Abstract
It is well known that ppGpp and DksA interact with bacterial RNA polymerase (RNAP) to alter promoter activity. This study suggests that GreA plays a major role and GreB plays a minor role in the ppGpp-DksA regulatory network. We present evidence that DksA and GreA/GreB are redundant and/or share similar functions: (i) on minimal medium GreA overproduction suppresses the growth defects of a dksA mutant; (ii) GreA and DksA overexpression partially suppresses the auxotrophy of a ppGpp-deficient strain; (iii) microarrays show that many genes are regulated similarly by GreA and DksA. We also find instances where GreA and DksA seem to act in opposition: (i) complete suppression of auxotrophy occurs by overexpression of GreA or DksA only in the absence of the other protein; (ii) PgadA and PgadE promoter fusions, along with many other genes, are dramatically affected in vivo by GreA overproduction only when DksA is absent; (iii) GreA and DksA show opposite regulation of a subset of genes. Mutations in key acidic residues of GreA and DksA suggest that properties seen here probably are not explained by known biochemical activities of these proteins. Our results indicate that the general pattern of gene expression and, in turn, the ability of Escherichia coli to grow under a defined condition are the result of a complex interplay between GreA, GreB, and DksA that also involves mutual control of their gene expression, competition for RNA polymerase binding, and similar or opposite action on RNA polymerase activity.
INTRODUCTION
GreA was first discovered by its ability to suppress a temperature-sensitive RNA polymerase (RNAP) mutant rpoB3595 [rpoB(Ts8)] when present in multicopy (45). At that time it was unknown whether this factor interacted directly with RNAP although it was likely since this particular mutant compromised phage lambda transcription antitermination (44). The name GreA was derived from its apparent ability to regulate growth (growth regulator). Currently, a large and still growing structural family of secondary channel proteins (GreA, GreB, DksA, RfaH, TraR, Gfh1, and eukaryotic TFIIS) are thought to interact with this RNAP channel for entry of substrates into the catalytic site and for extrusion of backtracked nascent RNA (6, 7, 13, 21, 31, 47). The functions of these proteins are different despite similar structures. For example, GreA and GreB participate in rescuing backtracked, arrested, or paused RNAP during elongation, but the lengths of their RNA cleavage products are different (19, 20, 22, 28). GreA was also found to have a role in transcriptional initiation (35). Unlike Gre factors, DksA is a pleiotropic regulator that functions mainly at the level of transcriptional initiation, and many of its regulatory effects require a nucleotide regulator, ppGpp. The ppGpp requirement is generally necessary for both negative and positive regulation exerted by DksA (29, 30, 32). However, ppGpp and DksA were shown to act antagonistically in several instances and independently in others (1, 23, 24).
It was shown in a previous study that the Escherichia coli rRNA promoter, rrnB P1, is stimulated by increased levels of GreA, barely stimulated by GreB, and inhibited by elevated DksA (35). Inhibition of this promoter by DksA was expected because of known synergy between DksA and ppGpp for negative regulation of ribosomal promoter initiation (29), but antagonistic regulatory effects of GreA and DksA were unexpected. Manipulation of DksA and/or ppGpp levels is now appreciated to give all possible classes of regulatory effects at the phenotypic level: parallel activation or inhibition, opposite regulation (here termed antagonistic), or independent (regulation by one but not the other). More recently, comparisons of DksA and ppGpp regulatory effects have been extended to the level of transcription profiling (1). In addition, antagonistic ppGpp-DksA regulatory effects on two genes, fliC and fimB, have also implicated roles for GreA (1, 2).
The starting point of this work was to ask whether the GreA, GreB, and DksA proteins, similarly induced, also display antagonistic regulatory patterns for a large class of promoters positively regulated by ppGpp. The existence of this class of promoters was initially inferred by acquisition of multiple amino acid requirements that accompany a complete deficiency of ppGpp (ppGpp0) due to deletions of relA and spoT (53). Additional evidence came from studies of the ppGpp dependence of individual amino acid biosynthetic promoters in vivo and in vitro (5, 9, 12). The ppGpp0 phenotype is easily scored by the failure of these strains to form colonies on minimal medium (M9 glucose) plates that lack amino acids. Inactivation of dksA in an otherwise wild-type (wt) ppGpp+ host has a potentially similar phenotype for this class of promoters because the specific amino acid requirements are a limited subset of those of ppGpp0 strains (8, 34, 39).
The inability of ppGpp0 strains to grow on M9 minimal medium has been used to isolate spontaneous mutants that suppress the auxotrophic phenotype, the so-called stringent or M+ mutants (27, 54). So far, these suppressors have been mapped exclusively within genes coding for the RNA polymerase β, β′, and σ subunits. The mutants are found in many structural regions, and their mechanisms of action are unclear. There is a variety of evidence that ppGpp interacts directly with RNA polymerase, but there is no agreement as to the binding site (3, 32, 52). It is nevertheless thought that these mutations induce conformational changes in RNA polymerase that mimic the presence of ppGpp. For example, the M+ mutants downregulate ribosomal promoters (9), activate a σ54-dependent promoter (48), and upregulate several promoters for amino acid synthesis (5).
We focus here on genetic and physiological approaches to assess covariation of amino acid auxotrophy displayed by ppGpp0 and dksA mutants and the ability of secondary channel proteins GreA, GreB, and DksA to suppress these regulatory defects. Since each of the factors varied in these experiments has pleiotropic regulatory effects, the interpretations of the results are highly qualified and probably mechanistically complex. We have therefore included limited studies of a mutant with two conserved acidic residue substitutions in the coiled-coil region of GreA, here termed GreA*, in order to help decide whether a particular regulatory activity is due to the ability of this factor to relieve arrested elongation complexes. An equivalent DksA* mutant was also used since the same acidic residues have been implicated in its synergistic effects on negative regulation by ppGpp (31).
We find various degrees of antagonism between the three factors in respect to ppGpp positively regulated processes, as reflected by altered growth and amino acid requirements. One explanation among many is the possibility of direct competition between the factors at the level of RNAP. Still, we also found instances of parallel regulation. This evidence comes from systematic genetic approaches in vivo and transcriptional microarrays. We find that when ppGpp is absent, regulation exerted by GreA, GreB, and DksA is more evident. These effects are even more dramatic when DksA and ppGpp are both removed. Our results indicate that growth of E. coli is the result of a complex interplay between these factors.
MATERIALS AND METHODS
Growth conditions.
All strains were grown in LB broth or synthetic M9 medium at 37°C with appropriate antibiotic concentrations, as follows: chloramphenicol, 20 μg/ml; kanamycin, 40 μg/ml; tetracycline, 20 μg/ml; ampicillin, 100 μg/ml; spectinomycin, 50 μg/ml. M9 medium was supplemented with thiamine (10 μg/ml), FeSO4 (0.5 μM), 0.2% glucose, and 0.2% Casamino Acids ([CAA] vitamin free) or single amino acids (100 μg/ml), when required.
We are aware that whenever cell growth is limited, selection for suppression mutations is evident. Therefore, we have taken precautions to minimize these complications. Frozen transductant stocks were prepared immediately after isolation; only fresh transformants were used for each experiment; overnight liquid cultures were avoided, and liquid growth was initiated only from fresh plates, as described earlier for ppGpp0 strains (34). In addition, several independent clones were systematically tested in most experiments.
Strains and plasmids.
The backgrounds, genotypes, and sources of the strains of E. coli used in this study are listed in Table S1 in the supplemental material. All strains used are derivatives of MG1655. Mutant alleles were introduced into this background via standard P1 transduction (26), selecting transductants with the appropriate antibiotic resistance. The vector control that we used was pHM1883 (35). Construction of the DksA overproducer pHM1506 was described previously (35). To construct plasmids pHM1873 (GreA overproducer, Spcr) and pHM1574 (GreB overproducer, Spcr), 2.2-kb AccI/HindIII fragments from pGF296 and pDNL278, respectively, were subcloned into pHM1883 cut with the same restriction enzymes. Plasmids pGreA* (pHM1854) and pDksA* (pHM1790) were obtained by site-directed mutagenesis of pHM1873 and pHM1506, respectively. Plasmid pETMfd, where mfd gene expression is under the control of its native promoter, was described previously (10).
To construct the transcriptional PgreA::lacZ, PgreB::lacZ, PdksA::lacZ, PgadA::lacZ, and PgadE::lacZ fusions, PCR amplifications of chromosomal DNA from the MG1655 strain were carried out using the primer pairs greAUP/greADO, greBUP/greBDO, dksAUP/dksADO, gadAUP/gadADO, and gadEUP/gadEDO, respectively (see Table S2 in the supplemental material). The PCR products were digested by EcoRI/BamHI and cloned into pRS415 (43). The fusions were introduced into the bacterial chromosome of strains CF7968 and CF12257 using the RS45 lambda phage, and we verified that the strains were monolysogens as described previously (36).
β-Galactosidase assays.
β-Galactosidase assays were performed as described previously (26).
Microarray analysis.
Strains were grown at 37°C in M9 minimal medium containing all the required amino acids at 100 μg/ml (Σ set, defined as DFHILQSTV), in the presence of isopropyl-β-d-thiogalactopyranoside (IPTG) at 0.1 mM and spectinomycin at 15 μg/ml. At an optical density at 600 nm (OD600) of ∼0.2, 20-ml samples were taken and added to an equal volume of ice-cold RNA-Later reagent (Ambion). The samples were then processed, hybridized with Affymetrix E. coli 2.0 arrays, and analyzed as described previously (33). We made the following direct comparisons between strains with the following characteristics: ppGpp0/vector and ppGpp0 dksA/vector; ppGpp0 dksA/vector and ppGpp0 dksA/pGreA; ppGpp0 dksA/vector and ppGpp0 dksA/pGreB; ppGpp0/vector and ppGpp0/pGreA; ppGpp0/vector and ppGpp0/pGreB; ppGpp0/pGreA and ppGpp0 dksA/pGreA; ppGpp0/pGreB and ppGpp0 dksA/pGreB.
Western blotting.
Western blot assays were performed with anti-DksA IgY as previously described (8) and with anti-GreA IgY (33). Despite several attempts with three different sources of anti-GreB antibodies (polyclonal IgY [US Biological] made to our own purified GreB protein, mouse monoclonal antibodies from NeoClone, and rabbit polyclonal antibodies from V. J. Hernandez), we were unable to detect GreB protein levels in any of our samples except in those carrying pGreB plasmid.
Microarray data accession number.
The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO series accession number GSE28795 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE28795).
RESULTS
GreA and DksA are functionally redundant in some instances of growth regulation.
As stated in the introduction, we were interested here in testing whether the apparent antagonism between GreA/GreB (GreA/B) and DksA at the ppGpp negatively regulated rRNA promoters also occurs for expression of pathways for amino acid biosynthesis which are positively controlled by ppGpp.
We first turned to DksA-deficient strains because they were reported to be completely or only partially auxotrophic for a set of amino acids that are similar to those required by the ppGpp0 strain (8, 34, 39). We discovered that the absence of DksA in fact only severely impaired growth on M9 glucose minimal medium plates (M9 minimal), and therefore the mutant is a slow-growing bradytroph instead of an auxotroph. The wild-type strain formed colonies after 24 h at 37°C, while the dksA mutant formed small colonies after 72 h (see Fig. S1 in the supplemental material). Addition of Casamino Acids (CAA) allowed the dksA mutant to form large colonies in 1 day, but these were still smaller than those of the wild type as if growth is not solely determined by amino acid availability. There were host-dependent variations in the dksA strain due to mutations whose functions are related to ppGpp metabolism. For example, RelA senses amino acid deprivation and responds with ppGpp synthesis. A dksA relA double deletion improved growth, shortening by half the time required to give equivalent colonies on M9 minimal plates. A complete deficiency of ppGpp, achieved by deleting relA and spoT (ppGpp0 dksA strain), gave colonies on M9 CAA plates that were twice the size of those formed by the ppGpp+ strain. Together, these observations suggest that ppGpp is detrimental for bacterial growth in the absence of DksA.
The dksA mutant grew after 3 days at 37°C on M9 minimal, but surprisingly the greA mutation completely abolished the growth of the dksA strain even for several more days whether GreB was present or not (Table 1, rows 1, 5, and 13). The greA mutation by itself did not inhibit colony growth in all media tested (Table 1, row 9, and data not shown). Thus, growth on M9 minimal medium was allowed in the absence of either DksA or GreA but not in the absence of both. This indicated a synthetic-lethal relationship and a functional redundancy between the two proteins. Since the absence of dksA but not greA impaired growth as mentioned above, the functional redundancy is only partial.
Table 1.
Effect of multicopy greA, greB, and dksA genes on growth of the ppGpp+ (spoT+relA+ or spoT+ ΔrelA) strains
| Row no. | Strain descriptionb | Plasmid | Growth on M9 platesa |
|
|---|---|---|---|---|
| − IPTG | + IPTG | |||
| 1 | Wild type (± greB) | Vectorc | + | + |
| 2 | pGreA | + | + (≪) | |
| 3 | pGreB | + | + (<) | |
| 4 | pDksA | + | + (≪) | |
| 5 | dksA (± greB) | Vector | + | + |
| 6 | pGreA | + | + (>) | |
| 7 | pGreB | + | + (≫) | |
| 8 | pDksA | + | + (≪) | |
| 9 | greA (± greB) | Vector | + | + |
| 10 | pGreA | + | + | |
| 11 | pGreB | + | + | |
| 12 | pDksA | + | + (≪) | |
| 13 | dksA greA (± greB) | Vector | − | − |
| 14 | pGreA | + | + (>) | |
| 15 | pGreB | + | + (≫) | |
| 16 | pDksA | + | + (≪) | |
Strains grown in LB medium supplemented with 50 μg/ml spectinomycin were diluted in 10 mM Mg2SO4 prior to plating on LB or M9 glucose minimal plates with (+) or without (−) 0.1 mM IPTG. Colony diameters were measured after 3-day incubations at 37°C on plates containing less than 100 colonies. +, the strain formed colonies; −, less than 1% of colonies were present compared to plating of the same dilution on LB medium. Symbols between parentheses denote colony diameters on IPTG-containing plates that were smaller (< and ≪) or larger (> and ≫) than the diameter of the colonies formed by the same strain on plates not supplemented with IPTG.
relA+ and ΔrelA strains yielded similar results. ± greB, similar results were obtained in the presence or absence of the greB gene.
The vector control was pHM1883.
This led us to test whether Gre factor overproduction could complement the extremely slow growth phenotype of the dksA mutant. Therefore, we transformed the dksA and dksA greA strains with the pGreA (pHM1873) or pGreB (pHM1874) plasmids. Unless noted otherwise, all plasmids used in this study are derivatives of the low-copy-number pGB2 vector (pSC101 origin) in which the expression of the cloned factors is under the control of the IPTG-inducible Ptac promoter. In the dksA mutant, both plasmids increased the colony size on minimal plates in the presence of 0.1 mM IPTG (Table 1, rows 6 and 7); this suggested that either GreA or GreB could replace the growth-promoting function of DksA. Transforming the dksA greA double mutant with any of the three plasmids (pGreA, pGreB, or pDksA [pHM1506]) was sufficient to allow growth (Table 1, rows 13 to 16), which reinforced this notion.
Conversely, overexpression of these three proteins inhibited growth to various extents under certain conditions. For example, IPTG-induced GreA, GreB, and DksA all reduced colony size on minimal plates in the wild-type strain, with GreB being the least inhibitory of the three (Table 1, rows 1 to 4). Such inhibition was also seen for pDksA in all of the other strains (Table 1, rows 5, 8, 9, 12, and 16).
Partial complementation of a ppGpp0 strain auxotrophies by overexpression of Gre factors or DksA.
Since growth on minimal plates requires ppGpp-dependent expression of a number of amino acid synthesis pathways, we asked whether multicopy greA, greB, or dksA genes could also suppress all auxotrophies of a ppGpp0 strain. Table 2, rows 1 to 4, shows that pGreA, pGreB, or pDksA was unable to allow the ppGpp0 strain to grow on minimal plates.
Table 2.
Effect of multicopy greA, greB, and dksA genes on the auxtrophies of ppGpp0 strains
| Row no. | Strain descriptionb | Plasmid | Growth on M9 platesa |
||
|---|---|---|---|---|---|
| Without amino acids |
With DQ |
||||
| − IPTG | + IPTG | + IPTG | |||
| 1 | Wild type (± greB) | Vectorc | − | − | − |
| 2 | pGreA | − | − | + | |
| 3 | pGreB | − | − | − | |
| 4 | pDksA | − | − | + | |
| 5 | dksA (± greB) | Vector | − | − | + |
| 6 | pGreA | − | + | + (≫) | |
| 7 | pGreB | − | − | + | |
| 8 | pDksA | − | − | + (≫) | |
| 9 | greA (± greB) | Vector | − | − | − |
| 10 | pGreA | − | − | + | |
| 11 | pGreB | − | − | − | |
| 12 | pDksA | − | + | + (≫) | |
| 13 | dksA greA (± greB) | Vector | − | − | − |
| 14 | pGreA | − | + | + (≫) | |
| 15 | pGreB | − | − | + | |
| 16 | pDksA | − | + | + (≫) | |
Surprisingly, IPTG induction of these plasmids revealed a special relationship between GreA and DksA. IPTG-induced GreA restored prototrophy only in the absence of DksA (Table 2, rows 2, 5, 6, 10, and 14). Conversely, DksA overexpression conferred prototrophy to a ppGpp0 strain only when the greA gene was inactivated (Table 2, rows 4, 8, 12, and 16). Thus, there seems to be an antagonism between GreA and DksA.
This special relationship does not apply to GreB because it did not restore prototrophy under any of these conditions (Table 2, rows 7 and 15). Yet earlier GreB overproduction improved growth of the ppGpp+ dksA strain and allowed growth of the ppGpp+ dksA greA mutants on M9 minimal plates (Table 1, rows 7 and 15). This could be viewed in two ways: either GreB requires ppGpp for its growth-promoting functions, or the growth defect is so severe in the absence of ppGpp that the weak GreB activity cannot overcome it.
The specific amino acids required by strains devoid of ppGpp and/or DksA can vary depending on the genetic background (34). In the genetic background we used (CF15615), a ppGpp0 strain formed colonies on minimal plates supplemented with a minimum set of amino acids (DQILVFHST), termed the Σ set. It should be mentioned that in this set, ILVFHST without DQ is sufficient for growth in liquid medium, but addition of DQ is necessary for plate growth (34). The ppGpp0 dksA and dksA greB mutants formed colonies on M9 glucose plates supplemented simply with DQ with or without the remaining Σ set (Table 2, rows 1 and 5). However, the single greA, single greB, the double dksA greA, and the triple dksA greA greB mutants all remained auxotrophic (Table 2, rows 1, 5, 9, and 13). The growth of strains listed in Table 2 that could not be rescued solely by DQ was also not restored by adding any other single amino acid from the Σ set (data not shown).
Next, we checked the effects of overproducing GreA, GreB, and DksA in the backgrounds described above. We found that both pGreA and pDksA were able to restore growth on minimal plates supplemented with DQ for all the strains tested (Table 2, rows 2, 4, 6, 8, 10, 12, 14, and 16). The case of pDksA in the ppGpp0 strain was especially puzzling, since Table 2, row 5, showed that the removal of dksA had the same effect, i.e., allowed growth in the presence of DQ only. Although there might be other explanations, this could imply that the growth phenotype does not simply rely on the presence or absence of each factor but that their concentration can be crucial. Also, the data obtained with the pGreB plasmid seemed to reinforce the idea of interactions between the factors, reminiscent of the special relationship between GreA and DksA. This is because, in the presence of DQ, pGreB was able to rescue the growth defect only in the absence of a chromosomal source of DksA, independent of whether the chromosomal wild-type greA gene was present or not (Table 2, rows 7 and 15), i.e., in the ppGpp0 dksA greA and ppGpp0 dksA greA greB mutants (Table 2, row 15). This could indicate that there is antagonism between GreA/B and DksA but not between GreA and GreB.
The suppression of the ppGpp0 strain auxotrophy by GreA and DksA overproduction may not involve the hydrolysis of backtracked RNA or inhibition of ribosomal promoters.
As mentioned in the introduction, acidic residue mutants of GreA at the tip of the coiled-coil structure are impaired in their ability to rescue arrested RNA polymerase. In order to differentiate if this property was involved in abolishing the amino acid requirements studied here, we employed a plasmid overproducing GreA* [GreA(D41A E44Y), pHM1854], the mutant version of GreA (31).
We found that GreA* did restore prototrophy on M9 minimal medium to the ppGpp0 strain but only in the absence of the chromosomal wild-type dksA gene (Table 3). The latter observation is of interest because it suggests that this pGreA* phenotype is recessive to a chromosomal copy of dksA, not greA, which provides another indication of a special relationship between GreA and DksA. Other investigators have suggested from studies of the PfimB or PfliC promoters that the absence of the highly abundant DksA protein facilitates the access of less abundant GreA to the secondary channel, thereby potentiating the ability of GreA to rescue arrested RNA polymerase (1, 2). This suggestion may well explain the need for a chromosomal dksA gene deletion to confer ppGpp0 prototrophy. However, it follows from the dispensability of the two conserved acidic residues that complementation of ppGpp0 auxotrophy is unlikely to be related to the hydrolysis of backtracked RNA or to the release of paused or arrested RNAP.
Table 3.
Effect of multicopy greA* and dksA* genes on the auxtrophies of the dksA and ppGpp0 strains
| Row no. | Strain description | Plasmid | Growth on M9 platesa |
|||
|---|---|---|---|---|---|---|
| ppGpp+ |
ppGpp0 |
|||||
| − IPTG | + IPTG | − IPTG | + IPTG | |||
| 1 | dksA+ | Vectorb | + | + | − | − |
| 2 | pGreA* | + | + (<) | − | − | |
| 3 | pDksA* | + | + (<) | − | − | |
| 4 | dksA | Vector | + | + | − | − |
| 5 | pGreA* | + | + (>) | − | + | |
| 6 | pDksA* | + | + | + | + (>) | |
Growth conditions and scoring were as described for Table 1.
The vector control was pHM1883.
We also examined effects of pDksA* [DksA(D71N D74N), pHM1790] carrying corresponding mutations in the acidic residues at the tip of the coiled coil. Although the role of these residues is less well documented than for GreA*, it has been reported that this mutant is defective in several DksA-dependent aspects of transcription in vitro although it is still able to bind to RNAP (31). Table 3 reveals several interesting features of DksA*: (i) most importantly, overexpression of the mutant protein was able to reverse the ppGpp0 auxotrophy; (ii) reversal occurred only when the wild-type chromosomal dksA was absent and then even without IPTG induction; (iii) reversal occurred despite the presence of a chromosomal wild-type greA gene, which was not true for pDksA (Table 2, rows 8 and 12). It thus appears that the D71N D74N mutations abolish or override the regulatory relationship of DksA and GreA described above.
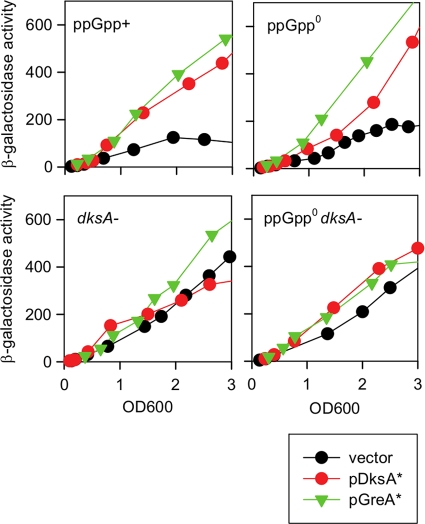
We next wondered if GreA* and DksA* effects on rRNA promoters might explain the complementation of ppGpp0 dksA strain auxotrophy. This is relevant to ideas of stimulation of amino acid synthesis by models involving indirect effects resulting from inhibition of ribosomal promoters by ppGpp (see Discussion). We have previously shown during growth in LB medium that the rrnB P1 promoter is activated 2-fold by GreA overproduction and repressed by DksA (35). In the experiment shown in Fig. 1, with cells grown in LB medium, pDksA* overproduction in both ppGpp+ and ppGpp0 strains activated the fusion instead of inhibiting it and delayed the onset of rrnB P1 repression that normally occurs late in exponential phase. Unlike complementation of ppGpp0 strain auxotrophy, the effect of pDksA* on rrnB P1 occurred despite the presence of a wild-type chromosomal dksA gene. Surprisingly, in a dksA mutant background whether ppGpp was present or not, the lacZ fusion activity was slightly lower at an OD600 of 3, late in growth. When the dksA gene was inactivated, the activity was almost the same as that of the vector control.
Fig 1.
Overproduction of GreA* and DksA* alters rrnB P1::lacZ activity. The effects of overproduced GreA* and DksA* on the rrnB P1 promoter were monitored in ppGpp+, ppGpp+ dksA, ppGpp0, and ppGpp0 dksA strains by β-galactosidase assessment throughout growth. All strains were grown at 37°C in LB medium; IPTG was added to 0.1 mM when the culture reached an OD600 of ∼ 0.1 to 0.15. Strains designated here as ppGpp+ correspond to a ΔrelA spoT+ strain. dksA−, dksA mutant.
In contrast, activation of the rrnB P1 promoter by GreA* induction differed depending on ppGpp and growth phase. Promoter activity was the same in early stationary phase (OD600 of 2) in both ppGpp0 and ppGpp+ strains, but thereafter (OD600 of >3) activity became limited when ppGpp was present (Fig. 1 and data not shown). In the dksA mutant the rrnB P1 fusion activity in the presence of GreA* was the same as in the dksA+ strain when ppGpp+ was present and slightly higher than that of the vector control. On the other hand, in the ppGpp0 dksA background, GreA* could not stimulate the fusion activity any further than already obtained in the vector control strain.
In another attempt to distinguish if GreA- and GreB-mediated antipausing activity was responsible for restoring prototrophy, we tested the effects of Mfd. This protein has been shown to also relieve RNAP pauses but by a mechanism unrelated to that of Gre factors (7, 41). It has been also proposed that DksA* has similar effects on transcriptional elongation as the Gre factors, and in doing so it rescues stalled replication forks in dksA mutant cells (49). We found that overproduction of Mfd from a multicopy plasmid did not reverse the slow-growth phenotype of the dksA mutant strains on M9 glucose plates, nor did it restore growth of the ppGpp0 dksA strain on those plates (data not shown). Likewise, we also found that Mfd overproduction in ppGpp0 dksA+ cells did not restore growth on minimal glucose plates whether supplemented with DQ or not (data not shown). These results support the conclusion that restoration of the prototrophy, in particular DQ auxotrophy, was not due to relief of transcriptional pauses.
Effect of GreA and GreB overproduction on global transcription.
To gain further insight in the mechanism of suppression of the ppGpp0 auxotrophy, we examined changes in global transcription profiles due to GreA or GreB factor overproduction in ppGpp0 and ppGpp0 dksA hosts. In order to grow the strains under defined conditions, M9 minimal medium was supplemented with the amino acids required for growth by the parental strains (Σ set, DQILVFHST). This necessity minimized the chance of seeing regulatory effects on the specific amino acid biosynthetic pathways satiated by these nine supplements but allowed a glimpse of effects on other pathways. IPTG was present at 0.1 mM throughout growth, and samples were collected at an OD600 of ∼0.2. Direct comparisons were made between data obtained from strains differing in only one variable (for example, the ppGpp0 and ppGpp0 dksA strains and the ppGpp0 dksA/vector and ppGpp0 dksA/pGreA strains; see Materials and Methods for details). The accepted significance level was set at a 2.5-fold difference in each comparison. In order to ensure that accepted ratios were not based on very low signal strengths (below 100), a proviso was added that one of the signals in a given ratio had to be at least 250 units.
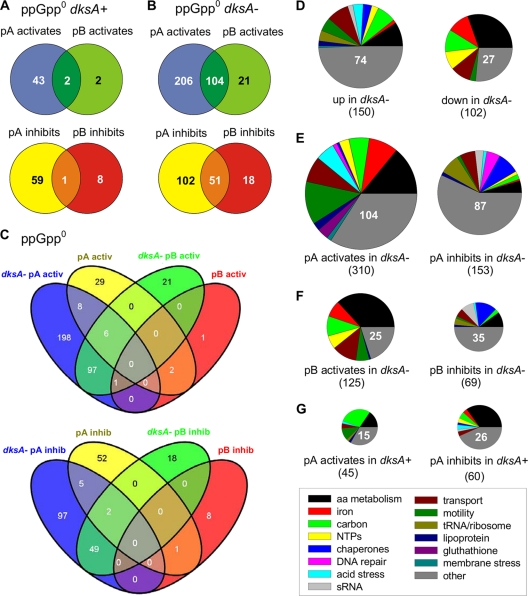
It would be expected from indications of competition from the growth experiments above that more genes would be affected by GreA or GreB overproduction in the absence of DksA than in its presence. If there was no competition, the number of genes affected should be similar regardless of the dksA allelic state. We found that when DksA was present, GreA overproduction affected only 105 genes (45 activated, 60 inhibited), while GreB affected 13 genes (4 activated, 9 inhibited) (Fig. 2A), for a total of 115. In a ppGpp0 dksA strain, a total of 501 genes were affected (Fig. 2B). There were only a few instances where the same gene was affected by GreA or GreB independent of dksA (Fig. 2C). Finding a nearly 5-to-1 ratio of genes affected in the absence of DksA strongly suggested an overall competition or antagonism between GreA/B and DksA.
Fig 2.
Transcription profiling indicates instances of both antagonism and parallel action between GreA, GreB, and DksA. All experiments were performed in a ppGpp0 strain background. (A) Comparison of genes activated or inhibited by overproduced GreA (pA) or GreB (pB). (B) Same experiment as shown in panel A, but for ppGpp0 dksA strains. (C) Comparison of genes either activated or inhibited by pA or pB in the ppGpp0 and ppGpp0 dksA backgrounds (comparison of data shown in A and B). The Venn diagrams presented in panels A, B, and C were made with the use of VENNY software (J. C. Oliveros, BioinfoGP, Madrid, Spain [http://bioinfogp.cnb.csic.es/tools/venny/index.html]) (D) Analysis of functional group of genes activated and inhibited by dksA deletion. (E and F) Same experiment as shown in panel D, but for GreA or GreB overproduction in a ppGpp0 dksA strain. (G) Same experiment as shown in panel D but for GreA overproduction in a ppGpp0 dksA+ strain. The sizes of the pie diagrams in panels D to F are roughly proportional to the number of genes affected and specified in parentheses. dksA+, dksA wild type; dksA−, dksA mutant.
In accordance with our earlier finding that GreA had a more potent effect on the growth phenotypes than GreB, profiling revealed that GreA overproduction affected a larger set of genes than GreB although there was a significant overlap. Specifically, in the dksA mutant background, GreA upregulated 310 genes, while GreB activated only 125 genes (with 104 overlapping genes). A similar disproportion was true for inhibitory effects: GreA downregulated 153 genes while GreB inhibited only 69 (with 51 overlapping genes) (Fig. 2B).
The next question we addressed was whether functional gene categories were preferentially affected among the different comparisons. Deleting dksA in the ppGpp0 background caused a significant increase in transcription of 150 genes and downregulation of 102 (Fig. 2D). Among the genes that were upregulated, the major categories were amino acid metabolism, carbon metabolism, transport, and motility. On the other hand, those that were repressed also included amino acid and carbon metabolism, in addition to iron and nucleotide metabolism. Clearly, individual genes within the same functional class were differentially regulated by DksA in a ppGpp0 strain. Profiling studies on the effects of dksA and ppGpp were performed by other investigators with similar results (1) (see Discussion).
When GreA was overexpressed in the dksA mutant, the major categories of the upregulated genes in this large group included amino acid metabolism, motility, iron metabolism, and transport. Those that were downregulated included chaperones, ribosome/tRNA-modifying enzymes, transport, and DNA repair (Fig. 2E). In the case of overexpressed GreB, the most activated genes again were those involved in amino acid metabolism, transport, and carbon and iron metabolism. The downregulated genes included chaperones, small RNAs, amino acid metabolism, and transport (Fig. 2F).
In the ppGpp0 dksA+ strain, pGreA activated the genes in the carbon, amino acid metabolism, and motility categories. The downregulated genes were predominantly in the amino acid metabolism group.
A particularly intriguing class was one whose genes were specifically affected in dksA mutant strain, but this effect was suppressed by overexpressed GreA and/or GreB (134 in total) (see Table S3 in the supplemental material). Of these, 76 genes were downregulated in the presence of greA and greB chromosomal copies only. This is interesting because genes downregulated in a dksA background are presumably activated by wild-type DksA. Reversal of such inhibition by GreA or GreB could mean that, in those instances, overexpressed GreA or GreB can function like DksA. Conversely, 58 genes were activated by a dksA deletion but inhibited by GreA or GreB. Again, this implies that overexpressed GreA and GreB can function similarly to DksA. It is striking that similarities of function are noted for both inhibition and stimulation; of course, we do not yet have direct evidence that such complementation occurs at the promoter level of any individual gene.
DksA and GreA/B effects in vivo on the expression of gadA and gadE genes.
Overall, the microarray analysis revealed that the most dramatically affected of all genes were gadA, gadB, gadC, and gadE (see Table S3 in the supplemental material). The gadE gene product is a luxR family transcriptional regulator of the gadABC operon (18, 37, 42). The gadA gene encodes glutamic acid decarboxylase; gadE-gadABC regulation is induced by acid and salt shock and is sensitive to RpoS, EvgAS, and H-NS regulation. Our profiling revealed that gadA was activated by 90-fold and that gadE was activated by 40-fold in the ppGpp0 dksA/pGreA strain compared to the level in the vector control. We therefore selected gadA and gadE for further study because this activation was predominantly achieved by pGreA induction and especially evident in a dksA strain; GreB overexpression had minor effects, as did the dksA gene deletion alone. Interestingly, these genes were also found to be significantly downregulated in the ppGpp0 strain compared to the wild type (K. Potrykus and M. Cashel, unpublished results).
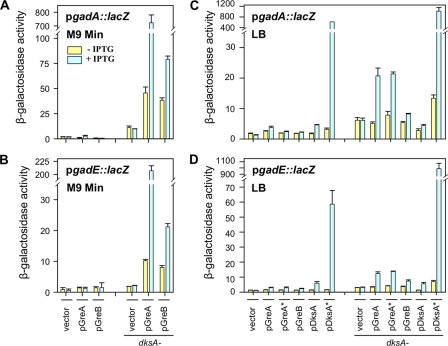
Therefore, PgadA::lacZ and PgadE::lacZ transcriptional fusions were constructed, and their activities were surveyed for effects of the dksA gene deletion and IPTG induction of pGreA and pGreB plasmids (Fig. 3A and B). The data verify transcriptional profiling results with respect to dramatic activation of gadA and gadE by GreA in the absence of dksA and furthermore localize the regulation to the complex promoter regions of these genes.
Fig 3.
PgadA::lacZ and PgadE::lacZ fusion activities during growth in minimal and LB media. (A and B) The effects of overproduced GreA and GreB on the gadA and gadE promoter fusion activities were monitored in ppGpp0 and ppGpp0 dksA strains at 37°C in M9 glucose medium. β-Galactosidase measurements were performed when cultures reached an OD600 of ∼0.2. (C and D) The effects of overproduced GreA, GreA*, GreB, DksA, and DksA* on the gadA and gadE promoter fusion activities were monitored in ppGpp0 and ppGpp0 dksA strains at 37°C in LB. β-Galactosidase measurements were performed before cultures reached an OD600 of ∼1.0. When added, IPTG was present at 0.1 mM.
We also wanted to compare the effects of GreA, GreA*, DksA, and DksA* proteins on these fusion activities. However, we found that some of these strains grew very poorly in M9 glucose liquid medium, even when the medium was supplemented with the Σ set and especially when GreA* and DksA* protein overproduction was induced by IPTG addition (data not shown). We therefore performed subsequent experiments in LB medium, where growth of all strains is robust.
Preliminary control experiments were done to ensure that our interpretations of reporter activities were not complicated by known complexities affecting PgadA and PgadE promoters. Studies with gadE and rpoS mutants verified that induction of each fusion was abolished in stationary phase (OD of 2) in the absence of GadE, RpoS, DksA, or ppGpp (data not shown). This verifies the known RpoS activation of GadE mentioned above. It also verified the dependence of PgadA and PgadE on DksA and ppGpp through RpoS; RpoS regulation by ppGpp and DksA has been demonstrated previously (8, 16). Thus, sampling during exponential phase of growth (OD of <1) in a ppGpp0 or ppGpp0 dksA host in our experiments ensured that indirect regulatory effects on these promoters were minimized.
Figure 3C shows that overexpressing GreA and GreA* in the absence of the wild-type dksA allele, but not in its presence, induced gadA about 4-fold. Inducing GreB in a dksA background had only about a 2-fold effect. Overproduction of DksA had an even more modest effect. On the other hand, DksA* without IPTG induction barely had an effect on gadA when the wild-type chromosomal dksA gene was present and a 5-fold effect when the gene was deleted. Overexpression of DksA* gave the most dramatic fusion activation. In the dksA+ strain activation reached about 600 Miller units (30-fold over the uninduced control) and 1,100 units (about 70-fold difference) in the absence of chromosomal dksA. Similar results can be seen in Fig. 3D with a PgadE::lacZ reporter fusion, with the exception that overexpression of DksA* in the presence of chromosomal dksA was only 1/10th the increase (60 Miller units) of when dksA was missing (600 units). Overall, the qualitative phenomenon still persisted in LB medium: the gadA and gadE genes were more actively induced when DksA was absent, and GreA overproduction had a stronger effect than GreB.
Effect of the different factors on the transcription of others: E. coli maintains a balance between the RNA polymerase-interacting factors.
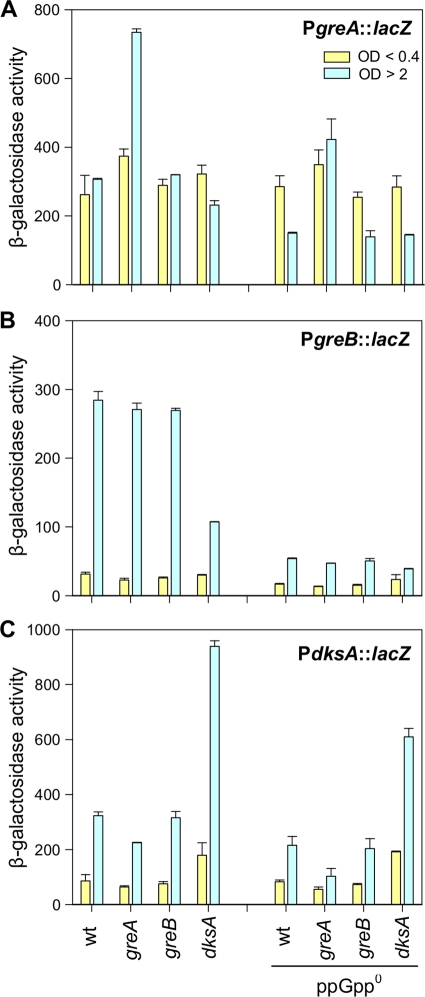
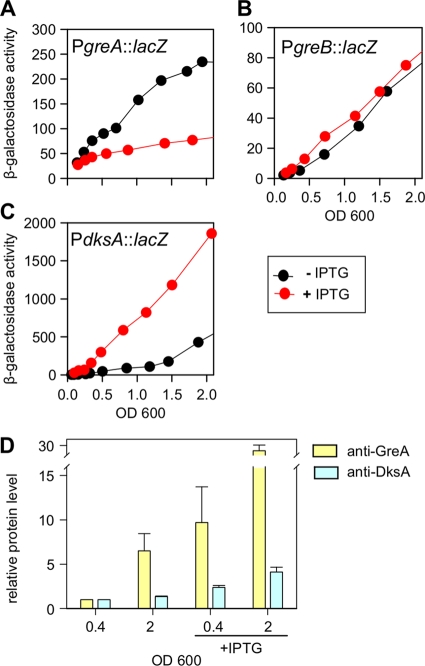
The apparent competition between the Gre and DksA proteins might be due to mutual regulatory effects that they could exert on each other. To test that hypothesis, we constructed single-copy transcriptional lacZ fusions of the greA, greB, and dksA promoter regions (Materials and Methods).
As seen in Fig. 4A, PgreA::lacZ fusion activities in exponential phase of growth were not appreciably affected by greA, greB, or dksA gene deletions, whether in the wild-type or ppGpp0 background. However, in the early stationary phase (OD of >2), the ppGpp+ greA mutant strain showed a 2-fold increase in the fusion activity, indicating previously reported greA autorepression (33). In the ppGpp0 greA mutant, fusion activity at a OD of >2 remained high, while activities in the other mutant strains dropped by half, again suggesting autorepression.
Fig 4.
PgreA::lacZ, PgreB::lacZ, and PdksA::lacZ activities in different backgrounds reveal complex regulation of the three factors. The experiments were performed in wild-type or ppGpp0 cells also carrying a greA, greB, or dksA deletion. Activities of the promoters, as indicated on the figure, were monitored by β-galactosidase assays at early exponential phase (OD600 of <0.4) and early stationary growth (OD600 of ≥2).
For the PgreB::lacZ fusion (Fig. 4B), there were only modest differences during exponential growth in ppGpp+ and ppGpp0 backgrounds. However, in the stationary phase we observed a 10-fold increase in activity of the wild type as well as in the greA and greB mutants and only a 3-fold induction in the dksA mutant. With the corresponding ppGpp0 strains there was only a 2- to 3-fold activation, indicating that greB transcription was stimulated by ppGpp and that this effect was largely due to DksA.
The activity of the PdksA::lacZ fusion (Fig. 4C) during exponential growth was increased by 2-fold by the dksA gene deletion, regardless of ppGpp; the greA or greB mutation had no effect. In the early stationary phase, activities of all fusions were increased 2.5- to 4-fold relative to exponential phase with the exception of the ppGpp0 greA mutant, where there was only a 2-fold increase. Still, the most predominant effect was observed in the dksA mutant, where the fusion activity was increased 5-fold in the presence of ppGpp and 3-fold activated in a ppGpp0 background. This suggested that dksA is also subject to autorepression, which is also consistent with recently published reports (11, 14).
RpoS does not seem to be involved in stationary phase induction of these fusions. The rpoS mutation had no effect on activities of the PgreA::lacZ or PdksA::lacZ fusions, while PgreB::lacZ fusion activity was slightly decreased, but activation in the stationary phase persisted (data not shown).
An imbalance caused by the deletion of one of the three factors might have a different effect than overexpression of that factor from an IPTG-inducible plasmid. Therefore, we analyzed the effects of overexpressing GreA or GreB in the ppGpp0 background. We found that overexpressed GreA caused an increase in the PdksA::lacZ fusion activity in both exponential and early stationary phases of growth. Western blot analysis confirmed an increase in the DksA protein level, predominantly in the stationary phase (Fig. 5). Overexpressed GreB had only minor effects (data not shown).
Fig 5.
Overproduced GreA activates PdksA::lacZ and increases DksA level in ppGpp0 cells. (A to C) PgreA::lacZ, PgreB::lacZ, and PdksA::lacZ activities in ppGpp0 cells carrying the pGreA plasmid were monitored by β-galactosidase assays throughout growth at 37°C in LB medium. When present, IPTG was added to 0.1 mM at the beginning of the experiment. (D) GreA and DksA protein levels assessed by Western blotting under the same conditions as in panels A to C. The samples for the Western analysis were collected at OD600 of 0.4 and 2. Protein levels were normalized for each blot separately to the level found in the sample obtained at an OD600 of 0.4 without IPTG; the alpha subunit of RNAP was used as the loading control.
Role of the Gre factors and DksA in the suppression by RNA polymerase mutants.
At the beginning of this work, we were interested in which factors would make a ppGpp0 strain regain prototrophy. Now, we wish to investigate whether any of these factors contribute to RNA polymerase M+ mutant phenotypes, i.e., whether the M+ phenotype is due to a change in the factor's affinity or interaction with the mutant RNAP.
The M+ mutants in the ppGpp0 background can be subdivided into three phenotypic classes depending on additional growth tests originally employed to discern between relA and ΔrelA ΔspoT (ppGpp0) mutants. All M+ mutants are able to grow in M9 minimal medium by definition (class I). Inability to grow on M9 minimal medium is taken to indicate a complete absence of ppGpp (53). A subset of M+ mutants (class II) is also able to grow on minimal plates containing Ser, Met, and Gly (SMG plates) (51). The SMG growth test relies on the ability of low levels of ppGpp to stimulate Ile synthesis in wild-type but not relA mutant strains; this test is specific for E. coli K-12 strains. Yet another subset of M+ mutants (class III) is resistant to 3-aminotriazole (AT plates). This growth test relies on the ability of ppGpp to override the inhibitory effect of AT on the HisG activity by activating the his operon promoter just upstream of hisG (40). This growth test can be modified to detect various levels of ppGpp by adjusting the 3-aminotriazole concentration present; 15 to 18 mM is typically used to discern between relA mutants (AT sensitive) and ΔrelA spoT point mutants with high basal levels of ppGpp during growth (40). A hierarchy seems to exist for the M+ phenotypic classes: all class III mutants meet the criteria of class II; all class II mutants meet the criteria of class I but not class III. It seems plausible that the existence of this hierarchy among M+ mutants represents a spectrum of responses to increasing ppGpp concentrations rather than random effects on different phenotypes. In contrast, the growth test responses with wild-type RNAP probably reflect regulatory responses rather than titrations of ppGpp levels alone.
We first introduced the greA, greB, and dksA deletions alone or in different combinations into the ppGpp0 strains carrying the rpoB (M+) mutations. We arbitrarily chose duplicate examples of class I [rpoB(S531F) and rpoB(G534C)], class II [(rpoB(L533P) and rpoB(L571P)], and class III [rpoB(A532Δ), which has a deletion of A532 in rpoB, and rpoB(T563P)] mutants, listed from top to bottom in Table 4. On minimal plates, none of the single deletions reversed prototrophy (Table 3). The double and triple mutants had no effect, except for the weak M+ suppressors (class I), where the greA and dksA gene deletions with or without greB deletion caused loss of the phenotype. This suggested that class I required either GreA or DksA to grow on minimal plates, reminiscent of synthetic lethality of wild-type strains (Table 1).
Table 4.
Role of GreA/B and DksA in the M+ mutant suppression of the ppGpp0 phenotype
| M+ mutant strain description | Additional mutation(s) | Growth on the indicated mediuma |
||
|---|---|---|---|---|
| M9 | SMG | AT | ||
| rpoB114(S531F) | None | + | − | − |
| greA | + | − | − | |
| greB | + | − | − | |
| dksA | + | − | − | |
| greA greB | + | − | − | |
| greA dksA | − | − | − | |
| greB dksA | + | − | − | |
| greA greB dksA | − | − | − | |
| rpoB(G534C) | None | + | − | − |
| greA | + | − | − | |
| greB | + | − | − | |
| dksA | + | − | − | |
| greA greB | + | − | − | |
| greA dksA | − | − | − | |
| greB dksA | + | − | − | |
| greA greB dksA | − | − | − | |
| rpoB3443(L533P) | None | + | + | − |
| greA | + | + | − | |
| greB | + | + | − | |
| dksA | + | + | − | |
| greA greB | + | + | − | |
| greA dksA | + | − | − | |
| greB dksA | + | + | − | |
| greA greB dksA | + | − | − | |
| rpoB(L571P) | None | + | + | − |
| greA | + | + | − | |
| greB | + | + | − | |
| dksA | + | + | − | |
| greA greB | + | + | − | |
| greA dksA | + | − | − | |
| greB dksA | + | + | − | |
| greA greB dksA | + | − | − | |
| rpoB3449(A532Δ) | None | + | + | + |
| greA | + | + | + | |
| greB | + | + | + | |
| dksA | + | + | + | |
| greA greB | + | + | + | |
| greA dksA | + | − | + | |
| greB dksA | + | + | + | |
| greA greB dksA | + | − | − | |
| rpoB3370(T563P) | None | + | + | + |
| greA | + | + | + | |
| greB | + | + | + | |
| dksA | + | + | + | |
| greA greB | + | + | + | |
| greA dksA | + | − | − | |
| greB dksA | + | + | + | |
| greA greB dksA | + | − | − | |
Growth was monitored by streaking cells on the appropriate medium and scored after 2 days at 37°C. Scoring is otherwise as described for Table 1. M, M9 minimal medium; SMG, 100 mM (each) serine, methionine, and glycine; AT, 18 mM 3-amino-1,2,4-triazole. The ppGpp0 btuB::Tn10 (ppGpp0) control strain does not grow on M, SMG, or AT medium.
The growth test that defines class II mutants demonstrated similar results. Class I mutants did not grow on SMG plates with any combination of deletions. Class II and III mutant phenotypes were reversed only by the simultaneous deletion of the greA and dksA genes, again with or without GreB present.
Resistance to aminotriazole showed a slightly different but more complex pattern. It is true that deletion of greA and dksA with the greB deletion blocked AT growth of class III mutants, but the difference is that the greA dksA strain did not allow growth of the rpoB(T563P) strain, whereas it allowed growth of the rpoB(A532Δ) strain. At this point we do not know how to explain this difference.
Overall, these comparisons suggest that the hierarchal behavior of the M+ mutants is generally preserved in respect to greA, greB, and dksA deletions. Additional complexities within classes II and III indicate that there might be different mechanisms to achieve the same result, i.e., growth.
DISCUSSION
The major finding of this work is that GreA/B factors provide major contributions to already complex regulation by DksA and ppGpp. It has been evident for a long time that a complete deficiency of ppGpp in E. coli confers multiple and complex requirements for many amino acids, usually detected as a failure to grow in M9 minimal medium. There are also indications that these requirements are not nearly as absolute as those generated by mutants in specific biosynthetic pathways. For example, the requirements of ppGpp0 strains were originally defined on drop-out media that had one amino acid missing from a full complement of 20. If histidine is the missing amino acid, ppGpp0 strains cannot grow for several days on plates, suggesting an absolute requirement (data not shown). However, in the presence of the Σ set of amino acids lacking histidine (DQILVFST), growth of ppGpp0 strains occurs after 2 days as if the requirement is leaky (data not shown).
Prototrophy of wild-type strains is thought to be due to the positive regulation of amino acid biosynthesis gene transcription by ppGpp and/or DksA, as well as effects of multiple indirect mechanisms. Many of these indirect models are ultimately based on direct inhibition of ribosomal transcription by ppGpp, which in turn would make more RNA polymerase available for transcription of amino acid biosynthesis genes (15, 30). It has been reported that GreA and perhaps TraR and GreB are RNAP secondary channel factors that can influence rRNA promoter activities in addition to DksA (6, 35, 38). Therefore, it seemed worthwhile to explore the effects of GreA, GreB, and DksA on ppGpp-dependent amino acid biosynthesis pathways. In our approach, we have used systematic genetic studies (an array of deletions combined with effects of overexpression) coupled with transcriptional profiling and analysis of RNA polymerase mutants that abolish the amino acid requirements of the ppGpp0 cells. This allowed us to find instances, both at the phenotypic level and at the level of individual gene expression, where Gre factors and DksA act in the same way and other instances where they seem to be antagonistic.
GreA/B and DksA can be redundant.
The first evidence showing that GreA/B and DksA can act in the same way came from the analysis of mutants in a ppGpp+ background: (i) the growth deficiency of a dksA mutant was complemented by multicopy greA and greB; (ii) prototrophy of a ppGpp+ strain required the activity of either GreA or DksA and was lost in the dksA greA double mutant. The factors thus appear to be redundant for growth in minimal medium. The conclusion can be extended to the ppGpp0 background: DksA and GreA overproduction suppressed the absence of growth of the ppGpp0 strain on minimal glucose with DQ; they conferred complete prototrophy (growth on minimal glucose) only when the other factor was absent.
It is difficult to propose a specific mechanism for the GreA/B complementation of the dksA mutant growth because a large number of genes is likely to be involved. This complementation could be indirect, with Gre factors acting on genes whose expression might not be compromised in the dksA mutant. The same difficulties apply for complementation of ppGpp0 auxotrophy by GreA/B or DksA overexpression (see below). However, our microarray studies reveal that there is a large group of genes regulated in the same way by GreA and DksA. Future analysis of the effects of Gre and DksA factors on transcription of these genes in vitro should help to determine whether regulation of this class occurs by similar or different mechanisms.
GreA/B and DksA can be antagonistic.
In parallel with evidence of redundancy or similar activity, we also found that GreA/B and DksA can have opposite effects. The antagonism is shown by the fact that complete prototrophy can be achieved in a ppGpp0 background by GreA or DksA overexpression only when the other is absent. Also, many more genes are affected by GreA/B overexpression, both negatively and positively, when DksA is absent (463 versus 105 for GreA; 194 versus 13 for GreB). A simple explanation is that GreA/B has enhanced access to the secondary channel of RNAP in the absence of the more abundant DksA protein. The Pgad promoters also provide examples of antagonism. Activation by GreA largely depended on dksA gene inactivation, but DksA appeared to have little effect on these promoters by itself.
Possible mechanisms of regulation.
Our studies with the conserved acidic residue mutants of GreA and DksA were undertaken to provide clues to the wild-type protein behavior. In the case of GreA*, complementation of amino acid requirements was the same as with wild-type GreA, suggesting that the ability of this factor to restore prototrophy is unrelated to its ability to rescue backtracked transcriptional complexes.
We also found that even moderate overproduction of DksA* (without IPTG induction) was able to complement amino acid requirements in the absence, but not the presence, of a chromosomal copy of wild-type dksA, as if the wild-type DksA was competing here with DksA* for essential targets. This competition could be direct or indirect. For example, it is possible that one of the targets could be the dksA promoter: DksA* would alter dksA autorepression and in turn change the DksA protein levels in the cell in the way that affects growth on minimal plates. On the other hand, DksA* action could rely on altering GreA regulation. Since GreA is able to promote growth on minimal plates only in the absence of dksA (Table 2), DksA* overproduction would be futile here.
Previous assays with rrnB P1::lacZ fusions indicated that DksA inhibited this ribosomal promoter, while GreA stimulated its activity at the level of transcription initiation (35). Here, we show with similar assays that DksA* lost the ability to inhibit ribosomal promoter activity. In contrast, the mutation in GreA* did not alter this protein's ability to activate the promoter. However, in the dksA mutant GreA* could not activate the promoter, unlike wt GreA, suggesting that GreA* activation was probably based only on its ability to exclude the inhibitory DksA from binding to RNA polymerase.
As already mentioned above, these observations are of great interest because many indirect mechanisms proposed for positive amino acid promoter regulation by ppGpp rely on inhibition of rRNA transcription leading to either excess core RNA polymerase or enhanced access of alternative sigma factors to the core. The apparent positive regulation by DksA* together with the lack of inhibition of rRNA synthesis and both the activation of ribosomal promoter and suppression of auxotrophy by GreA* contradicts these models and favors more direct mechanisms. In the future, these in vivo results should be verified in vitro to ascertain the direct activation by DksA* and GreA* of the amino acid promoters, as has been already done with wild-type DksA (30).
Also, there have been reports of involvement of DksA, GreA, Mfd, and ppGpp in clearing the stalled arrays of backed-up RNAP complexes that would otherwise prevent replication fork progression. This phenomenon is evident in cells subject to DNA damage and deficient in DNA repair pathways (for example, ruvABC, recN, uvrD, and rep) (4, 25, 50). Since the strains used here are wild type for these pathways, then this phenomenon is unlikely to explain the growth defects we observe.
Recently, it has been suggested that the amino acid requirements of the ppGpp+ dksA strain are a consequence of replication fork collapse (49). This potentially could affect our interpretations. However, it is easily understood how replication fork collapse could impair growth but not how it could result in specific amino acid requirements or specific changes in metabolism noted for dksA mutants.
Studies of the effects of greA, greB, and dksA deletions on the RNA polymerase M+ mutants also reveal perturbation of growth phenotypes. One way of thinking about the M+ mutants is that the RNAP is locked in the conformation that mimics the presence of ppGpp. These can represent polymerases that are more sensitive than normal to ppGpp or that bypass the need for ppGpp (17). Nevertheless, deletion of both greA and dksA resulted in loss of the M+ class-defining phenotype, with few exceptions (Table 4). This indicates that the growth phenotype of the M+ mutants still relies on the interplay between the RNAP secondary channel factors and polymerase, as in the case of wild-type RNAP.
Hypothesis: how GreA or DksA overexpression results in ppGpp0 prototrophy.
We now come back to the question as to what are the ways that GreA or DksA overexpression could reverse a ppGpp0 auxotrophy? It could be a response to changes in absolute quantities in GreA or DksA or be due to changes in the ratio between these factors. Overexpression could compensate for the absence of ppGpp without necessarily involving perturbation of the ratios of the secondary channel proteins; for example, GreA has a direct effect on the RNAP-DNA open complex formation (35). Alternatively, reversal of auxotrophy could be due to changes in ratios independent of overexpression, as is the case for deletions. Considering only results obtained on minimal glucose DQ plates, it seems that the increase of the GreA/DksA ratio is sufficient to complement the growth defect of a ppGpp0 strain, as observed for ppGpp0 dksA and ppGpp0/pGreA strains.
We can use the promoter fusions of greA, greB, and dksA to speculate on differences in the expression of the three factors in ppGpp0 and wt strains. In a comparison of the wt and ppGpp0 strains, there is no difference in the expression of the three factors in exponential phase. In stationary phase there is less expression in the ppGpp0 strain than in the wild type, especially in the case of PgreB::lacZ. Still, GreB overexpression is the least potent factor in rescuing ppGpp auxotrophy. Therefore, it seems unlikely the auxotrophies of a ppGpp0 strain are due to a deficiency in one of the three proteins.
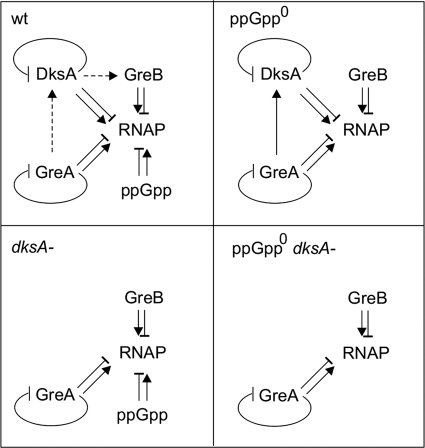
The regulatory circuits that are deduced in strains devoid of ppGpp, dksA, or both are depicted in Fig. 6. It should be noted that the evidence for the relationships represented with dashed lines is based solely on promoter fusions. We found that there is evidence of GreA and DksA autorepression, consistent with recently published data (11, 14, 33). Also, we found evidence that GreA increases DksA levels in the ppGpp0 cells and increases dksA promoter activity in the presence of ppGpp. This last result might explain the lack of growth on minimal glucose of the ppGpp0/pGreA strain; the chromosomal copy of dksA whose transcription is activated by GreA overexpression might not allow a sufficient increase of the GreA/DksA ratio.
Fig 6.
Interaction network of GreA, GreB, DksA, ppGpp, and RNA polymerase. The four panels depict changes in interactions of all five components when ppGpp, DksA, or both are missing. Arrows represent activation; bars represent inhibition. Dashed lines between the factors are interactions deduced from promoter transcriptional fusions only, whereas the solid lines depict evidence from both promoter fusions and Western blot assays. Interactions with RNAP come from microarray data and growth complementation studies.
Concluding remarks.
Finally, our experiments provide a comparison of interactions between Gre factors and DksA in a ppGpp0 background that potentiate regulation. We conclude that there are many varied interactions, including competition for RNAP. This strengthens conclusions drawn in another gene profiling study, where the authors showed similar and divergent effects of ppGpp and DksA deficiencies in comparison to a wild-type strain (1). Although GreA and GreB effects were not measured directly in that study, it was noted that a subset of 39 genes whose expression was altered in the DksA-deficient strain (311 in total) were the same as those identified previously as differentially regulated by overexpression of GreA in an otherwise wild-type background (46). Aberg et al. (1) had also directly verified this relationship for the fliC gene. Moreover, their earlier study had explored the difference between in vitro and in vivo regulation of the fimB promoter, and they found that GreA and GreB were able to stimulate transcription from this promoter only in the absence of DksA (2). This comprises direct evidence for antagonism of DksA and Gre factors at a promoter that is quite distinct from rrnB P1 (35). Overall, although our growth conditions differ from those of Aberg et al. (1), both studies strongly support the hypothesis that DksA and Gre factors compete directly or indirectly for the interactions with RNAP. Yet it has been published that GreA, GreB, and DksA levels remain remarkably constant throughout all phases of growth (38). We would like to argue that constant levels of these proteins are achieved through mutual regulatory effects of one factor on another, and their mutual regulation serves to maintain the balance in the cell. Also, these interactions can be complex because they could involve passive or active binding to RNAP and possibly displacement of previously bound factor. Individual promoter structures, sigma factors, DNA recognition proteins, and growth conditions add to this complexity. Remarkably, this was imagined nearly 20 years ago when multicopy GreA was found to suppress the temperature sensitivity of an RNAP mutant and alter phage lambda antitermination and rho termination. “This alteration, we imagine, might make this site on polymerase receptive to some factors but repulsive to others” (44).
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard D'Ari for critical reading of the manuscript. The Friday Seminar group again provided constructive discussions and encouragement. Nigel Savery and Patrice Moreau kindly and promptly provided the Mfd plasmids and the Keio strains, respectively. We also thank Marty Blum for medium preparation. Miho Matsuda (NIH) on short notice kindly guided us through the intricacies of colony microscopy.
This work was supported by the intramural program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health.
Footnotes
Published ahead of print 4 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Aberg A, Fernandez-Vazquez J, Cabrer-Panes JD, Sanchez A, Balsalobre C. 2009. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J. Bacteriol. 191:3226–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aberg A, Shingler V, Balsalobre C. 2008. Regulation of the fimB promoter: a case of differential regulation by ppGpp and DksA in vivo. Mol. Microbiol. 67:1223–1241 [DOI] [PubMed] [Google Scholar]
- 3. Artsimovitch I, et al. 2004. Structural basis for transcription regulation by alarmone ppGpp. Cell 117:299–310 [DOI] [PubMed] [Google Scholar]
- 4. Baharoglu Z, Lestini R, Duigou S, Michel B. 2010. RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol. Microbiol. 77:324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barker MM, Gaal T, Gourse RL. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305:689–702 [DOI] [PubMed] [Google Scholar]
- 6. Blankschien MD, et al. 2009. TraR, a homolog of a RNAP secondary channel interactor, modulates transcription. PLoS Genet. 5:e1000345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borukhov S, Lee J, Laptenko O. 2005. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 55:1315–1324 [DOI] [PubMed] [Google Scholar]
- 8. Brown L, Gentry D, Elliott T, Cashel M. 2002. DksA affects ppGpp induction of RpoS at a translational level. J. Bacteriol. 184:4455–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cashel M, Gentry DR, Hernandez VJ, Vinella D. 1996. The stringent response, p. 1458–1496 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed, vol 1 American Society for Microbiology, Washington, DC [Google Scholar]
- 10. Chambers AL, Smith AJ, Savery NJ. 2003. A DNA translocation motif in the bacterial transcription-repair coupling factor, Mfd. Nucleic Acids Res. 31:6409–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chandrangsu P, Lemke JJ, Gourse RL. 2011. The dksA promoter is negatively feedback regulated by DksA and ppGpp. Mol. Microbiol. 80:1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Choy HE. 2000. The study of guanosine 5′-diphosphate 3′-diphosphate-mediated transcription regulation in vitro using a coupled transcription-translation system. J. Biol. Chem. 275:6783–6789 [DOI] [PubMed] [Google Scholar]
- 13. Deighan P, Hochschild A. 2006. Conformational toggle triggers a modulator of RNA polymerase activity. Trends Biochem. Sci. 31:424–426 [DOI] [PubMed] [Google Scholar]
- 14. Edwards AN, et al. 2011. Circuitry linking the Csr and stringent response global regulatory systems. Mol. Microbiol. 80:1561–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gummesson B, et al. 2009. Increased RNA polymerase availability directs resources towards growth at the expense of maintenance. EMBO J. 28:2209–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harinarayanan R, Murphy H, Cashel M. 2008. Synthetic growth phenotypes of Escherichia coli lacking ppGpp and transketolase A (tktA) are due to ppGpp-mediated transcriptional regulation of tktB. Mol. Microbiol. 69:882–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hernandez VJ, Cashel M. 1995. Changes in conserved region 3 of Escherichia coli sigma 70 mediate ppGpp-dependent functions in vivo. J. Mol. Biol. 252:536–549 [DOI] [PubMed] [Google Scholar]
- 18. Itou J, Eguchi Y, Utsumi R. 2009. Molecular mechanism of transcriptional cascade initiated by the EvgS/EvgA system in Escherichia coli K-12. Biosci. Biotechnol. Biochem. 73:870–878 [DOI] [PubMed] [Google Scholar]
- 19. Komissarova N, Kashlev M. 1997. RNA polymerase switches between inactivated and activated states by translocating back and forth along the DNA and the RNA. J. Biol. Chem. 272:15329–15338 [DOI] [PubMed] [Google Scholar]
- 20. Komissarova N, Kashlev M. 1997. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc. Natl. Acad. Sci. U. S. A. 94:1755–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lamour V, Hogan BP, Erie DA, Darst SA. 2006. Crystal structure of Thermus aquaticus Gfh1, a Gre-factor paralog that inhibits rather than stimulates transcript cleavage. J. Mol. Biol. 356:179–188 [DOI] [PubMed] [Google Scholar]
- 22. Laptenko O, Lee J, Lomakin I, Borukhov S. 2003. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 22:6322–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lyzen R, Kochanowska M, Wegrzyn G, Szalewska-Palasz A. 2009. Transcription from bacteriophage lambda pR promoter is regulated independently and antagonistically by DksA and ppGpp. Nucleic Acids Res. 37:6655–6664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magnusson LU, Gummesson B, Joksimovic P, Farewell A, Nystrom T. 2007. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J. Bacteriol. 189:5193–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meddows TR, Savory AP, Grove JI, Moore T, Lloyd RG. 2005. RecN protein and transcription factor DksA combine to promote faithful recombinational repair of DNA double-strand breaks. Mol. Microbiol. 57:97–110 [DOI] [PubMed] [Google Scholar]
- 26. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Murphy H, Cashel M. 2003. Isolation of RNA polymerase suppressors of a (p)ppGpp deficiency. Methods Enzymol. 371:596–601 [DOI] [PubMed] [Google Scholar]
- 28. Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. 1995. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 92:4596–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paul BJ, et al. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322 [DOI] [PubMed] [Google Scholar]
- 30. Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc. Natl. Acad. Sci. U. S. A. 102:7823–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Perederina A, et al. 2004. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell 118:297–309 [DOI] [PubMed] [Google Scholar]
- 32. Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 33. Potrykus K, Murphy H, Chen X, Epstein JA, Cashel M. 2010. Imprecise transcription termination within Escherichia coli greA leader gives rise to an array of short transcripts, GraL. Nucleic Acids Res. 38:1636–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Potrykus K, Murphy H, Philippe N, Cashel M. 2011. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13:563–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Potrykus K, et al. 2006. Antagonistic regulation of Escherichia coli ribosomal RNA rrnB P1 promoter activity by GreA and DksA. J. Biol. Chem. 281:15238–15248 [DOI] [PubMed] [Google Scholar]
- 36. Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ruiz C, McMurry LM, Levy SB. 2008. Role of the multidrug resistance regulator MarA in global regulation of the hdeAB acid resistance operon in Escherichia coli. J. Bacteriol. 190:1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rutherford ST, et al. 2007. Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J. Mol. Biol. 366:1243–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rutherford ST, Villers CL, Lee JH, Ross W, Gourse RL. 2009. Allosteric control of Escherichia coli rRNA promoter complexes by DksA. Genes Dev. 23:236–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sarubbi E, Rudd KE, Cashel M. 1988. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol. Gen. Genet. 213:214–222 [DOI] [PubMed] [Google Scholar]
- 41. Savery NJ. 2007. The molecular mechanism of transcription-coupled DNA repair. Trends Microbiol. 15:326–333 [DOI] [PubMed] [Google Scholar]
- 42. Sayed AK, Foster JW. 2009. A 750 bp sensory integration region directs global control of the Escherichia coli GadE acid resistance regulator. Mol. Microbiol. 71:1435–1450 [DOI] [PubMed] [Google Scholar]
- 43. Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 44. Sparkowski J, Das A. 1992. Simultaneous gain and loss of functions caused by a single amino acid substitution in the beta subunit of Escherichia coli RNA polymerase: suppression of nusA and rho mutations and conditional lethality. Genetics 130:411–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sparkowski J, Das A. 1990. The nucleotide sequence of greA, a suppressor gene that restores growth of an Escherichia coli RNA polymerase mutant at high temperature. Nucleic Acids Res. 18:6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stepanova E, et al. 2007. Analysis of promoter targets for Escherichia coli transcription elongation factor GreA in vivo and in vitro. J. Bacteriol. 189:8772–8785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Svetlov V, Belogurov GA, Shabrova E, Vassylyev DG, Artsimovitch I. 2007. Allosteric control of the RNA polymerase by the elongation factor RfaH. Nucleic Acids Res. 35:5694–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Szalewska-Palasz A, et al. 2007. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on sigma54. J. Biol. Chem. 282:18046–18056 [DOI] [PubMed] [Google Scholar]
- 49. Tehranchi AK, et al. 2010. The transcription factor DksA prevents conflicts between DNA replication and transcription machinery. Cell 141:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG. 2005. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol. Cell 19:247–258 [DOI] [PubMed] [Google Scholar]
- 51. Uzan M, Danchin A. 1976. A rapid test for the relA mutation in E. coli. Biochem. Biophys. Res. Commun. 69:751–758 [DOI] [PubMed] [Google Scholar]
- 52. Vrentas CE, et al. 2008. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J. Mol. Biol. 377:551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xiao H, et al. 1991. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266:5980–5990 [PubMed] [Google Scholar]
- 54. Zhou YN, Jin DJ. 1998. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 95:2908–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.