Abstract
In Xanthomonas campestris pv. campestris, SoxR likely functions as a sensor of redox-cycling drugs and as a transcriptional regulator. Oxidized SoxR binds directly to its target site and activates the expression of xcc0300, a gene that has protective roles against the toxicity of redox-cycling compounds. In addition, SoxR acts as a noninducible repressor of its own expression. X. campestris pv. campestris requires SoxR both for protection against redox-cycling drugs and for full virulence on a host plant. The X. campestris model of the gene regulation and physiological roles of SoxR represents a novel variant of existing bacterial SoxR models.
INTRODUCTION
SoxR, a homodimeric protein that belongs to the MerR family of transcriptional regulators, senses superoxide-generating compounds via the one-electron oxidation of its [2Fe-2S] cluster. In Escherichia coli, SoxR is thought to have a role in the sensing of superoxide stress. Its sole target is soxS, a transcriptional regulator of the AraC/XylS family, which it upregulates. The newly synthesized SoxS then induces the expressions of many genes in the regulon involved in superoxide stress protection and repair (7, 8, 46). However, a recent report has shown that E. coli SoxR directly senses redox-cycling drugs rather than superoxide anions (12). Additionally, other reactive radicals, such as reactive nitrogen species, guanine radicals, and pyocyanin, have been shown to activate SoxR (25, 28). Recent observations indicated that the E. coli SoxR paradigm does not apply for many bacteria. In the plant-pathogenic bacterium Agrobacterium tumefaciens, SoxR directly binds to and activates the transcription of target genes upon exposure to superoxide anions. It is also involved in adaptive protection against superoxide stress through the regulation of sodBII, which encodes iron superoxide dismutase (SOD) (11). In Pseudomonas, SoxR directly binds to and alters the transcription of its target genes but has a limited role in the sensing of superoxide anions (9, 21). Moreover, genes in the pseudomonad SoxR regulon have only minor physiological roles in protection against superoxide stress, and their expressions are induced by phenazines, which are redox-active antibiotics (10, 21, 29, 31). For bacterial interactions with a host, soxR has been shown to be essential for virulence and pathogenicity in some bacteria (13, 19).
Xanthomonas campestris pv. campestris is a plant-pathogenic bacterium causing black rot in crucifers (45). The accumulation of reactive oxygen species (ROS), including superoxide anions, and the production of redox-active compounds are parts of the defensive response of plants to pathogenic microbes (23). The roles of SoxR in X. campestris pv. campestris as a stress sensor, transcriptional regulator, and virulence factor were evaluated. Our findings reveal a novel variation of the SoxR model for gene regulation and its physiological roles.
MATERIALS AND METHODS
Bacterial growth conditions.
The bacterial strains used in this study are listed in Table 1. Xanthomonas strains were grown aerobically in Silva-Buddenhagen (SB) medium (5) at 28°C with continuous shaking at 150 rpm. Routinely, a culture grown overnight was inoculated into SB medium to give an optical density at 600 nm (OD600) of about 0.1. Exponential-phase (OD600 of about 0.5, after 4 h of growth) cells were used in all experiments, unless stated otherwise. The oxidant induction experiments were conducted with cells treated for 30 min with 100 μM menadione (MD), paraquat (PQ), cumene hydroperoxide (CHP), or hydrogen peroxide (H2O2).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or characteristic(s) | Source |
|---|---|---|
| Strains | ||
| X. campestris pv. campestris | ||
| Wild type | X. campestris pv. campestris ATCC 33913 | 6 |
| soxR mutant | ATCC 33913 soxR::pKNOCK-Km; Kmr | This study |
| xcc0300 mutant | ATCC 33913 xcc0300::pKNOCK-Km; Kmr | S. Mongkolsuk et al., unpublished data |
| E. coli BL21(DE3) | F−dcmompT hsdSB(rB− mB−) gal λ (DE3) | Novagen |
| Plasmids | ||
| pKNOCK | Suicide vector; RP4 oriT R6K γ-ori | 1 |
| pBBR1MCS-4 | Broad-host-range cloning vector; rep mob lacZα Apr | 22 |
| pBBR1MCS-5 | Broad-host-range cloning vector; rep mob lacZα Gmr | 22 |
| pSoxR | pBBR1MCS-5 carrying X. campestris pv. campestris soxR | This study |
| pSodBI | pBBR1MCS-4 carrying A. tumefacienssodBI | 37 |
| pXcc0300 | pBBR1MCS-4 carrying X. campestris pv. campestris xcc0300 | This study |
| pDrive | A-T cloning vector; Kmr Apr pUC origin T7 SP6 lacZα+ | Qiagen, Germany |
| pETBlue-2 | Expression vector; Apr pUC origin f1 origin T7 lacZα+ | Novagen |
Molecular biology techniques.
Common molecular genetic techniques, including genomic DNA, plasmid, and RNA preparations; restriction endonuclease digestion; DNA ligation; transformation of E. coli; gel electrophoresis; and blotting analysis, were performed by using standard protocols (39). The transformation of plasmid DNA into X. campestris pv. campestris was performed by electroporation under previously described conditions (32). DNA sequences were determined by using an ABI 310 automated DNA sequencer (Applied Biosystems). Plasmids used in this study are described in Table 1.
Purification of the SoxR protein.
The untagged SoxR protein was heterologously expressed in E. coli cells by using the pETBlue-2 expression vector (Novagen) as previously described (11). Full-length soxR was PCR amplified from X. campestris pv. campestris genomic DNA by using primers BT2690 (5′-GGCCATGGAGCGTGAGTTGT-3′) and BT556 (5′-CGCTCAGCCGCCGACAGT-3′) prior to cloning into NcoI/HincII-cut pETBlue-2, generating pETsoxR. A culture of E. coli BL21(DE3) cells harboring pETsoxR was induced with isopropyl-β-d-thiogalactopyranoside (IPTG), and SoxR in the crude lysate was purified aerobically through an equilibrated Whatman P-11 phosphocellulose column. The purified SoxR protein was eluted by using a step gradient of 0.2 to 1.0 M KCl. SoxR was detected by its reddish-brown color. Fractions containing the purified SoxR protein were analyzed by SDS-PAGE.
Construction of the soxR mutant and pSoxR.
The soxR mutant was constructed by gene inactivation mediated by the pKNOCK suicide plasmid. The soxR DNA fragment was amplified from X. campestris pv. campestris genomic DNA by using primers BT572 (5′-GCCGATACAGTCGGTGAG-3′) and BT573 (5′-CGGCGGATCGCGGTGATC-3′). The PCR product was ligated into the pDrive vector (Qiagen, Germany), and an EcoRI fragment was subcloned into pKNOCK-Km. The recombinant plasmid was then transformed into wild-type X. campestris pv. campestris cells, and clones were selected for kanamycin resistance. The mutation was verified by Southern blot analysis.
Plasmid pSoxR was constructed for the ectopic expression of soxR. The full-length gene was excised by first digesting pETsoxR with BglII and blunt ending the product by treatment with Klenow fragment. The blunt-ended product was then digested with HindIII, and the soxR fragment was cloned into broad-host-range plasmid pBBR1MCS-5 (22), generating pSoxR.
Primer extension.
Primer extension experiments were performed by using 32P-labeled primers BT571 (5′-CAACGCCACCATGCCCA-3′) for soxR and BT2740 (5′-CACGATAGAAGCGCAGGGTG-3′) for xcc0300. These primers were end labeled by using [γ-32P]ATP and T4 DNA kinase. The labeled primer was incubated with 10 μg of total DNase I-treated RNA at 65°C for 15 min and at 25°C for 5 min, after which Superscript III reverse transcriptase (RT) was added, and the reaction mixture was incubated at 55°C for 60 min. The extension products were analyzed on a sequencing gel (8% polyacrylamide–7 M urea) along with a DNA ladder.
Cloning of the soxR promoter and site-directed mutagenesis.
The putative soxR promoter region was amplified by using primers BT570 (5′-CGCTCGTAGAAATGCAAC-3′) and BT571 and X. campestris pv. campestris genomic DNA as the template. The 285-bp soxR promoter fragment was cloned into the pDrive vector (Qiagen, Germany), generating pDrivePsoxR. The nucleotide sequence was determined to ensure that no mutations had occurred.
Site-directed mutagenesis of the soxR promoter to change the putative SoxR box was performed by using a PCR-based method described previously (30). A mutagenic forward primer (BT3139 [5′-GGGGGTTAAAAAAGGTCAAGGCAATGC-3′]) was used with primer M13-reverse (5′-CAGGAAACAGCTATGACC-3′), and primer M13-forward (5′-GTAAAACGACGGCCAGTG-3′) was used with a mutagenic reverse primer (BT3140 [5′-TTTTTAACCCCCAGGCAGGATGCTGCG-3′]), in a PCR amplification reaction with pDrivePsoxR as the template. PCR products were cloned into SmaI-cut pUC18 prior to DNA sequencing.
Gel mobility shift assay.
Primer BT570 and 32P-labeled primer BT571 were used to amplify the putative soxR promoter by using pDrivePsoxR as a template. The PCR product (20 ng) was incubated with increasing concentrations of purified SoxR protein in 1× binding buffer (300 μg ml−1 bovine serum albumin [BSA], 12% glycerol, 10 mM KCl, 1 mM dithiothreitol [DTT], and 12 mM HEPES-NaOH buffer [pH 7.9]) at 30°C for 30 min. The migration differences of the protein-DNA complex and free probe were analyzed by native 5% polyacrylamide gel electrophoresis.
RT-PCR.
First-strand cDNA synthesis was carried out by using 2 μg of DNase-treated RNA, hexaoligonucleotide random primers, and RevertAid Moloney murine leukemia virus (M-MuLV) reverse transcriptase (Fermentas, Lithuania) according to the manufacturer's instructions. Endpoint RT-PCR was performed to determine the expression level of xcc0300 by using primers BT2679 (5′-GCGTGCATCTGGCCTTCAA-3′) and BT2680 (5′-CAGTCAGTTCGAGCACGGC-3′) for 25 cycles as follows: 95°C for 20 s, 55°C for 20 s, and 72°C for 30 s. In order to normalize cDNA samples, 16S rRNA was RT-PCR amplified by using primers BT2781 (5′-GCCCGCACAAGCGGTGGAG-3′) and BT2782 (5′-ACGTCATCCCCACCTTCCT-3′) and was used as an internal control. To measure the expression level of soxR in wild-type X. campestris pv. campestris, a soxR mutant, and a soxR complemented strain, the cDNA was PCR amplified with BT3650 (5′-CCAAGGTTGAGGTCAA-3′) and BT3651 (5′-GCAGCACATCACGC-3′) for 30 cycles, as follows: 95°C for 20 s, 55°C for 20 s, and 72°C for 30 s. The PCR products were analyzed by 1.8% agarose gel electrophoresis.
qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) was conducted to measure the transcript level of xcc0300 in wild-type X. campestris pv. campestris harboring either pBBR1MCS-4 or pSodBI, grown with or without induction with 100 μM MD for 15 min. First-strand cDNA synthesis was done as described above for RT-PCR. Real-time PCR was conducted by using 20 ng cDNA, a specific primer pair (BT2679 and BT2680 for xcc0300 and BT2781 and BT2782 for the 16S rRNA gene, which was used for normalization), and SYBR green PCRMasterMix (Applied Biosystems). Reaction mixtures were run on an Applied Biosystems StepOne Plus thermocycler for 40 cycles under the following conditions: a denaturation step at 95°C for 30 s, an annealing step at 55°C for 30 s, and an extension step at 72°C for 30 s. Relative expression levels were determined by using StepOne software v2.1 and expressed as the fold change in the expression level relative to the uninduced level. Experiments were repeated independently three times.
Determination of oxidant resistance levels.
The resistance levels of X. campestris pv. campestris strains were determined by using a plate sensitivity assay as previously described (34). The exponential-phase cultures were 10-fold serially diluted into fresh SB medium, and 10 μl of each dilution was spotted onto an SB agar plate containing the appropriate concentration of the oxidant. The plates were incubated at 28°C for 48 h before bacterial colonies were scored.
SOD activity assay.
The total superoxide dismutase (SOD) activity was monitored on the basis of the xanthine-xanthine oxidase-coupled reduction of cytochrome c (26). One unit of SOD activity refers to the amount of enzyme required to inhibit the rate of reduction of cytochrome c by 50%.
Virulence test for X. campestris pv. campestris strains.
The virulence of X. campestris pv. campestris strains was assessed by using the leaf-clipping method on a compatible host plant, Chinese radish (Raphanus sativus), as previously described (4). Briefly, a bacterial inoculum from a culture of a given test strain grown overnight was adjusted to a final OD600 of 1.0. Three leaves per plant were randomly inoculated with the tested strains by leaf clipping, and each bacterial strain was used to inoculate five leaves. The lengths of the lesions on the infected leaves were measured at 14 days postinoculation. The lower detection limit for lesion size was less than 1 mm. Experiments were performed in triplicate.
RESULTS AND DISCUSSION
X. campestris pv. campestris soxR.
Analysis of the genomic sequence of X. campestris pv. campestris ATCC 33913 (6) revealed xcc2831, an annotated coding sequence (CDS) with a high level of homology to SoxR. This CDS is located 708 bases upstream of the divergently transcribed xcc2832 gene, which encodes a major facilitator superfamily (MFS) transporter with an unassigned function. A putative SoxR box, 5′-CCTCAACCAAGGTTGAGG-3′, which has a high level of sequence identity to the E. coli SoxR box consensus sequence, 5′-CCTCAAGTTAACTTGAGG-3′ (17), was identified 3 bases upstream from the putative soxR initiation codon (ATG). The soxR genomic organization, in which soxR is located adjacent to a putative MFS gene, is conserved in all Xanthomonas spp. whose genomic sequences have been determined and possess soxR (6, 24, 27, 35, 38, 41–43). It is noteworthy that no putative soxR was identified in the genome sequence of X. albilineans, a xylem-limited sugarcane pathogen with a reduced genome (33). The deduced amino acid sequence of X. campestris pv. campestris SoxR shares 57.4%, 56.8%, and 56.8% identity with the Pseudomonas aeruginosa, E. coli, and A. tumefaciens SoxR proteins, respectively. All amino acid residues previously identified as important for SoxR function, including the DNA-binding and iron-sulfur cluster-binding (CX2CXCX5C motif) domains and the two residues R48 and W84 (which correspond to E. coli SoxR R55 and W91, residues that are involved in redox signaling activity), are conserved (44). This finding suggests that X. campestris pv. campestris SoxR probably has a role in the sensing of redox-active compounds and/or superoxide anions. A BLASTP algorithm search was used to search the genome of X. campestris pv. campestris with the E. coli SoxS sequence as the protein query. No X. campestris pv. campestris CDSs with significant sequence homology to SoxS were found.
SoxR binds directly to the binding site of the target gene xcc0300.
The absence of a soxS homolog in the X. campestris pv. campestris genome suggests that X. campestris SoxR functions differently from E. coli SoxR. However, it could function similarly to the P. aeruginosa and A. tumefaciens SoxR proteins, which interact directly with the binding sites of target genes. Analyses of the SoxR-binding sites from different bacteria revealed a high level of conservation (11, 17). Thus, the consensus sequence for an E. coli SoxR box (5′-CCTCAAGTTAACTTGAGG-3′) (17) was used to search the X. campestris pv. campestris genome (6) using the MAST program (2). Several regions with homology to the E. coli SoxR box were identified; however, these boxes were positioned in regions unlikely to be involved in the regulation of nearby genes. Nonetheless, one putative SoxR box (5′-CCTCAACCATGCTTTAGG-3′) located 38 bases upstream of the translation initiation codon (ATG) of xcc0300 was identified, and the position of the putative SoxR box suggested that xcc0300 could be regulated directly by SoxR. A gel mobility shift assay was performed by using purified SoxR and a 219-bp DNA fragment from the xcc0300 regulatory region. The untagged Xanthomonas SoxR protein was purified from a high-level-expression E. coli strain as described in Materials and Methods. The spectrum obtained from spectrophotometric analyses suggested that purified SoxR contains an oxidized iron-sulfur cluster (data not shown). Our results show that SoxR binds specifically to the xcc0300 regulatory DNA fragments (Fig. 1A). Excess cold probe (CP), but not unrelated DNA (UD), competed with the labeled probe in the SoxR-binding complexes. No binding complexes were detected when an unrelated protein (UP), bovine serum albumin (BSA), was added to the labeled probe (Fig. 1A). These findings support the hypothesis that oxidized X. campestris pv. campestris SoxR regulates its target gene by binding directly to a site upstream of its target gene.
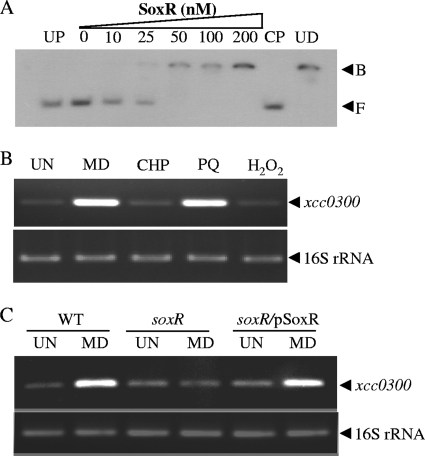
Fig 1.
Expression analysis of the xcc0300 gene. (A) A gel retardation assay was conducted by using purified SoxR protein and a 32P-labeled 219-bp xcc0300 promoter fragment. UP signifies an unrelated protein (1 μM BSA). CP and UD represent the cold probe (100 ng unlabeled promoter fragment) and unrelated DNA (1 μg plasmid pUC18), respectively, which were added to the binding reaction mixture along with 200 nM SoxR. F and B represent the free and bound probes, respectively. (B) xcc0300 transcript levels in X. campestris pv. campestris wild-type cells cultivated under uninduced (UN) or 100 μM MD-, PQ-, CHP-, or H2O2-induced conditions were measured by using endpoint RT-PCR. (C) xcc0300 transcript levels in X. campestris pv. campestris wild-type (WT), soxR mutant, and complemented soxR mutant (soxR/pSoxR) strains grown under UN or 100 μM MD-induced conditions were determined by using endpoint RT-PCR.
SoxR-regulated MD-induced transcription of xcc0300.
We extended our investigation by determining the patterns of xcc0300 expression in response to chemicals and stresses and establishing the role of SoxR in the regulation of stress-induced xcc0300 expression. X. campestris cultures were treated with 100 μM menadione (MD), paraquat (PQ), hydrogen peroxide (H2O2), or cumene hydroperoxide (CHP). MD and PQ are redox-cycling drugs that generate intracellular superoxide anions during aerobic metabolism. Endpoint RT-PCR results clearly indicated that only MD and PQ treatments induced xcc0300 expression, while the other oxidants had no effect on expression (Fig. 1B). The pattern of xcc0300 expression resembles patterns of other bacterial SoxR-regulated genes that are strongly induced by redox-cycling drugs. A soxR mutant was constructed and used to test the role of SoxR in the MD-mediated induction of xcc0300. The basal expression levels of xcc0300 in wild-type, soxR mutant, and complemented soxR mutant (soxR/pSoxR) strains were not significantly different (Fig. 1C). However, the MD-mediated induction of xcc0300 expression seen for the wild-type strain was abolished in the soxR mutant (Fig. 1C). The wild-type pattern of MD-mediated induction of xcc0300 expression was restored in the complemented soxR strain (soxR/pSoxR) (Fig. 1C). Similar results were obtained when PQ was used instead of MD (data not shown).
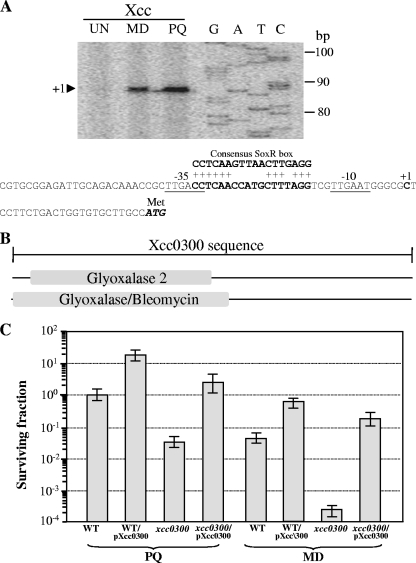
The proposed mechanism of the transcriptional activation of a target gene by SoxR involves the oxidation of the [2Fe-2S] cluster of SoxR and subsequent binding to the SoxR box (16). The SoxR-DNA complex then induces an alteration in the DNA conformation and aligns promoter elements that facilitate increased transcription by RNA polymerase (17). To determine the architecture of the xcc0300 promoter in relation to the SoxR box, total RNA samples were prepared from a wild-type culture induced with 100 μM MD or PQ and used in primer extension experiments. The 88-bp extension products were detected in both the MD- and PQ-induced samples (Fig. 2A). The 5′ end of xcc0300 corresponding to the transcription start site (position +1) was mapped to a C residue located 24 bases upstream of the putative ATG codon. The putative −35 and −10 regions were identified as TTGACC and TTGAAT, respectively, and were separated by 19 bp. The putative SoxR box (5′-CCTCAACCATGCTTTAGG-3′) was typically located between the −35 and −10 promoter motifs (14). The binding of oxidized SoxR to this region likely twists the DNA and alters the promoter structure to resemble a more favorable 17-bp spacing between the consensus promoter elements, which facilitates the binding of RNA polymerase and thus results in transcription activation.
Fig 2.
Characterization of the xcc0300 promoter and analysis of physiological roles of xcc0300. (A) Primer extension was performed to localize the 5′ ends of the xcc0300 transcripts. The primer extension products from a reaction mixture containing 32P-labeled primer BT2740 and 10 μg RNA extracted from wild-type X. campestris pv. campestris cells cultivated under uninduced (UN) or MD- or PQ-induced conditions were separated on a sequencing gel. A DNA ladder (G, A, T, and C) was prepared by using a sequencing kit with a labeled pUC/M13 forward primer and pGEM-3Zf as the template. Numbers to the left indicate DNA sizes in base pairs. An arrowhead represents the putative soxR transcription start site (position +1). The putative −10 and −35 elements of the xcc0300 promoter are underlined. The consensus E. coli SoxR-binding box is aligned above the xcc0300 promoter sequence, and the conserved residues are indicated by a plus sign. (B) Domain structure of the putative Xcc0300 protein. The 225-amino-acid sequence of Xcc0300 was analyzed by using the InterProScan algorithm (36). Glyoxalase/bleomycin represents the glyoxalase/bleomycin resistance protein/dihydroxybiphenyl dioxygenase superfamily. (C) The X. campestris pv. campestris wild-type strain harboring either the pBBR1MCS-4 vector control (WT) or pXcc0300 (WT/pXcc0300) and the xcc0300 mutant strain harboring either pBBR1MCS-4 (xcc0300) or pXcc0300 (xcc0300/pXcc0300) were grown to the exponential phase. A plate sensitivity assay was then performed by using SB agar plates containing either 1 mM PQ or 250 μM MD. The surviving fraction was calculated by dividing the number of CFU on plates containing an oxidant by the number of CFU on plates lacking an oxidant. Experiments were performed in triplicate, and the means ± standard deviations are shown.
When X. campestris is growing under physiological conditions (i.e., is uninduced), the reduced SoxR probably does not bind to the xcc0300 SoxR box and represses the expression of the gene. This speculation is supported by observations that neither the inactivation of soxR in the mutant nor the overexpression of soxR from pSoxR has much of an effect on uninduced xcc0300 expression levels. However, more experimental evidence is required to conclusively confirm this speculation. Upon exposure to MD, reduced SoxR becomes oxidized and subsequently binds to the SoxR box and activates the transcription of xcc0300.
SoxR senses redox-cycling drugs.
SoxR was proposed previously to sense different types of chemicals and stresses (7, 12). Originally, SoxR was thought to be a sensor of superoxide anions generated by redox-cycling drugs. Recent findings showed that SoxR directly senses the redox-cycling drugs and not the superoxide anions (12). The question then arises of whether the MD-induced SoxR-mediated expression of xcc0300 results from the oxidation of SoxR by superoxide radicals generated by MD from the redox-cycling reaction or from the direct oxidation of SoxR by MD. We hypothesized that if superoxide radicals are involved in the activation of SoxR, either the performance of the experiment anaerobically or the high-level expression of superoxide dismutase (SOD) (a superoxide-scavenging enzyme) should lower the magnitude of MD-induced xcc0300 expression. Similar rationales have been used with other bacteria to test whether SoxR senses either superoxide anions or the redox-cycling drugs themselves (12, 18). X. campestris is an obligate aerobe (3). We therefore performed induction experiments under anaerobic conditions using aerobically grown cells to test this hypothesis. While anaerobic conditions inhibited X. campestris growth for the duration of the experiment (up to 3 h), with less than a 10-fold decrease in cell viability, the data from anaerobic MD induction experiments monitoring xcc0300 transcription were inconclusive. Clearly, such conditions could not be compared to those of aerobically grown X. campestris cells. We therefore adopted a second rationale, positing that if superoxide levels are responsible for the SoxR-mediated induction of xcc0300, then an increased level of the superoxide scavenger superoxide dismutase should dampen the induction of xcc0300 transcription in response to MD. Experiments were repeated with a wild-type strain of X. campestris pv. campestris harboring pSodBI (37) to allow a high level of expression of Agrobacterium tumefaciens sodBI (an iron-containing SOD); this strain produced increased levels of SOD (10.1 ± 1.4 U mg−1 protein) compared to those of the wild-type strain (3.5 ± 0.1 U mg−1). The fold induction of xcc0300 transcription relative to the uninduced level was measured by using quantitative real-time RT-PCR. For X. campestris pv. campestris carrying pSodBI, treatment with 100 μM MD induced xcc0300 transcription by 153.6-fold ± 19.0-fold, which is comparable to the level of the wild-type strain carrying the pBBR1MCS control (166.4-fold ± 20.9-fold). These data suggest that superoxide radicals are unlikely to be responsible for the MD-induced SoxR-mediated expression of xcc0300. Thus, X. campestris pv. campestris SoxR likely senses the redox-cycling drug MD and not the superoxide anions generated by MD treatment.
xcc0300 has roles in protection against redox-cycling drugs.
InterProScan protein sequence analysis (36) of Xcc0300 indicated that the protein has domains homologous to proteins belonging to the glyoxalase 2 and glyoxalase/bleomycin resistance protein/dihydroxybiphenyl dioxygenase superfamilies (Fig. 2B). CDSs that share a high level of identity (>50%) with Xcc0300 were found in some xanthomonads (X. axonopodis, X. citri, and X. euvesicatoria) and in certain bacterial species, i.e., Stenotrophomonas maltophilia and Burkholderia sp. We tested the resistance of an xcc0300 mutant to redox-cycling drugs and oxidants and showed that the inactivation of this gene resulted in reduced resistance to MD and PQ but not to other oxidants tested, including H2O2 and CHP (Fig. 2C and data not shown). The phenotype could be complemented by the expression of xcc0300 in trans from an expression vector (Fig. 2C). At present, the biochemical mechanism of Xcc0300 and its direct physiological role in protecting bacteria against redox-cycling drug toxicities are not known; however, they are being further investigated for X. campestris pv. campestris.
The data regarding the physiological roles and regulation of expression of xcc0300 support the idea that X. campestris pv. campestris SoxR acts both as a sensor for redox-cycling drugs and as a transcriptional regulator. The oxidation of SoxR leads to the direct transcriptional activation of xcc0300, a gene involved in protection against redox-cycling drugs. A similar regulation of gene expression by SoxR has been observed for the alphaproteobacterium A. tumefaciens. In this bacterium, treatment with superoxide/redox-cycling drugs induces the SoxR-dependent activation of sodBII, which encodes Fe-SOD, and atu5152, a gene encoding an protein of unknown function (11, 37).
soxR expression is autoregulated.
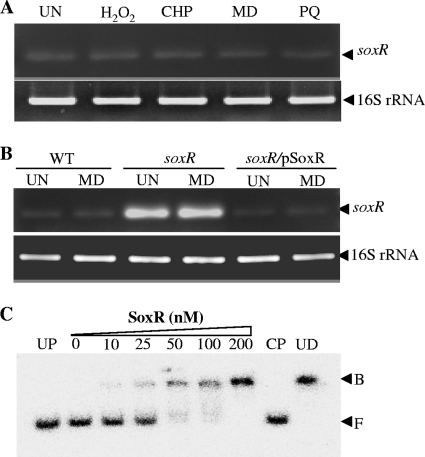
SoxR belongs to the MerR family of transcriptional regulators. The genes in this family typically regulate their own expressions. For several bacteria, soxR expression has been shown to be autoregulated and induced by superoxide generators (11, 13). Thus, the regulation of X. campestris pv. campestris soxR expression was investigated. Bacterial cultures were challenged with redox-cycling drugs and other oxidants. Total RNA was extracted from these cultures and used as a template for endpoint RT-PCR amplification using soxR-specific primers (BT3650 and BT3651). The results shown in Fig. 3A indicated that the treatment of the bacterial cultures with either redox-cycling drugs (MD and PQ) or peroxides (H2O2 and CHP) did not induce soxR expression. Unlike the expression of the soxR genes from A. tumefaciens and P. aeruginosa, X. campestris pv. campestris soxR expression could not be induced by the exposure of bacterial cultures to redox-cycling drugs (11, 13). This raises the question of whether X. campestris soxR is autoregulated. Thus, the level of soxR transcription was determined by using endpoint RT-PCR with three strains: the wild type, a soxR mutant, and a soxR complemented strain. Primers BT3650 and BT3651 were used in RT-PCRs to measure the levels of the 5′ ends of the soxR transcripts. As illustrated in Fig. 3B, the uninduced level of soxR transcripts was considerably higher (25-fold, based on densitometric analysis) for the soxR mutant than for the wild-type strain. The increased level of expression of soxR in the soxR mutant could be reduced to the wild-type level by complementation with pSoxR (a functional copy of soxR in an expression vector) in the soxR/pSoxR strain. Furthermore, the treatment of the bacterial cultures with MD did not alter the patterns or levels of soxR expression in these strains. These data indicate that reduced and oxidized SoxR functions as a transcriptional repressor of its own gene. This feature was also seen in a previous study regarding the regulation of E. coli SoxR (17).
Fig 3.
Expression analysis of soxR. (A) soxR transcript levels in the X. campestris pv. campestris wild-type strain were determined under uninducing (UN) conditions or following induction with 100 μM H2O2, CHP, PQ, or MD using endpoint RT-PCR. (B) soxR transcript levels in X. campestris pv. campestris wild-type, soxR mutant, and soxR/pSoxR strains cultivated under UN conditions or following MD induction were determined by using endpoint RT-PCR. (C) A gel retardation assay was conducted by using purified SoxR protein and a 32P-labeled 285-bp soxR promoter fragment. CP and UD represent the cold probe and unrelated DNA, respectively. F and B indicate the free and bound probes, respectively.
Characterization of the soxR promoter.
The ability of SoxR to bind to the putative SoxR box located at the 5′ end of soxR was investigated by using a gel mobility shift assay. The 285-bp soxR promoter fragment spanning the putative SoxR box was PCR amplified. Binding reactions were performed by using increasing concentrations of purified SoxR (mostly in the oxidized form) and 32P-labeled promoter region fragments. The results showed that the SoxR promoter-binding complexes could be detected at a SoxR concentration of 10 nM. The ability of the CP, but not UD, to compete with the labeled probe in a binding complex and the lack of binding of a UP to the probe indicated the probe's in vitro binding specificity for purified SoxR (Fig. 3C) and showed that oxidized SoxR could bind to the SoxR box located upstream of the gene. Next, the contribution of the sequence of the putative SoxR box to the binding of SoxR was assessed. PCR-based site-directed mutagenesis was performed to change the inverted repeat of the putative SoxR box by replacing 5′-CCTCAACC—GGTTGAGG-3′ with 5′-CCTGGGGG—AAAAAAGG-3′. The binding reaction was then repeated with the mutated SoxR box. No binding complex including purified SoxR was detected, even at high concentrations of the SoxR protein (data not shown). This finding confirms that the X. campestris SoxR box is required for the binding of the SoxR regulator.
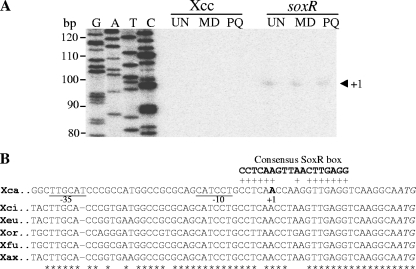
The promoter architecture of a SoxR-regulated gene plays an important role in the function of SoxR as a transcription regulator (15). Therefore, we determined the sequences of the 5′ ends of soxR transcripts using primer extension experiments. Total RNA samples were extracted from cultures of wild-type X. campestris pv. campestris and the soxR mutant strain grown with or without exposure to redox-cycling drugs (MD or PQ). Primer extension experiments were performed by using 32P-labeled primer BT573. The 98-bp primer extension products were observed only for RNA samples from the soxR mutant (Fig. 4A). These results are consistent with those from RT-PCR in that the soxR expression level in the mutant was constitutively high. The transcriptional start site (position +1) was mapped to an A residue located 20 bases upstream of a putative soxR ATG translation start codon (Fig. 4). Analysis of the soxR promoter showed TTGCAT and CATCCT motifs corresponding to −35 and −10 promoter elements separated by 18 bp (Fig. 4B). These two motifs are analogous to the X. campestris consensus σ70 promoter sequences (TTGTNN for the −35 element and [T/A]ATNA[A/T] for the −10 element) (20). The putative SoxR-binding site, 5′-CCTCAACCAAGGTTGAGG-3′, is located near the +1 site (Fig. 4B). The position of this SoxR box is unusual in comparison with those of all other known SoxR-regulated genes. Generally, the SoxR box lies between the −35 and −10 promoter motifs (11, 29).
Fig 4.
Characterization of the soxR promoter. (A) Primer extension was performed to localize the 5′ end of the soxR transcripts. Total RNA was prepared from the wild-type or soxR mutant X. campestris pv. campestris strain grown under uninduced (UN) or MD- or PQ-induced conditions. Reverse transcription was then performed with 10 μg of this RNA using 32P-labeled primer BT571, and the products were separated on a sequencing gel. A DNA ladder (G, A, T, and C) was prepared by using a sequencing kit with labeled pUC/M13 forward primer and pGEM-3Zf as the template. Numbers to the left indicate DNA sizes in base pairs. An arrowhead represents the putative soxR transcription start site (position +1). (B) Alignment of putative soxR promoter sequences from X. campestris pv. campestris with the promoter sequences from X. citri (Xci), X. euvesicatoria (Xeu), X. oryzae pv. oryzae (Xor), X. axonopodis pv. citrumelo (Xax), and X. fuscans subsp. aurantifolii (Xfu). The transcription start site (position +1) and putative −10 and −35 elements of the X. campestris pv. campestris soxR promoter are in boldface type and underlined, respectively. The consensus E. coli SoxR-binding box is aligned above the X. campestris pv. campestris promoter sequences, and the conserved residues are indicated by a plus sign. The conserved bases among all promoter sequences are indicated by asterisks.
SoxR senses redox-cycling drugs via the oxidation of its iron-sulfur cluster, which results in a protein conformational change (16). Typically, the binding of reduced SoxR hinders the binding of RNA polymerase, thereby repressing transcription, while the binding of oxidized SoxR to a target promoter aligns the promoter structures that facilitate RNA polymerase binding, thus leading to transcription initiation. For X. campestris pv. campestris, data from expression analysis of the mutant, gel mobility shift assays, and primer extension experiments suggest that both reduced and oxidized forms of SoxR bind to the target site located near the gene transcription initiation site and occlude the binding of RNA polymerase to the promoter. This accounts for the observed low levels of noninducible expression of soxR. While similar to E. coli in this respect, X. campestris pv. campestris is nevertheless different from other bacteria characterized thus far, in which superoxide anions/redox-cycling drugs induce soxR expression (11, 13).
Further analysis of putative soxR promoters from publicly available completed Xanthomonas genome sequences, i.e., X. citri (formerly X. axonopodis pv. citri) (6), X. euvesicatoria (formerly X. campestris pv. vesicatoria) (42), X. oryzae pv. oryzae (38), X. axonopodis pv. citrumelo (GenBank accession number CP002914), and X. fuscans subsp. aurantifolii (27), indicated that the atypical regulation of soxR observed for X. campestris pv. campestris is not a strain-specific phenomenon. The data show that the putative SoxR-binding sites in all Xanthomonas spp. are located in the same positions as that in X. campestris pv. campestris (Fig. 4B). We therefore assessed the expression profiles of soxR in response to treatment with MD in these Xanthomonas species using endpoint RT-PCR. In all of the strains tested, soxR expression was not inducible by MD treatment, suggesting that this pattern of soxR expression is common among xanthomonads (data not shown).
Physiological roles of soxR in X. campestris pv. campestris.
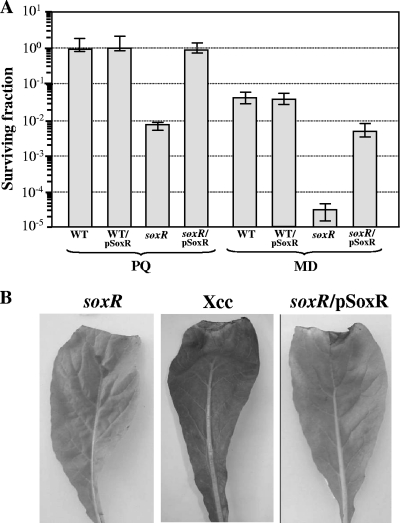
soxR has diverse physiological roles in different bacteria, including a role as an adaptive defense against superoxide stress/redox-cycling drugs. Therefore, the resistance of the soxR mutant to various oxidative stress-generating agents was determined by using a plate sensitivity assay. The mutant was >100-fold more sensitive to redox-cycling substances like MD (250 μM) and PQ (1 mM) than the parental strain (Fig. 5A). This altered phenotype can be complemented by the plasmid-borne expression of soxR from pSoxR. Meanwhile, no significant difference in resistance to organic hydroperoxides (CHP and t-butyl hydroperoxides) or H2O2 was observed between the soxR mutant and a wild-type strain (data not shown). Preliminary expression analyses of sod genes (xcc0190, xcc0191, xcc0395, xcc2278, and xcc2501) in X. campestris pv. campestris indicated that SoxR is not involved in the regulation of these genes (P. Vattanaviboon et al., unpublished data). Furthermore, the total SOD activity of the soxR mutant (3.4 ± 0.5 U mg−1 protein) was not significantly different from that of the isogenic wild-type strain (3.5 ± 0.2 U mg−1 protein). Exposure to 100 μM MD or PQ did not result in increased levels of total SOD activity in either the wild type or the soxR mutant strain (data not shown). Thus, the lower level of resistance of the soxR mutant to redox-cycling drugs is not due to an altered regulation of the sod genes. This further supports the idea that Xanthomonas SoxR has not evolved to sense and respond to superoxide anions. However, SoxR has physiological roles in protecting X. campestris pv. campestris from redox-cycling drugs, at least in part, via its regulation of xcc0300. X. campestris belongs to a group of soil and plant-pathogenic bacteria that are likely to encounter redox-cycling compounds both in the environment (e.g., generated by microbes and plants) and during interactions with host plants.
Fig 5.
Analysis of the physiological roles of soxR. (A) Determination of resistance to superoxide generators in X. campestris pv. campestris strains. A plate sensitivity assay was performed by using SB agar plates containing either 1 mM PQ or 250 μM MD. Exponential-phase cultures of the X. campestris pv. campestris wild-type strain harboring the pBBR1MCS-4 vector control (WT) or pSoxR (WT/pSoxR) and the soxR mutant strain harboring pBBR1MCS-4 (soxR) or pSoxR (soxR/pSoxR) were used in these experiments. The surviving fraction was calculating by dividing the number of CFU on plates containing an oxidant by the number CFU on plates lacking an oxidant. Experiments were performed in triplicate, and the means ± standard deviations are shown. (B) Virulence assay of X. campestris pv. campestris strains. The virulences of the X. campestris pv. campestris wild-type (WT), soxR mutant (soxR), and complemented (soxR/pSoxR) strains were determined by using the leaf-clipping method (4) on Chinese radish (Raphanus sativus), a compatible host plant. The length of the black rot lesion was then measured at 14 days postinoculation.
X. campestris pv. campestris causes black rot disease in a variety of cruciferous crops worldwide. The data presented here suggest that SoxR senses and responds to redox-cycling drugs. The roles of redox-active compounds in host-microbe interactions are not clear, but previous reports of P. aeruginosa revealed that soxR is important for the infection process (13, 29). The effect of soxR inactivation on the virulence of X. campestris pv. campestris in a susceptible host plant was evaluated. The soxR mutant, the isogenic wild-type strain, and the soxR mutant harboring plasmid pSoxR (soxR/pSoxR) were inoculated into Chinese radish (Raphanus sativus) leaves using a leaf-clipping method described previously (4). The length of the black rot lesion was measured at 14 days postinoculation. In all experiments, no lesions were detected in leaves treated with the soxR mutant, while treatment with X. campestris pv. campestris and the soxR-complemented strain (soxR/pSoxR) resulted in lesion lengths of 13.0 ± 1.0 mm and 11.7 ± 1.2 mm, respectively (Fig. 5B). Plants are known to produce many redox-cycling compounds, but the roles of these compounds in physiological processes have not been clearly established. During interactions with a host plant, bacteria probably encounter plant-secreted redox-cycling quinones such as plumbagin and juglone, which are toxic to the bacteria (40). These host-generated redox-cycling compounds could cross the bacterial membrane and react with the reactive centers or various intracellular components, either directly or indirectly, via the generation of superoxide anions in redox-cycling reactions, leading to growth inhibition and cell death. The SoxR system has evolved to sense the presence of redox-cycling compounds and to activate genes involved in defense against the toxicity of these compounds. Although the production and accumulation of superoxide anions associated with the plant host defense response to microbial invasion have been observed (23), due to its negative charge, extracellularly generated superoxide anions could not cross the cytoplasmic membrane and cause intracellular damage to the bacteria. However, extracellular superoxide anions could cause damage to the membrane and to reactive components residing in the periplasmic space. In X. campestris, SoxR has not evolved to sense superoxide anions. It is clear from the analysis of interactions of a soxR mutant with a host plant that the gene is required for full bacterial virulence. Thus, the ability to sense and respond to redox-cycling compounds is essential for the virulence of X. campestris pv. campestris in a radish host.
Conclusion.
The X. campestris pv. campestris SoxR model involves SoxR acting as a noninducible repressor of its own expression while having the ability to sense redox-cycling drugs such as MD and PQ. The oxidized form of SoxR binds directly to the promoter and upregulates the expression of xcc0300 (a gene implicated in protection against redox-cycling drugs). Physiologically, soxR has a role in protecting the bacteria from the toxicity of redox-cycling drugs, partly through its ability to upregulate xcc0300 in response to these drugs. We also showed that the ability to sense redox-active drugs is important for the bacteria because a soxR mutant is less virulent on a host plant. The autoregulation of soxR and the sensing of redox-cycling drugs share similarities with the E. coli model, while the direct binding and activation of a target gene have aspects in common with the P. aeruginosa and A. tumefaciens models. The X. campestris pv. campestris SoxR model of gene regulation and its physiological roles are a hybrid of the existing models and represent a novel variant.
ACKNOWLEDGMENTS
The research was supported by grants from the National Center for Genetic Engineering and Biotechnology (BTB-01-PG-14-5112), the Chulabhorn Research Institute, the Toray Science Foundation, and Mahidol University. A.M. was supported by a Golden Jubilee scholarship, PHD/0142/2548, from the Thailand Research Fund. S.C and P.N. were supported by scholarships from the Chulabhorn Graduate Institute.
Parts of this work are from the dissertation of A.M. submitted for a Ph.D. degree from Mahidol University.
Footnotes
Published ahead of print 4 November 2011
REFERENCES
- 1. Alexeyev MF. 1999. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques 26:824–826, 828 [DOI] [PubMed] [Google Scholar]
- 2. Bailey TL, Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54 [DOI] [PubMed] [Google Scholar]
- 3. Bergey D, Holt J. 1994. Bergey's manual of determinative bacteriology, 9th ed. William & Wilkins, Baltimore, MD. [Google Scholar]
- 4. Charoenlap N, et al. 2011. Evaluation of the virulence of Xanthomonas campestris pv. campestris mutant strains lacking functional genes in the OxyR regulon. Curr. Microbiol. 63:232–237 [DOI] [PubMed] [Google Scholar]
- 5. Chauvatcharin N, et al. 2005. Genetic and physiological analysis of the major OxyR-regulated katA from Xanthomonas campestris pv. phaseoli. Microbiology 151:597–605 [DOI] [PubMed] [Google Scholar]
- 6. da Silva AC, et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459–463 [DOI] [PubMed] [Google Scholar]
- 7. Demple B, Ding H, Jorgensen M. 2002. Escherichia coli SoxR protein: sensor/transducer of oxidative stress and nitric oxide. Methods Enzymol. 348:355–364 [DOI] [PubMed] [Google Scholar]
- 8. Demple B, Hidalgo E, Ding H. 1999. Transcriptional regulation via redox-sensitive iron-sulphur centres in an oxidative stress response. Biochem. Soc. Symp. 64:119–128 [PubMed] [Google Scholar]
- 9. Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol. Microbiol. 61:1308–1321 [DOI] [PubMed] [Google Scholar]
- 10. Dietrich LE, Teal TK, Price-Whelan A, Newman DK. 2008. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321:1203–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eiamphungporn W, Charoenlap N, Vattanaviboon P, Mongkolsuk S. 2006. Agrobacterium tumefaciens soxR is involved in superoxide stress protection and also directly regulates superoxide-inducible expression of itself and a target gene. J. Bacteriol. 188:8669–8673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gu M, Imlay JA. 2011. The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Mol. Microbiol. 79:1136–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ha U, Jin S. 1999. Expression of the soxR gene of Pseudomonas aeruginosa is inducible during infection of burn wounds in mice and is required to cause efficient bacteremia. Infect. Immun. 67:5324–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hidalgo E, Demple B. 1994. An iron-sulfur center essential for transcriptional activation by the redox-sensing SoxR protein. EMBO J. 13:138–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hidalgo E, Demple B. 1997. Spacing of promoter elements regulates the basal expression of the soxS gene and converts SoxR from a transcriptional activator into a repressor. EMBO J. 16:1056–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hidalgo E, Ding H, Demple B. 1997. Redox signal transduction via iron-sulfur clusters in the SoxR transcription activator. Trends Biochem. Sci. 22:207–210 [DOI] [PubMed] [Google Scholar]
- 17. Hidalgo E, Leautaud V, Demple B. 1998. The redox-regulated SoxR protein acts from a single DNA site as a repressor and an allosteric activator. EMBO J. 17:2629–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Imlay J, Gu M. 2011. Many plants and bacteria excrete redox-cycling compounds. Free Radic. Biol. Med. 50:1814–1815 [DOI] [PubMed] [Google Scholar]
- 19. Kang IH, Kim JS, Lee JK. 2007. The virulence of Vibrio vulnificus is affected by the cellular level of superoxide dismutase activity. J. Microbiol. Biotechnol. 17:1399–1402 [PubMed] [Google Scholar]
- 20. Katzen F, Becker A, Zorreguieta A, Puhler A, Ielpi L. 1996. Promoter analysis of the Xanthomonas campestris pv. campestris gum operon directing biosynthesis of the xanthan polysaccharide. J. Bacteriol. 178:4313–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kobayashi K, Tagawa S. 2004. Activation of SoxR-dependent transcription in Pseudomonas aeruginosa. J. Biochem. 136:607–615 [DOI] [PubMed] [Google Scholar]
- 22. Kovach ME, et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 23. Lamb C, Dixon RA. 1997. The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:251–275 [DOI] [PubMed] [Google Scholar]
- 24. Lee BM, et al. 2005. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 33:577–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee PE, Demple B, Barton JK. 2009. DNA-mediated redox signaling for transcriptional activation of SoxR. Proc. Natl. Acad. Sci. U. S. A. 106:13164–13168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McCord JM, Fridovich I. 1969. Superoxide dismutase: an enzymatic function for erythrocuprein. J. Biol. Chem. 244:6049–6055 [PubMed] [Google Scholar]
- 27. Moreira LM, et al. 2010. Novel insights into the genomic basis of citrus canker based on the genome sequences of two strains of Xanthomonas fuscans subsp. aurantifolii. BMC Genomics 11:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, Storz G. 2004. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc. Natl. Acad. Sci. U. S. A. 101:745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palma M, et al. 2005. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect. Immun. 73:2958–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Panmanee W, et al. 2002. OhrR, a transcription repressor that senses and responds to changes in organic peroxide levels in Xanthomonas campestris pv. phaseoli. Mol. Microbiol. 45:1647–1654 [DOI] [PubMed] [Google Scholar]
- 31. Park W, Pena-Llopis S, Lee Y, Demple B. 2006. Regulation of superoxide stress in Pseudomonas putida KT2440 is different from the SoxR paradigm in Escherichia coli. Biochem. Biophys. Res. Commun. 341:51–56 [DOI] [PubMed] [Google Scholar]
- 32. Patikarnmonthon N, et al. 2010. Copper ions potentiate organic hydroperoxide and hydrogen peroxide toxicity through different mechanisms in Xanthomonas campestris pv. campestris. FEMS Microbiol. Lett. 313:75–80 [DOI] [PubMed] [Google Scholar]
- 33. Pieretti I, et al. 2009. The complete genome sequence of Xanthomonas albilineans provides new insights into the reductive genome evolution of the xylem-limited Xanthomonadaceae. BMC Genomics 10:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Prapagdee B, Vattanaviboon P, Mongkolsuk S. 2004. The role of a bifunctional catalase-peroxidase KatA in protection of Agrobacterium tumefaciens from menadione toxicity. FEMS Microbiol. Lett. 232:217–223 [DOI] [PubMed] [Google Scholar]
- 35. Qian W, et al. 2005. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 15:757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Quevillon E, et al. 2005. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Saenkham P, Eiamphungporn W, Farrand SK, Vattanaviboon P, Mongkolsuk S. 2007. Multiple superoxide dismutases in Agrobacterium tumefaciens: functional analysis, gene regulation, and influence on tumorigenesis. J. Bacteriol. 189:8807–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Salzberg SL, et al. 2008. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics 9:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 40. Soballe B, Poole RK. 1999. Microbial ubiquinones: multiple roles in respiration, gene regulation and oxidative stress management. Microbiology 145:1817–1830 [DOI] [PubMed] [Google Scholar]
- 41. Studholme DJ, et al. 2010. Genome-wide sequencing data reveals virulence factors implicated in banana Xanthomonas wilt. FEMS Microbiol. Lett. 310:182–192 [DOI] [PubMed] [Google Scholar]
- 42. Thieme F, et al. 2005. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J. Bacteriol. 187:7254–7266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vorholter FJ, et al. 2008. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 134:33–45 [DOI] [PubMed] [Google Scholar]
- 44. Watanabe S, Kita A, Kobayashi K, Miki K. 2008. Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA. Proc. Natl. Acad. Sci. U. S. A. 105:4121–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Williams PH. 1980. Black rot: a continuing threat to world crucifers. Plant Dis. 64:736–742 [Google Scholar]
- 46. Wu J, Weiss B. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]