration, 3 μl of derivatized sample was injected into an Agilent 6890-5973 GC-MS system with an HP-5MS column (30 m by 0.25 mm by 0.25 μm). GC oven temperature was programmed from 60°C to180°C at 5°C per min and from 180°C to 260°C at 10°C per min. The flow rate of carrier gas (helium) was set at 1 ml min−1. The mass spectrometer was operated in the electron impact (EI) mode at 70 eV.
Metabolic flux ratio analysis.
The GC-MS data were analyzed as described previously (33). Briefly, the mass isotopomer distribution vectors (MDV) of alanine, glycine, valine, proline, serine, threonine, phenylalanine, aspartate, glutamate, histidine, tyrosine, glycerol, and glucose were determined from the respective mass spectra and were corrected for the natural abundance of all stable isotopes including 13C, 29Si, 30Si, 15N, and 18O. From the MDV of the amino acids, glycerol, and glucose, the MDV of their respective precursor intermediates, including glucose-6-P, glyceraldehyde-3-P, 3-P-glycerate, phosphoenolpyruvate (PEP), pyruvate, pentose-5-P, oxaloacetate, and α-ketoglutarate, could be easily derived. The intermediate metabolite MDV were then used to calculate the fractional contributions of different pathways to a target metabolite pool based on a set of algebraic equations. This calculation was achieved by developing a computer algorithm using MATLAB, version 6.0 (Mathworks). In order to identify the metabolic network structure of R. sphaeroides during photoheterotrophic growth on glucose, we developed equations that are required to assess the activities of the CBB cycle, EMP pathway, ED pathway, malic enzyme, and pyruvate carboxylase.
(i) PEP formed via the CBB cycle.
If the CBB cycle is active, the C-1–C-2 carbon bonds in PEP are cleaved by the CBB cycle, yielding equation 1:
| (1) |
In equation 1, fPEP←CBB, fPEP←GLC, and 1 − fPEP←CBB − fPEP←GLC are the fractions of PEP (or 3-P-glycerate) molecules derived through the CBB cycle, from glucose through the EMP or ED pathway and from oxaloacetate via the PEP carboxykinase, respectively. PEP12 and OAA12 are the MDV of fragments 1 to 2 of PEP and oxaloacetate (OAA), respectively. GLC1U and GLC2U are uniformly 13C-labeled 1- and 2-carbon glucose fragments, respectively (33). GLC1U-CO2 represents the combination of two metabolites, as described previously (33). Here, the full equilibration between 3-P-glycerate and PEP pools was assumed because the labeling patterns of fragments 1 to 2 of serine and phenylalanine are identical in both [U-13C]glucose and [1-13C]glucose experiments. Although the labeling pattern of pentose-5-P fragments 3 to 4 was unknown, analysis of the mass distribution of pentose-5-P fragments 1 to 5 showed that pentose-5-P was generated solely from fructose-6-P and glyceraldehyde-3-P. Therefore, the lower bound (lb) for PEP formed via the CBB cycle was derived under the assumption that pentose-5-P fragments 3 to 4 originated from glyceraldehyde-3-P (or 3-P-glycerate) fragments 1 to 2 through the CBB cycle (equation 2), while the upper bound (ub) was derived by assuming that the C-3–C-4 carbon bonds in pentose-5-P were not cleaved (equation 3):
| (2) |
| (3) |
| (4) |
In equations 2 and 3, the division is a left-hand matrix division. Ser12 is the MDV of fragments 1 to 2 of serine, and T represents the transposition of matrices.
(ii) 3-P-glycerate formed via the EMP pathway.
The in vivo activity of the EMP pathway was assessed from the [1-13C]glucose labeling experiment. The EMP pathway yields 50% unlabeled and 50% 3-P-glycerate (serine) that is 13C labeled at the C-3 position, whereas the ED pathway yields unlabeled 3-P-glycerate. Thus, equation 5 was obtained:
| (5) |
In equation 5, 13C is the natural abundance of 13C (i.e., 0.011), dCO2 is the labeling degree of CO2, d1 is the labeled fraction of [1-13C]glucose (d1 = 1), and p1 is the purity of [1-13C]glucose (p1 = 0.99). P5P15 and OAA24 are pentose-5-P fragments 1 to 5 and oxaloacetate fragments 2 to 4, respectively. Ser13 is the MDV of fragments 1 to 3 of serine. SFL is the summed fractional labeling of a fragment as described previously (16). The values of fPEP←CBB, fPEP←GLC, and fPEP←OAA can be calculated with equations 2 to 4. Thus, the fraction of 3-P-glycerate molecules formed via the EMP pathway can be quantified by using equation 6:
| (6) |
(iii) Pyruvate formed via the ED pathway.
If the ED pathway is active, excess intact C-1–C-2 fragments may be introduced into the pyruvate pool via the ED pathway, yielding equation 7:
| (7) |
In equation 7, fPYR←ED and fPYR←MAL are the fractions of pyruvate molecules derived through the ED pathway and from malate via the malic enzyme, respectively. MAL12 is the MDV of malate fragments 1 to 2. By assuming that malate is entirely synthesized from oxaloacetate, the lower bound for pyruvate formed via the ED pathway, and the upper bound for pyruvate from malate can be calculated (equation 8):
| (8) |
(iv) Oxaloacetate formed from pyruvate.
The derivation for the fraction of oxaloacetate formed from PEP in Escherichia coli has been described previously (33). Similarly, the fraction of oxaloacetate molecules originating from pyruvate via the pyruvate carboxylase can be calculated by using equation 9:
| (9) |
In equation 9, the division is a left-hand matrix division. OAA14, AKG25, and PYR13 are the MDV of oxaloacetate fragments 1 to 4, α-ketoglutarate fragments 2 to 5, and pyruvate fragments 1 to 3, respectively. This equation gives the least squares solutions of fOAA←PYR and dCO2 (i.e., labeling degree of CO2).
Bioreaction network and net flux analysis.
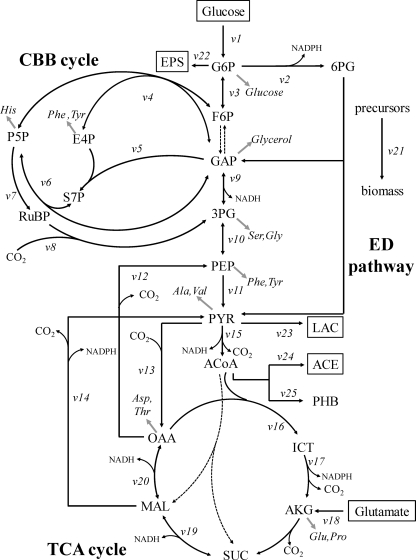
For quantification of carbon fluxes in the central metabolism, a bioreaction network was constructed based on the R. sphaeroides KD131 genome sequence (27), as shown in Fig. 1 (also see Table S1 in the supplemental material). This network included the reactions of the CBB cycle, EMP and ED pathways, and TCA cycle, as well as the reactions catalyzed by pyruvate carboxylase, PEP carboxykinase, and malic enzyme. The oxidative branch of the PP pathway was excluded due to the absence of the 6-P-gluconate dehydrogenase. The network of active pathways identified by flux ratio analysis was used for flux quantification (see Results).
Fig 1.
Bioreaction network of R. sphaeroides central carbon metabolism. Metabolites in boxes are extracellular substrates or products. Dashed arrows indicate reactions or pathways identified to be inactive in this study. Double-headed arrows indicate reactions assumed to be reversible. Gray arrows indicate precursor withdrawal for the amino acid analyzed by GC-MS. Abbreviations: G6P, glucose-6-P; 6PG, 6-P-gluconate; F6P, fructose-6-P; GAP, glyceraldehyde-3-P; Ru5P, ribulose-5-P; E4P, erythrose-4-P; S7P, sedoheptulose-7-P; RuBP, ribulose-1,5-bisphosphate; 3PG, 3-P-glycerate; PYR, pyruvate; ACoA, acetyl-CoA; ICT, isocitrate; AKG, α-ketoglutarate; SUC, succinate; MAL, malate; OAA, oxaloacetate; EPS, extracellular polysaccharide; LAC, lactate; ACE, acetate.
From the bioreaction network, a stoichiometric matrix containing 25 unknown fluxes (Fig. 1, v1 to v25) and 22 metabolite balances was constructed. Net fluxes were then calculated based on three different data sets: (i) substrate uptake and product formation rates, (ii) macromolecular biomass composition, and (iii) the calculated flux ratios. Specifically, the following flux ratios were used: the lower and upper bounds of PEP derived through the Calvin cycle, PEP originating from oxaloacetate, the lower bound of pyruvate derived through the ED pathway, the upper bound of pyruvate originating from malate, and oxaloacetate originating from pyruvate. The precursor requirements for biomass formation were derived from the biochemical information concerning biosynthetic pathways in R. sphaeroides (available at http://www.genome.jp/kegg/) and the experimentally determined macromolecular composition. The amino acid composition of a protein was assumed to be the same in R. sphaeroides mutant strains used in this study and was taken from Zhuang et al. (44). Similarly, the lipid composition was taken from Imhoff and Bias-Imhoff (20). The carbon flux distribution in the bioreaction network was determined by minimizing the sum of the weighted square residuals of the constraints from both metabolite balances and flux ratios as described previously (12). This calculation was achieved by developing a computer algorithm using MATLAB, version 6.0.
NADH balancing.
NADH formation and consumption from the metabolism were quantified using the intracellular flux values obtained from 13C labeling experiments. NADH formation was determined from the carbon fluxes through the NAD+-dependent dehydrogenases in central metabolism. The cofactor specificities of individual redox enzymes in R. sphaeroides central carbon metabolism have been reported previously (14). The glucose-6-P dehydrogenase was found to accept NAD+ as well as NADP+, approximately 16% of the total activity under quasi in vivo conditions. The glyceraldehyde-3-P dehydrogenase and malate dehydrogenase are entirely NAD+ dependent, while the isocitrate dehydrogenase is specific for NADP+. The pyruvate dehydrogenase (PDH) complex and α-ketoglutarate dehydrogenase are highly specific for NAD+ since both contain the NAD+-specific dihydrolipoamide dehydrogenase. The malic enzyme was assumed to be NADP+ dependent based on annotation and prediction from the amino acid sequence. Succinate dehydrogenase delivers electrons to ubiquinone, which could generate NADH by reverse electron flow via complex I. NADH consumption was calculated from the measured H2 evolution rate and the carbon fluxes through NADH-oxidizing reactions. The dehydrogenase in the nitrogenase complex was assumed to be specific for NADH since specificity for NADPH has not been described so far. Based on analysis of protein sequences, the 3-hydroxybutyryl-CoA dehydrogenase involved in PHB synthesis and the lactate dehydrogenase were assumed to be specific for the cofactors NADPH and NADH, respectively. NADH formation through the transhydrogenase was not considered because R. sphaeroides possesses only the membrane-bound NADPH-forming transhydrogenase (14).
RESULTS
Growth parameters.
To investigate the photoheterotrophic metabolism in H2-producing R. sphaeroides, we grew R. sphaeroides KD131 anaerobically under continuous illumination. Glucose and glutamate were supplied as the carbon and nitrogen sources, respectively, and bicarbonate was added to maintain the pH of the culture. The physiological parameters were determined for the R. sphaeroides KD131 wild type, a PHB synthase knockout mutant (ΔphbC strain), an H2-uptake hydrogenase knockout mutant (ΔhupSL strain), and a double-knockout mutant (ΔhupSL ΔphbC strain) (Table 1).
Table 1.
Growth parameters of photoheterotrophic cultures of R. sphaeroides KD131 wild-type, ΔphbC, ΔhupSL, and double mutant ΔhupSL ΔphbC strainsa
| Strain | Biomass yield (g of CDW/g of glucose) | Specific rate (per g of CDW/h) |
Carbon balance (%)c | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glucose uptake (μmol) | Glutamate uptake (μmol) | EPS formation (μmol) | Lactate secretion (μmol) | Acetate secretion (μmol) | PHB formation (μmol) | Net CO2 formation (mmol)b | Hydrogen evolution (mmol) | |||
| Wild type | 0.95 ± 0.01 | 291 ± 10 | 185 ± 20 | 29 ± 1 | 8 ± 1 | 16 ± 1 | 2 ± 1 | 0.59 ± 0.05 | 1.02 ± 0.10 | 105 |
| ΔphbC strain | 0.91 ± 0.03 | 305 ± 15 | 236 ± 17 | 62 ± 2 | 20 ± 1 | 11 ± 1 | 0 ± 0 | 0.88 ± 0.09 | 1.94 ± 0.17 | 109 |
| ΔhupSL strain | 0.76 ± 0.02 | 368 ± 20 | 273 ± 10 | 65 ± 3 | 7 ± 1 | 9 ± 1 | 18 ± 1 | 1.38 ± 0.15 | 2.47 ± 0.21 | 105 |
| ΔhupSL ΔphbC strain | 0.72 ± 0.02 | 386 ± 11 | 285 ± 13 | 59 ± 3 | 15 ± 1 | 8 ± 1 | 0 ± 0 | 1.68 ± 0.22 | 2.60 ± 0.23 | 105 |
Cells were grown anaerobically under continuous illumination on the glucose minimal medium supplemented with glutamate and bicarbonate.
The net CO2 formation rate was calculated based on the measurements of the CO2 concentration in the headspace and the bicarbonate concentration in the medium of the cultures.
The biomass elemental composition was taken from reference 4.
During photoheterotrophic growth, wild-type R. sphaeroides converted glucose and glutamate mainly to biomass and CO2 and formed a small amount of metabolic by-products including acetate, lactate, extracellular polysaccharides (EPS), and PHB (Table 1). H2 was accumulated during the mid-exponential and early stationary growth phases, and the molar yield of H2 on glucose was 4.28 mol mol−1. Compared to the wild type, the H2 evolution rate was increased 1.9-, 2.4-, and 2.6-fold for the ΔphbC, ΔhupSL, and double mutant ΔhupSL ΔphbC strains, respectively (Table 1). The three mutant strains exhibited higher uptake rates of glucose and glutamate and lower biomass yields than the wild type (Fig. 2). The CO2 evolution rate was also increased for the ΔphbC and ΔhupSL mutants, and the highest rate was observed for the double mutant. That is, a higher fraction of the substrate (glucose and glutamate) carbon was released as CO2, and less carbon was incorporated into cell material in the mutants than in the wild type. The PHB contents were similar in the wild-type and ΔhupSL mutant strains, while PHB was not detected in the ΔphbC strain and the double mutants. In addition, to obtain accurate information on the specific precursor requirements for subsequent flux analysis, we determined the relative fractions of the major biomass components of R. sphaeroides: protein, lipids, RNA, and DNA (see Table S2 in the supplemental material).
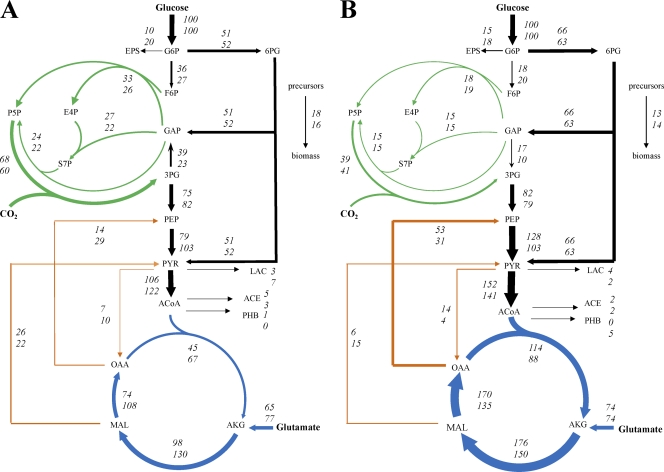
Fig 2.
In vivo carbon flux distribution in H2-producing R. sphaeroides KD131 wild-type (top numbers) and ΔphbC mutant (bottom numbers) strains (A) and in the ΔhupSL ΔphbC double mutant (top numbers) and ΔhupSL mutant (bottom numbers) (B). All strains were grown photoheterotrophically on glucose and glutamate. The flux values are expressed relative to the specific glucose uptake rate of the respective strain. Arrows indicate the direction of the net fluxes determined, and their widths are scaled to the fluxes in the wild-type strain in panel A and the double mutant in panel B. The 95% confidence intervals were less than 10% for all the fluxes.
Identification of network structure.
The network structure of glucose metabolism in photoheterotrophic R. sphaeroides has remained unclear although the ED pathway was reported to be present based on 14C-radiolabeled experiments (5). To identify the network of active pathways, we used 13C-based metabolic flux ratio analysis that relies on the [U-13C]glucose and [1-13C]glucose tracer experiments and GC-MS analysis of mass isotopomer patterns in cellular amino acids, glycerol, and glucose (see Table S3 in the supplemental material). To assess if the isotopic steady state was achieved, cell aliquots were harvested from 13C-labeled experiments at different time points during the late exponential growth phase. The determined mass isotopomer distributions of key amino acids were almost unchanged with the time of harvest, which is consistent with previous reports that have showed that a (quasi-)steady state can be reached during the exponential growth phase in batch culture (10, 36). From the GC-MS data, the labeling patterns of the precursor metabolites were derived based on biochemical information concerning biosynthetic pathways in R. sphaeroides (see Table S4 in the supplemental material). The origins of key metabolites in the central metabolism were then quantitatively determined (Table 2), which allowed us to identify the network of active pathways.
Table 2.
Origins of metabolic intermediates determined by flux ratio analysis of experiments with 100% [1-13C]glucose, 30% [U-13C]glucose, and 70% naturally labeled glucose
| Metabolitea | % of total pool in R. sphaeroides strain |
|||
|---|---|---|---|---|
| Wild type | ΔphbC strain | ΔhupSL strain | ΔhupSL ΔphbC strain | |
| PEP through CBB cycle (lb) | 64 ± 3 | 53 ± 3 | 38 ± 4 | 28 ± 4 |
| PEP through CBB cycle (ub) | 97 ± 3 | 75 ± 4 | 50 ± 6 | 35 ± 5 |
| PEP from glucose (ub) | 26 ± 2 | 31 ± 2 | 35 ± 2 | 37 ± 2 |
| 3PG through EMP pathwayb | 0 ± 1 | 0 ± 1 | 0 ± 1 | 0 ± 1 |
| PYR through ED pathway (lb) | 21 ± 4 | 23 ± 1 | 26 ± 1 | 30 ± 1 |
| OAA from PYR | 5 ± 1 | 7 ± 1 | 2 ± 1 | 7 ± 1 |
| PEP from OAA | 10 ± 1 | 15 ± 1 | 18 ± 1 | 27 ± 1 |
| PYR from MAL (ub) | 32 ± 4 | 16 ± 2 | 13 ± 2 | 5 ± 1 |
For metabolite abbreviations, see the legend to Fig. 1. lb, lower bound; ub, upper bound.
Split ratios obtained from experiments with 100% [1-13C]glucose.
The flux ratio analysis of R. sphaeroides showed that the ED pathway was active during anaerobic photoheterotrophic growth on glucose. The summed fractional labeling of pyruvate fragments 1 to 2 (24%; detected in valine) was much higher than that of PEP fragments 1 to 2 (3%; detected in phenylalanine and tyrosine) in the [1-13C]glucose labeling experiment, which directly demonstrated the use of the ED pathway for glucose catabolism. The activity of the EMP pathway could not be assessed simply by detecting the 13C label at the C-3 position of pyruvate in the [1-13C]glucose labeling experiment because the CBB cycle also yields pyruvate that is 13C labeled at the C-3 position. Based on the 13C labeling patterns of serine and phenylalanine from both [U-13C]glucose and [1-13C]glucose labeling experiments, the fractional contributions of alternative pathways to 3-P-glycerate and PEP pools were quantitatively analyzed, and the EMP pathway flux was found to be absent (Table 2). Consistently, in vitro enzyme activity analysis also confirmed that the R. sphaeroides KD131 strain lacks the 6-phosphofructokinase activity under anaerobic photoheterotrophic conditions. In addition, because the labeled fraction of glucose-6-P (i.e., 30%) was almost identical to that of the input glucose and was not diluted by other intermediate metabolites such as glyceraldehyde-3-P, no activity of fructose-1,6-bisphosphatase was detected.
The activity of the CBB cycle was revealed by the 13C labeling pattern of phenylalanine fragments 1 to 2 from the [U-13C]glucose experiment (see Table S3 in the supplemental material). A significant fraction of C-1–C-2 carbon bonds in PEP were cleaved due to the action of the CBB cycle. The flux ratio analysis showed that more than 64% of PEP molecules originated through the CBB cycle in the wild-type R. sphaeroides (Table 2). The absence of the oxidative branch of the PP pathway was verified from the labeling data of pentose-5-P (detected in histidine) found in the [U-13C]glucose experiment, which were consistent with the data expected when pentose-5-P was generated entirely from fructose-6-P and glyceraldehyde-3-P through the CBB cycle. In agreement with this result, no activity of 6-P-gluconate dehydrogenase was detected from in vitro enzyme activity analysis.
The flux ratio analysis of R. sphaeroides showed that a small fraction of oxaloacetate molecules were derived from pyruvate via the anaplerotic reaction catalyzed by pyruvate carboxylase (Table 2). The 13C label of TCA cycle intermediates such as α-ketoglutarate (detected in glutamate and proline) and oxaloacetate (detected in aspartate and threonine) was significantly diluted (see Table S4 in the supplemental material) since glutamate was utilized as the nitrogen source for nitrogenase-mediated hydrogen formation. Based on the analysis of labeling patterns of α-ketoglutarate and oxaloacetate, no activity was found for the ethylmalonyl-CoA pathway (9) that would result in a marked increase in the 13C label of oxaloacetate fragments 1 to 2. Moreover, the flux ratio analysis of R. sphaeroides showed the activities of malic enzyme and PEP carboxykinase that catalyze the gluconeogenic conversion of malate to pyruvate and the conversion of oxaloacetate to PEP, respectively (Table 2).
Comparing the results of flux ratio analysis of the R. sphaeroides wild-type, ΔphbC, ΔhupSL, and double mutant ΔhupSL ΔphbC strains, different flux ratio patterns were found (Table 2). (i) The fraction of PEP molecules derived through the CBB cycle was largest in the wild-type strain and smallest in the double mutant, whereas the contribution of the ED pathway to pyruvate synthesis was increased in the mutants. (ii) The fraction of pyruvate molecules originating from malate via malic enzyme was decreased in ΔphbC and ΔhupSL mutants and smallest in the double mutant. (iii) A gradual increase in the amount of PEP molecules arising from oxaloacetate via PEP carboxykinase was observed in the following order: ΔphbC strain, ΔhupSL strain, and the ΔhupSL ΔphbC double mutant.
Metabolic net fluxes.
To quantitatively compare in vivo enzyme or pathway activities in R. sphaeroides wild-type and mutants, the absolute net fluxes were quantified by combining the physiological data (Table 1), the biomass composition data (see Table S2 in the supplemental material), and the flux ratios (Table 2). Based on the results of flux ratio analysis, the reactions catalyzed by 6-phosphofructokinase and 6-P-gluconate dehydrogenase, fructose-1,6-bisphosphatase, and the ethylmalonyl-CoA pathway were omitted from the bioreaction network for net flux analysis (Fig. 1). The intracellular net fluxes were then determined as the best fit to the available data sets using a parameter-fitting approach.
The flux distribution in wild-type R. sphaeroides grown under anaerobic photoheterotrophic conditions was very different from that reported previously for the aerobic chemoheterotrophic culture (13). During anaerobic growth on glucose with light, 51% of the glucose molecules were routed through the ED pathway, 36% converted to fructose-6-P that entered the CBB cycle, and the rest was used for synthesis of biomass and extracellular polysaccharides (Fig. 2). The ED pathway also supplied another key intermediate, glyceraldehyde-3-P, for the CBB cycle. The CBB cycle activity was very high; the flux of CO2 fixation catalyzed by ribulose-1,5-P2 carboxylase was 66% relative to the glucose uptake rate and fixed 24% of the CO2 that was liberated through the TCA cycle and pyruvate dehydrogenase (PDH). The flux between 3-P-glycerate and glyceraldehyde-3-P carried by glyceraldehyde-3-P dehydrogenase (GAPDH) was operated in the CBB cycle (gluconeogenic) direction. More than half of the 3-P-glycerate molecules synthesized through the CBB cycle were fed into the glycolytic pathway and the TCA cycle for complete oxidation, with only slight by-product formation. The backward fluxes from the TCA cycle to glycolysis carried by malic enzyme and PEP carboxykinase were 26% and 14%, respectively, relative to the glucose uptake rate.
The PHB synthase knockout mutant, the ΔphbC strain, exhibited flux distributions similar to those of the wild type, except the increased flux through the TCA cycle (Fig. 2A). Interestingly, the inactivation of the H2-uptake hydrogenase extensively altered the flux distribution (Fig. 2B). The flux through the ED pathway was increased, and the GAPDH flux favored the glycolytic direction, while a lower fraction of glucose molecules was channeled into the CBB cycle in the ΔhupSL strain and ΔhupSL ΔphbC double mutant. The TCA cycle flux, particularly the flux through the malate dehydrogenase, as well as the PDH flux was increased remarkably in both mutants. An increase in the flux converting PEP to pyruvate in both mutants could be contributed by higher activity of pyruvate kinase or the use of a phosphotransferase system for glucose uptake (37). Moreover, these two mutants, especially the ΔhupSL ΔphbC double mutant, exhibited a significantly higher flux through the PEP carboxykinase and a lower flux through the malic enzyme than the wild type.
NADH balancing.
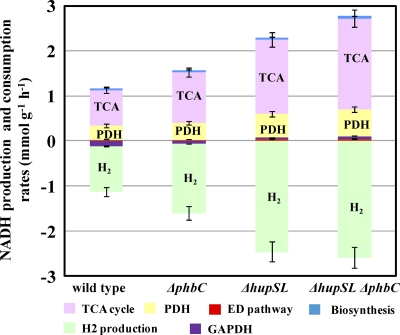
From the intracellular carbon fluxes determined from 13C labeling experiments, the rates of NADH formation and consumption from carbon metabolism were quantified (Fig. 3). The NADH consumption for H2 production via the nitrogenase complex was calculated using the measured H2 evolution rate. The NADH balance was achieved within 6% for all four stains. In wild-type R. sphaeroides, about 90% of the NADH generated during glucose oxidation was used for H2 production, and the rest (10%) was oxidized by GAPDH in the CBB cycle (Fig. 3). In hydrogenase-deficient mutants (ΔhupSL and ΔhupSL ΔphbC strains), NADH oxidation was accomplished solely by H2 production. Therefore, H2 production played a critical role in maintaining redox balance in photoheterotrophically grown R. sphaeroides. In all four strains studied, the TCA cycle and PDH and were the major producers of NADH. The increase in the TCA cycle and PDH fluxes in the three mutants led to the formation of a larger amount of NADH than in the wild type. Compared to that in the wild-type strain, the NADH production rate was increased 1.4-, 1.7-, and 2.0-fold in the ΔphbC, ΔhupSL, and double mutant ΔhupSL ΔphbC strains, respectively. This increase was contributed mostly by the higher fluxes through the TCA cycle and PDH in the mutants, which accounted for 82 to 88% of the increase in NADH formation. The change in GAPDH flux also made a minor contribution (12 to 18%) to the increased NADH production. Thus, the increase in the TCA cycle and PDH fluxes accounted for most of the electrons needed for the increased H2 production in the three mutants.
Fig 3.
NADH balancing based on metabolic flux distributions. NADH formation was contributed by PDH, the TCA cycle, the ED pathway, and biomass synthesis. NADH was consumed via H2 production. The GAPDH reaction in the CBB cycle could contribute to NADH formation or consumption.
Enzyme activities and intracellular NADH and NAD+ concentrations.
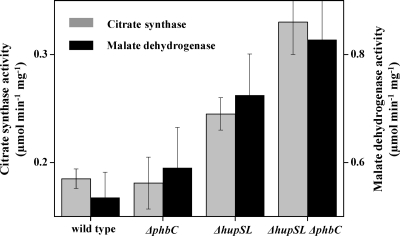
Our flux data showed that compared to the wild-type, the TCA cycle flux was increased remarkably in the three mutants, particularly in hydrogenase-deficient mutants (ΔhupSL and ΔhupSL ΔphbC strains). To investigate the regulatory mechanisms involved in this flux change, in vitro activities of malate dehydrogenase and citrate synthase in the TCA cycle were determined for all four strains. As shown in Fig. 4, the specific activities of both enzymes were increased in the three mutants, and the ΔhupSL ΔphbC double mutant exhibited about 1.7-fold higher activities of malate dehydrogenase and citrate synthase than the wild type. To investigate the correlations of in vivo TCA cycle enzyme activities with the pool sizes of intracellular redox equivalents, we determined the intracellular concentrations of NADH and NAD+ (Table 3). In accordance with previous findings (22), the NAD(H) pool was largely in the reduced form, NADH, under anaerobic photoheterotrophic conditions. An increase in the sizes of both NADH and NAD+ pools in the three mutants was observed. From these data, the NADH/NAD+ concentration ratio was calculated, which was decreased significantly in the three mutants and lowest in the ΔhupSL ΔphbC double mutant (Table 3). These results suggested that the was more oxidized in the mutants, which may be related to the increase in the TCA cycle flux.
Fig 4.
In vitro activities of citrate synthase and malate dehydrogenase in crude cell extracts. Activities with standard deviations are from at least triplicate experiments.
Table 3.
Intracellular NADH and NAD+ concentrationsa
| Strain | NADH (μmol/g of CDW) | NAD+ (μmol/g of CDW) | NADH/NAD+ ratio |
|---|---|---|---|
| Wild type | 2.33 ± 0.28 | 0.31 ± 0.03 | 7.52 ± 0.90 |
| ΔphbC strain | 2.53 ± 0.23 | 0.67 ± 0.09 | 3.78 ± 0.51 |
| ΔhupSL strain | 3.81 ± 0.32 | 2.22 ± 0.19 | 1.72 ± 0.15 |
| ΔhupSL ΔphbC strain | 3.82 ± 0.44 | 2.44 ± 0.26 | 1.57 ± 0.18 |
Concentrations with standard deviations are from at least triplicate experiments.
DISCUSSION
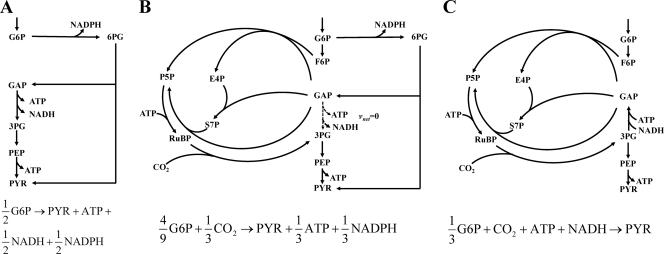
The primary objective of this study was to quantitatively elucidate how photoheterotrophic R. sphaeroides manipulates the metabolic network and redistributes intracellular fluxes for increased H2 production. To do this, we identified the network of glucose metabolism and quantified the carbon fluxes in photoheterotrophic R. sphaeroides by using 13C-constrained metabolic flux analysis. The flux results showed that the ED pathway was the major glucose catabolic pathway, while the complete EMP and PP pathways were absent. The in vivo activity of the CBB cycle was very high, which diverts fructose-6-P and glyceraldehyde-3-P to synthesize ribulose-1,5-diphosphate (ribulose-1,5-P2), then fix CO2, and finally generate 3-P-glycerate. In combination with the glycolytic reactions converting 3-P-glycerate to pyruvate, the extensive activity of the CBB cycle constitutes a catabolic route of glucose. As shown in Fig. 5, we analyzed the stoichiometries of three catabolic routes: the ED pathway only, the combination of the ED pathway and CBB cycle with the net flux through GAPDH of zero, and the CBB cycle only. The use of the CO2-fixing CBB cycle for glucose catabolism leads to increased yields of pyruvate molecules from glucose. Although the CBB cycle dissipates ATP, it does not seem to be of major consequence since the cyclic photophosphorylation generates a huge amount of ATP (>10 mmol g−1 h−1), based on our calculation using the reported model of the electron transport chain of purple nonsulfur bacteria (24, 40). Another big difference between the three catabolic routes is the formation of the reducing equivalents NADH and NADPH, partly because of the different directions of the GAPDH reaction (Fig. 5). The ED pathway generates one NADH and one NADPH molecule per molecule of glucose catabolized. The combined use of the ED pathway and CBB cycle significantly reduces the formation of the reducing equivalents, especially NADH. If the CBB cycle is the sole catabolic pathway, one NADH molecule is consumed per molecule of pyruvate synthesized. Our flux results showed that the contribution of the ED pathway and CBB cycle to glucose catabolism varied significantly and that the flux through GAPDH was operated in opposite directions between the wild-type and hydrogenase-deficient R. sphaeroides strains. This flux change contributed about 17% to the increased NADH production in ΔhupSL and double mutant ΔhupSL ΔphbC strains.
Fig 5.
Stoichiometries of three alternative routes for glucose catabolism. The ED pathway only (A), the combination of ED pathway and CBB cycle with the net flux from glyceraldehyde-3-P to 3-P-glycerate of zero (B), and the CBB cycle only (C) are shown.
The major contributor to the increased NADH production in the mutants was the higher flux through PDH and the TCA cycle. We attempted to analyze the regulatory mechanisms involved in this flux change by measuring the in vitro activities of TCA cycle enzymes and intracellular concentrations of NADH and NAD+. A significant decrease in the NADH/NAD+ concentration ratio was observed for the three mutants, especially the ΔhupSL ΔphbC double mutant. It has been reported that the activity of citrate synthase from Rhodobacter capsulatus was increased under “low reducing power” conditions (i.e., low NADH/NAD+ ratio) (8). Therefore, our results suggested that the increase in the TCA cycle flux in the mutants could result from activity-level regulation via the changes in the NADH/NAD+ ratio. These redox cofactors could also regulate the in vivo activities of CBB cycle enzymes such as ribulose-5-P kinase (34).
Significant changes in the fluxes converting C4 metabolites of the TCA cycle to C3 intermediates of glycolysis were found in response to knockout of PHB synthase or hydrogenase. In the three mutants, especially the ΔhupSL ΔphbC double mutant, the flux through the malic enzyme catalyzing the conversion of malate to pyruvate was reduced significantly, whereas the fluxes from malate to oxaloacetate to PEP to pyruvate, which are catalyzed by malate dehydrogenase, PEP carboxykinase, and pyruvate kinase, are increased remarkably. The malate dehydrogenase was reported to be entirely NAD+ dependent (14), and the malic enzyme was suggested to be NADP+ dependent based on annotation and prediction from the protein sequence. Thus, this flux change may result in increased formation of NADH rather than NADPH although the underlying regulatory mechanisms remain unclear. Considering that NADPH is mainly used for biomass formation, the biosynthetic requirements of NADPH were reduced for the mutant strains due to the lower biomass yields. Therefore, by changing the route of malate-to-pyruvate conversion, more NADH but less NADPH could be produced in the mutant strains than in the wild type.
In summary, this study provides quantitative insights into how the intracellular fluxes in photoheterotrophic cells are redistributed in response to knockout mutations to generate more electrons for increased H2 production. By using a similar method, a recent study has analyzed the factors affecting H2 yield in Rhodopseudomonas palustris during photoheterotrophic growth on ammonia and various carbon sources (31). Both studies provide quantitative knowledge of how purple nonsulfur bacteria flexibly select and operate various metabolic pathways in response to genetic or environmental perturbations. This information will be useful for metabolic engineering of this group of bacteria to improve photobiological production of H2.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Basic Research Program of China (973: 2012CB721101), the National Natural Science Foundation of China (30970035 and 31121001), and the Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-G-5). Y.T. and C.Y. were supported by SA_SIBS Scholarships.
Footnotes
Published ahead of print 4 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Babul J. 1978. Phosphofructokinases from Escherichia coli. Purification and characterization of the nonallosteric isozyme. J. Biol. Chem. 253: 4350–4355 [PubMed] [Google Scholar]
- 2. Benthin S, Nielsen J, Villadsen J. 1991. A simple and reliable method for the determination of cellular RNA content. Biotechnol. Technol. 5: 39–42 [Google Scholar]
- 3. Braunegg G, Sonnleitner B, Lafferty RM. 1978. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Appl. Microbiol. Biotechnol. 6: 29–37 [Google Scholar]
- 4. Carlozzi P, Sacchi A. 2001. Biomass production and studies on Rhodopseudomonas palustris grown in an outdoor, temperature controlled, underwater tubular photobioreactor. J. Biotechnol. 88: 239–249 [DOI] [PubMed] [Google Scholar]
- 5. Conrad R, Schlegel HG. 1977. Influence of aerobic and phototrophic growth conditions on the distribution of glucose and fructose carbon into the Entner-Doudoroff and Embden-Meyerhof pathways in Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 101: 277–290 [Google Scholar]
- 6. Dauner M, et al. 2002. Intracellular carbon fluxes in riboflavin-producing Bacillus subtilis during growth on two-carbon substrate mixtures. Appl. Environ. Microbiol. 68: 1760–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Silva AO, Fraenkel DG. 1979. The 6-phosphogluconate dehydrogenase reaction in Escherichia coli. J. Biol. Chem. 254: 10237–10242 [PubMed] [Google Scholar]
- 8. Eidels L, Preiss J. 1970. Citrate synthase. A regulatory enzyme from Rhodopseudomonas capsulata. J. Biol. Chem. 245: 2937–2945 [PubMed] [Google Scholar]
- 9. Erb TJ, et al. 2007. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc. Natl. Acad. Sci. U. S. A. 104: 10631–10636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fischer E, Sauer U. 2003. Metabolic flux profiling of Escherichia coli mutants in central carbon metabolism using GC-MS. Eur. J. Biochem. 270: 880–891 [DOI] [PubMed] [Google Scholar]
- 11. Fischer E, Sauer U. 2003. A novel metabolic cycle catalyzes glucose oxidation and anaplerosis in hungry Escherichia coli. J. Biol. Chem. 278: 46446–46451 [DOI] [PubMed] [Google Scholar]
- 12. Fischer E, Zamboni N, Sauer U. 2004. High-throughput metabolic flux analysis based on gas chromatography-mass spectrometry derived 13C constraints. Anal. Biochem. 325: 308–316 [DOI] [PubMed] [Google Scholar]
- 13. Fuhrer T, Fischer E, Sauer U. 2005. Experimental identification and quantification of glucose metabolism in seven bacterial species. J. Bacteriol. 187: 1581–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuhrer T, Sauer U. 2009. Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J. Bacteriol. 191: 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Golomysova A, Gomelsky M, Ivanov PS. 2010. Flux balance analysis of photoheterotrophic growth of purple nonsulfur bacteria relevant to biohydrogen production. Int. J. Hydrogen Energy 35: 12751–12760 [Google Scholar]
- 16. Gombert AK, Santos MM, Christensen B, Nielsen J. 2001. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183: 1441–1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Herbert D. 1971. Chemical analysis of microbial cells. Academic press, London, United Kingdom [Google Scholar]
- 18. Hillmer P, Gest H. 1977. H2 metabolism in the photosynthetic bacterium Rhodopseudomonas capsulata: H2 production by growing cultures. J. Bacteriol. 129: 724–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hua Q, Yang C, Baba T, Mori H, Shimizu K. 2003. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J. Bacteriol. 185: 7053–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Imhoff J, Bias-lmhoff U. 1995. Lipids, quinones and fatty acids of anoxygenic phototrophic bacteria. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 21. Izard J, Limberger RJ. 2003. Rapid screening method for quantitation of bacterial cell lipids from whole cells. J. Microbiol. Methods 55: 411–418 [DOI] [PubMed] [Google Scholar]
- 22. Jackson JB, Crofts AR. 1968. Energy-linked reduction of nicotinamide adenine dinucleotides in cells of Rhodospirillum rubrum. Biochem. Biophys. Res. Commun. 32: 908–915 [DOI] [PubMed] [Google Scholar]
- 23. Kim MS, Baek JS, Lee JK. 2006. Comparison of H2 accumulation by Rhodobacter sphaeroides KD131 and its uptake hydrogenase and PHB synthase deficient mutant. Inter. J. Hydrogen Energy 31: 121–127 [Google Scholar]
- 24. Klamt S, Grammel H, Straube R, Ghosh R, Gilles ED. 2008. Modeling the electron transport chain of purple non-sulfur bacteria. Mol. Syst. Biol. 4: 156–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee IH, Park JY, Kho DH, Kim MS, Lee JK. 2002. Reductive effect of H2 uptake and poly-β-hydroxybutyrate formation on nitrogenase-mediated H2 accumulation of Rhodobacter sphaeroides according to light intensity. Appl. Microbiol. Biotechnol. 60: 147–153 [DOI] [PubMed] [Google Scholar]
- 26. Li X, Liu T, Wu Y, Zhao G, Zhou Z. 2010. Derepressive effect of NH4+ on hydrogen production by deleting the glnA1 gene in Rhodobacter sphaeroides. Biotechnol. Bioeng. 106: 564–572 [DOI] [PubMed] [Google Scholar]
- 27. Lim SK, et al. 2009. Complete genome sequence of Rhodobacter sphaeroides KD131. J. Bacteriol. 191: 1118–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mackenzie C, et al. 2001. The home stretch, a first analysis of the nearly completed genome of Rhodobacter sphaeroides 2.4.1. Photosynth. Res. 70: 19–41 [DOI] [PubMed] [Google Scholar]
- 29. Mackenzie C, et al. 2007. Postgenomic adventures with Rhodobacter sphaeroides. Annu. Rev. Microbiol. 61: 283–307 [DOI] [PubMed] [Google Scholar]
- 30. Marco-Urrea E, et al. 2011. Identification and characterization of a re-citrate synthase in Dehalococcoides strain CBDB1. J. Bacteriol. 193: 5171–5178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McKinlay JB, Harwood CS. 2011. Calvin cycle flux, pathway constraints, and substrate oxidation state together determine the H2 biofuel yield in photoheterotrophic bacteria. mBio 2 (2): e00323–10 doi:10.1128/mBio.00323-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McKinlay JB, Harwood CS. 2010. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc. Natl. Acad. Sci. U. S. A. 107: 11669–11675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nanchen A, Fuhrer T, Sauer U. 2007. Determination of metabolic flux ratios from 13C-experiments and gas chromatography-mass spectrometry data: protocol and principles. Methods Mol. Biol. 358: 177–197 [DOI] [PubMed] [Google Scholar]
- 34. Rindt KP, Ohmann E. 1969. NADH and AMP as allosteric effectors of ribulose-5-phosphate kinase in Rhodopseudomonas spheroides. Biochem. Biophys. Res. Commun. 36: 357–364 [DOI] [PubMed] [Google Scholar]
- 35. Sauer U. 2006. Metabolic networks in motion: 13C-based flux analysis. Mol. Syst. Biol. 2: 62–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sauer U, et al. 1999. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181: 6679–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Snyder MA, Kaczorowski GJ, Barnes JEM, Walsh C. 1976. Inactivation of the phosphoenolpyruvate-dependent phosphotransferase system in various species of bacteria by vinylglycolic acid. J. Bacteriol. 127: 671–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tao YZ, et al. 2008. Characteristics of a new photosynthetic bacterial strain for hydrogen production and its application in wastewater treatment. Inter. J. Hydrogen Energy 33: 963–973 [Google Scholar]
- 39. Wall JD, Weaver PF, Gest H. 1975. Genetic transfer of nitrogenase-hydrogenase activity in Rhodopseudomonas capsulata. Nature 258: 630–631 [DOI] [PubMed] [Google Scholar]
- 40. Warthmann R, Pfennig N, Cypionka H. 1993. The quantum requirement for H2 production by anoxygenic phototrophic bacteria. Appl. Microbiol. Biotechnol. 39: 358–362 [Google Scholar]
- 41. Wiechert W. 2001. 13C metabolic flux analysis. Metab. Eng. 3: 195–206 [DOI] [PubMed] [Google Scholar]
- 42. Wright SK, Viola RE. 2001. Alteration of the specificity of malate dehydrogenase by chemical modulation of an active site arginine. J. Biol. Chem. 276: 31151–31155 [DOI] [PubMed] [Google Scholar]
- 43. Zamboni N, Fendt SM, Ruhl M, Sauer U. 2009. 13C-based metabolic flux analysis. Nat. Protoc. 4: 878–892 [DOI] [PubMed] [Google Scholar]
- 44. Zhuang CD, Wang SB, Dan H, LL, Jia YD. 2007. Analysis of nutrient composition in different photosynthetic bacteria strains. China Feed 2007 (12): 33–35 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.