Abstract
There are two closely related hik31 operons involved in signal transduction on the chromosome and the pSYSX plasmid in the cyanobacterium Synechocystis sp. strain PCC 6803. We studied the growth, cell morphology, and gene expression in operon and hik mutants for both copies, under different growth conditions, to examine whether the duplicated copies have the same or different functions and gene targets and whether they are similarly regulated. Phenotype analysis suggested that both operons regulated common and separate targets in the light and the dark. The chromosomal operon was involved in the negative control of autotrophic events, whereas the plasmid operon was involved in the positive control of heterotrophic events. Both the plasmid and double operon mutant cells were larger and had division defects. The growth data also showed a regulatory role for the chromosomal hik gene under high-CO2 conditions and the plasmid operon under low-O2 conditions. Metal stress experiments indicated a role for the chromosomal hik gene and operon in mediating Zn and Cd tolerance, the plasmid operon in Co tolerance, and the chromosomal operon and plasmid hik gene in Ni tolerance. We conclude that both operons are differentially and temporally regulated. We suggest that the chromosomal operon is the primarily expressed copy and the plasmid operon acts as a backup to maintain appropriate gene dosages. Both operons share an integrated regulatory relationship and are induced in high light, in glucose, and in active cell growth. Additionally, the plasmid operon is induced in the dark with or without glucose.
INTRODUCTION
Bacteria use several devices to monitor their environment and coordinate appropriate adaptive changes to maximize survival. These include chemotaxis receptors, sigma factors, Ser/Thr protein kinases, and two-component systems (2CSs) (25). The prototypical 2CS consists of a histidine kinase (Hik) sensor that is a transmembrane protein and a response regulator (Rre) that usually binds to DNA and acts as a transcription factor, either activating or repressing the target genes or both. Each protein has two or more domains that perform the various functions and participate in phosphotransfer reactions and can be classified into different types (3, 7, 8, 42). Higher-order 2CSs can have a more complex interaction with combinations of domains and cross talk between different partner 2CSs (7).
Signal transduction systems in the freshwater model cyanobacterium Synechocystis sp. strain PCC 6803 (here Synechocystis) are important for sensing, responding to, and adapting to different environmental changes. The Synechocystis genome includes about 47 Hik proteins and 45 Rre proteins, and these make up >2.5% of the genome. Although most of these are located on the chromosome, 3 each of the Hik proteins and Rre proteins are found on plasmids pSYSX and pSYSM. Unlike in other bacteria, the positions of these genes are scattered throughout the genome, and only 14 sets or 32 open reading frames are in close proximity to each other. The domains of these 2CS proteins have been compiled, and the functions of some of them have been determined but the partners for many Hik proteins have not been identified. The previously studied 2CSs have involved the chromosomally located genes in Synechocystis (22, 27, 29, 30, 40). Studies of 2CS genes on plasmids or duplicated regulatory genes in cyanobacteria have not been undertaken.
This study was concerned with the closely related hik31 operons (sll0788 to sll0790) on the chromosome (C3) and on the plasmid (P3) pSYSX (slr6039 to slr6041). They each contain a histidine kinase sensor (Hik), a response regulator (Rre), and a hypothetical protein (Hypo) in the same order, hinting at segmental duplications. The Hik proteins and the Rre proteins are about 96% identical in sequence at the protein level, and the Hypos are 99% identical. The Hypos have two DUF305 (domain of unknown function) domains that contain a double histidine (HH) motif that is presumed to be functionally important. Some of these proteins in other bacteria can bind to Fe2+, Zn2+, ethanediol, and Cl− (33). This is the only duplicated 2CS present in Synechocystis and the only cluster to be present on both the chromosome and the plasmid among the cyanobacteria that have been sequenced. The plasmid localization of this 2CS gene duplication is unusual, and to our knowledge, this is the only such system studied in bacteria.
The presence of two copies of this gene cluster raises interesting questions as to their function and the selective pressures that allow both copies to be stably maintained in the same cell. There are four main outcomes arising from gene duplications that explain the fate of duplicated genes—nonfunctionalization or loss of the duplicate, subfunctionalization to retain complementary functions, neofunctionalization to develop new functions, and differential regulation of duplicates similar in sequence and function (19). We consider the last possibility the most likely for the hik31 2CSs, since the upstream regions of the hik31 operons are only identical for 72 bp and there are many differences further upstream. Such an arrangement might enable coordinated, as well as differential, regulation of both operons. Several studies conclude that changes in regulation are more common than biochemical changes and that nonessential duplicate genes are retained by the cell. The duplication and location of the plasmid copy may also ease selection pressure, allowing gradual diversity in function for the two copies (20, 26). Varied environmental conditions could enable retention by altering gene dosages and lead to nonredundant network interactions with other proteins (15, 21).
These genes were first identified because they were actively expressed under several growth and stress conditions in microarray studies. The genes were upregulated under salt and osmotic stress (13), redox stress with DBMIB (10), pH 10 (35, 37), in the dark (38), and downregulated under heat shock (39), peroxide stress (14), and stationary phase (A. K. Singh and L. A. Sherman, unpublished data) in the wild type (WT) and specific mutants. The arrays for these experiments only had the chromosomal gene probes; however, once the plasmids were sequenced, and with the realization that the two operons were nearly identical, it was not known which copy was expressed. In one experiment with WT cells and the new Agilent microarrays that contained probes for both operon gene copies, it was determined that growth under low-O2 conditions (36; T. C. Summerfield and L. A. Sherman, unpublished data) led to the transcription of all 6 genes in the dark but only the chromosomal genes in the light. Thus, there may be temporal regulation of these genes with one of the copies sufficient in light but both copies needed in the dark for increased protein dosage. A putative double hik31 mutant has been implicated in the response to glucose (11). Also, a chromosomal Δhik31 mutant has implicated Hik31 as a transcriptional repressor under low-O2 conditions affecting photosynthetic and ribosomal genes (36). Additionally, both sets of operons are close to genes predicted to be Zn2+/Co2+ cation transporters, suggesting that these 2CSs may control the transduction of cation signals (12).
The presence of the Hypo closely associated with the 2CS genes indicates that it could be involved in signal transduction as an extra nonkinase receptor activating the Hik, making the operon a three-component system (3CS). Topology and domain predictions (using the SMART domain prediction tool and TOPCONS) indicate that both the DUF305 domains of the hypothetical protein and the N-terminal part of Hik31 are periplasmic. 3CSs are rare, and they enhance sensitivity, range of signal detection, and adaptability for the cell (4). Domain analysis of the hik31 operons against the well-studied EnvZ/OmpR proteins in Escherichia coli indicated that the operon arrangement and length of the proteins, domains, and secondary structures, as well as the topology, were very similar. It is thought that EnvZ, which does not have a defined sensor region, may sense osmotic changes indirectly through interactions with other integral membrane proteins or monitor membrane tension (42). Since neither Hik has domains for detecting light, O2, redox, or metals, it is possible that Hik31 may sense changes in a similar manner or through interactions with the Hypo.
The occurrence of two copies of the Hik and Rre raises questions of how specificity is maintained in phosphotransfer for both cognate sets of proteins. Both Hik31 proteins are identical in their functional domains and differ mainly in the β7 sheet and beyond by 19 residues in the C termini of the proteins. The Rre proteins differ in the receiver α1 region by 1 residue, in the output domain between β1 and β4 by 7 residues, and in the recognition helix by 1 residue. The α1 residue has also been shown to be a coevolving residue for the EnvZ-OmpR pair that affects specificity for the 2CS (5). Taken together, these changes may affect the way that both receiver and output parts of the Rre protein fold together so that the Hik proteins can bind selectively or in a hierarchical manner to transfer the phosphate (∼P) to their own partner Rre first. Moreover, homologs for the hik31 operon genes exist in 14 other cyanobacteria with these genes conserved and unique to cyanobaceria. Many of these homologs have chromosomal duplicates only, some do not contain the Hypo, and only two of them have plasmid copies.
In this study, we have explored the role of both hik31 operons in metabolism and tested for functions in response to different growth conditions using mutants in both copies. We examined the growth, ultrastructural properties, and gene expression to environmental parameters involving light, dark, and carbon sources. Our results indicate that both operons are involved in common and separate functions, are temporally and differentially regulated, and also share an integrated regulatory relationship.
MATERIALS AND METHODS
Cyanobacterial strains and growth conditions.
WT glucose-tolerant Synechocystis sp. strain PCC 6803 and mutants thereof were grown in BG-11 medium at 30°C under different conditions of light, dark, air composition, and metal salts. During the course of these studies, cells were grown under photoautotrophic (PA), mixotrophic (MT), and heterotrophic (HT) conditions with different durations of light and dark (LD) and in high continuous light (HL) for 6 days. High CO2 (HC) growth was performed in continuous light (LL) both with and without glucose for 2 days. Light intensities of 30 to 200 μE m−2 s−1 were used for different growth conditions, and the medium was supplemented with 5 mM glucose for MT and HT growth. Liquid cultures were grown in BG-11 medium buffered with 25 to 40 mM HEPES-NaOH (pH 7.5) in 250-ml Erlenmeyer flasks. Low-O2 growth experiments were performed with a 6-liter bioreactor (BioFlo 3000), and high-CO2 experiments were performed with 750-ml Cytolifts (Kontes, Inc.) (36). Spectinomycin and kanamycin (25 μg/ml) and chloramphenicol (10 μg/ml) were added to the medium for specific mutant strains. Growth was assayed by both absorbance at 730 nm (Perkin-Elmer UV-Vis λ 40 spectrophotometer) and cell counts in a Petroff-Hausser counting chamber. Doubling times were calculated for cultures grown for 3 days under PA, MT, and HT conditions and for 2 days under HC and low-O2 conditions. Growth on solid medium was performed in duplicate by spotting 5 μl of 4-fold serial dilutions on BG-11 plates containing appropriate antibiotics and 5 mM glucose as needed. Plates were incubated for 8 to 11 days in LL, 12L/12D, and 6L/18D with and without glucose and exposed to both 30 and 60 μmol photons/m2/s of light. Light-activated HT growth (LAHG) was tested on plates with 5 mM glucose exposed to 15 min of light (60 μmol photons/m2/s) per day for 15 days (31). These plates were maintained in the dark otherwise. For the metal tolerance experiment, 20 μM NiCl2 and 10 μM (each) CoCl2, ZnCl2, and CdCl2 were used. Spectral analysis of pigment composition was estimated from whole-cell absorption values (17). Statistical analysis of Table 1 doubling times was carried out through Tukey's test of comparison using SAS software and a significance level of α = 0.05 in collaboration with Zhuo Chen and Thomas Kuczek of the Statistical Consulting Service at Purdue University.
TABLE 1.
Growth properties of the WT strain and the operon and hik mutants under different trophic conditions of light, dark, and carbon
| Strain | Doubling time (h)a |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PA |
MTd |
HT, 0L/24Dd | HCe |
||||||||
| 24L/0D | 12L/12D | 6L/18D | 24L/0D HLb | 24L/0D | 12L/12D | 6L/18D | 24L/0D HLb | 24L/0D | 24L/0Dd | ||
| WT | 18.99 ± 0.03 | 40.87 ± 1.47 | 120.23 ± 9.53 | 35.62 ± 3.62 | 17.50 ± 0.19 | 16.73 ± 0.58 | 18.41 ± 0.06 | 13.72 ± 0.03 | 32.1 ± 2.28 | 14.47 ± 0.22 | 11.45 ± 0.17 |
| ΔC3 mutant | 19.00 ± 0.09 | 35.40 ± 0.87 | 88.18 ± 9.47 | 31.54 ± 1.13 | 19.20 ± 0.19f | 17.41 ± 0.32 | 19.01 ± 0.18 | 19.06 ± 0.70f | 29.91 ± 1.08 | 16.04 ± 0.34 | 10.05 ± 0.26 |
| ΔP3 mutant | 20.19 ± 0.20f | 54.12 ± 1.85 | 231.17 ± 4.45f | NGc,f | 16.70 ± 0.07 | 36.08 ± 0.97f | 37.82 ± 1.33f | 14.48 ± 0.05 | 47.50 ± 0.85f | 15.95 ± 0.63 | 14.68 ± 0.52f |
| ΔC3P3 mutant | 29.12 ± 0.07f | 85.02 ± 6.99f | 234.58 ± 11.03f | NGc,f | 24.10 ± 0.50f | 18.60 ± 0.69 | 21.64 ± 0.18f | 17.50 ± 0.04f | 50.99 ± 0.96f | 13.91 ± 0.53 | 13.29 ± 0.61 |
| ΔhikC mutant | 16.62 ± 0.08f | 55.34 ± 2.35 | 139.81 ± 19.94 | 38.78 ± 2.60 | 17.02 ± 0.02 | 17.71 ± 0.20 | 17.65 ± 0.19 | 13.58 ± 0.76 | 34.13 ± 0.87 | 15.80 ± 0.69 | 16.63 ± 0.39f |
| ΔhikP mutant | 19.45 ± 0.11 | 65.06 ± 3.90f | 140.99 ± 14.93 | 34.91 ± 1.89 | 17.80 ± 0.03 | 17.46 ± 0.14 | 18.87 ± 0.12 | 14.48 ± 0.10 | 30.70 ± 1.14 | 21.65 ± 2.38 | 10.22 ± 0.32 |
| ΔhikCP mutant | 19.30 ± 0.00 | 80.48 ± 6.54f | 139.70 ± 9.01 | 40.42 ± 3.89 | 19.20 ± 0.03f | 19.72 ± 0.32f | 18.48 ± 0.10 | 17.16 ± 0.25f | 28.96 ± 1.37 | 28.43 ± 4.36f | 15.82 ± 0.49f |
Doubling times of all strains are means ± standard errors of cell counts in a Petroff-Hausser counter for n = ≥3. Light at 30 μE m−2 s−1 was used for all conditions except high light (HL) and high CO2 (HC).
Light at 150 to 200 μE m−2 s−1 was used for HL growth.
NG, no growth.
Glucose was added to 5 mM.
A 3% CO2 concentration and light at 50 μE m−2 s−1 were used for HC growth. Bold font indicates a defect in pigment content.
α = 0.05, Tukey's comparison for each column separately using the WT as the standard.
Construction of mutants.
Six deletion mutants that involved hik31 were constructed. These included the entire hik31 operon located on the chromosome (the ΔC3 strain) or the plasmid (the ΔP3 strain), as well as both operons (the ΔC3P3 strain). In addition, hik31 genes were also deleted from the chromosome (ΔhikC strain), the plasmid (ΔhikP strain), and both positions (ΔhikCP strain). Synechocystis sp. strain PCC 6803 genomic DNA was used to amplify the plasmid operon (2,634 bp) along with flanking regions using the forward primer 5′-CTTCATGATGTGACTGTC-3′ and reverse primer 5′-ATGACAATGGTGCCATCG-3′ to yield a 4,524-bp PCR product that was cloned into the pGEM-T vector. Similarly, the chromosomal operon (2,658 bp) was amplified with forward primer 5′-CGGGATCCACTAACATGCTCTTGACAGACTCG-3′ and reverse primer 5′-CGGGATCCATCCCATTCCACTCATCCCATTGC-3′ containing a BamHI site (underlined) to yield a 4,180-bp PCR product that was then cloned into the pUC19 vector. Specific deletions were made using restriction sites in the coding region of the operons and the hik genes as outlined in Fig. 1A. The deleted portions were then replaced with various antibiotic resistance cassettes—spectinomycin (ΔC3 and ΔhikP strains), kanamycin (ΔP3 strain), and chloramphenicol (ΔhikC strain)—and used to transform WT Synechocystis sp. strain PCC 6803. The double mutants had different antibiotic resistance cassettes (spectinomycin and kanamycin for the ΔC3P3 strain and chloramphenicol and spectinomycin for ΔhikCP) replacing each deleted portion. Transformed colonies were selected on antibiotic plates and transferred over 2 to 4 months for segregation. Full segregation of the mutants was confirmed by PCR (Fig. 1B) and at regular intervals thereafter.
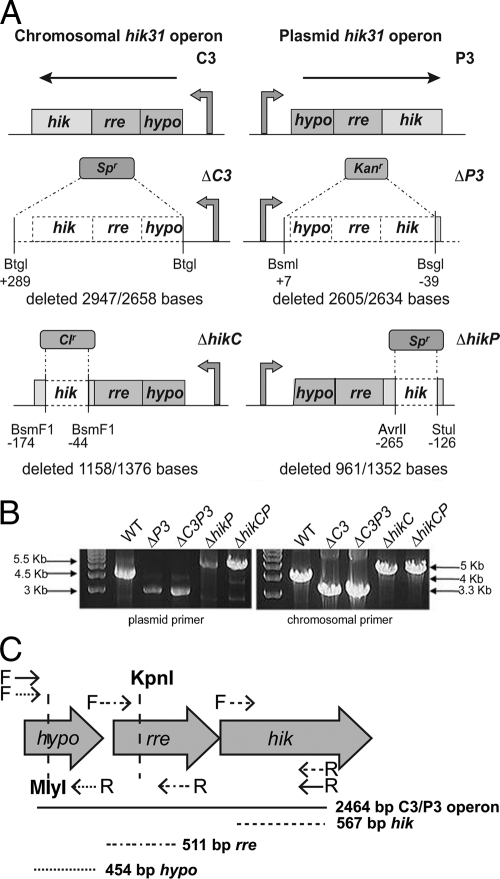
FIG 1.
Construction of the operon and hik mutants and separation of transcripts for the two operons. (A) Diagrammatic representation of the construction of hik31 mutants. We constructed deletion mutants that lacked all three genes in the operon on either the chromosome (the ΔC3 mutant) or the plasmid (the ΔP3 mutant) and on both the chromosome and the plasmid (the ΔC3P3 mutant). The dotted portions were deleted and replaced with various antibiotic resistance cassettes—spectinomycin (Spr), kanamycin (Kanr), and chloramphenicol (Clr). Similarly, we also made deletion mutants that lack hik31 alone on the chromosome (the ΔhikC mutant), on the plasmid (the ΔhikP mutant), and on both the chromosome and the plasmid (the ΔhikCP mutant). Shown are the portions of the genes that were replaced (dotted clear area), deleted extra (+), and left behind (−), as well as the restriction enzymes used. The promoter and the direction of transcription are indicated for each construct. (B) Gels showing PCR confirmation of the mutations and complete segregation. Separate primers were used to selectively amplify the chromosomal and plasmid copies. Sizes of the bands are indicated for the WT and mutants. (C) PCR primer and restriction digest design to differentiate the expression of the two operons. The restriction enzymes KpnI and MlyI cut at unique sites in the plasmid rre and hypo genes, respectively.
Cell morphology and electron microscopy.
Cell sizes and shapes were evaluated by light microscopy with a VWR Vista Vision camera. Cells for electron microscopy were prepared by microwave chemical fixation and sectioned, stained, and imaged in an FEI Philips CM-100 electron microscope as previously described (34). Cell sizes were measured by selecting cells in Photoshop and calculating the area on an Apple Macintosh computer using Ivision software.
RNA extraction and semiquantitative reverse transcription (RT)-PCR.
The experiments depicted in Fig. 6 and 7 were performed independently with different batches of WT and mutant cells. For monitoring of growth phase induction of the hik31 operon genes, WT cells were grown in PA 24L/0D for 6 days and cells from the end of each day were stored in STET buffer. For testing of induction of the hik31 operon genes by glucose in LL, WT cells were grown until about mid-log phase (30 h) and 5 mM glucose was added. Cells grown for 2, 6, 18, 24, and 48 h after the addition of glucose were stored in STET. For checking of induction of the hik31 operon genes by glucose under 12 L/12D conditions, WT cells were grown until about mid-log phase (30 h) and 5 mM glucose was added. Cells grown for 2 and 6 h in the light and in the dark after the addition of glucose were stored in STET. To evaluate transcription of the hik31 operon genes, the WT and mutants were grown for about 3 days in PA 24L/0D and 12L/12D, as well as in MT 24L/0D and 12L/12D; centrifuged at 8000 × g; and stored in STET buffer at −80°C. Cells were grown for 2 days under high-CO2 conditions before being stored as described above. For high-light experiments, cells were grown until about mid-log phase (26 h) at 30 μE m−2 s−1 and then exposed to 150 to 200 μE m−2 s−1 of light both with and without 5 mM glucose for 2 h before being treated as described above. Total RNA from two biological replicates for each condition was extracted and purified using Tri-reagent (Ambion). RNA was treated using DNase I (Invitrogen amplification grade) for 15 min and confirmed for reaction success through PCR. RNA was subsequently reverse transcribed using Superscript II (Invitrogen) and random primers. RT-PCR was then performed to amplify the transcripts of the C3 and P3 operons (2,464 bp), the rre C/P genes (511 bp) and the hypo C/P genes (454 bp) using the following common primers for both copies on the chromosome and the plasmid (shown in 5′-to-3′ orientation): C3/P3 operon, CAGCGGCTGGGGTAACAGCG (forward) and TGGCAAGGCCTAATCCTGCC (reverse); rre C/P, GGGTGCAGGACGGCAAACCTA (forward) and AAACGCACCTGGGCCGCTAC (reverse); hypo C/P, CAGCGGCTGGGGTAACAGCG (forward) and TCCATCTCCGGCCGTTCCGT (reverse). In order to measure the transcript levels of both individual operons, we used the scheme in Fig. 1C. Because the two operons are so similar, we could not use quantitative RT-PCR to analyze their individual transcription patterns and thus used the more qualitative RT-PCR. However, we took great care to identify appropriate conditions for amplification and separated the transcripts for both operons through restriction digests. The operon and rre amplicons were digested with KpnI at a unique restriction site in the plasmid rre gene to result in two fragments of 1,749 and 715 bp and 425 and 86 bp, respectively. The hypo transcripts were differentiated by digestion with MlyI, which cuts the plasmid hypo gene at a unique site to result in two fragments of 353 and 101 bp (Fig. 1C). These sites are not present on the chromosomal genes, and consequently, we could separate the chromosomal and plasmid transcripts for these genes. For simplicity, we have shown only the larger of the two digest pieces in the gel pictures in Fig. 6 and 7. We used sufficiently different primers (shown in 5′-to-3′ orientation) for both hik genes (567 bp) to tell their transcripts apart: hikC, GCTGGATCAAGAGCTTAC (forward) and GGTGTACGTAATTCGTGG (reverse); hikP, GCTGGATCAAGAATTAAC (forward) and GGTGTACGTAATTCGTGG (reverse). RT-PCR was performed at 94°C for 1 min and 30 to 40 cycles of 94°C for 15 s, 54°C for 30 s, and 68 to 72°C for 60 to 180 s, depending on amplicon size and abundance. The rnpB gene was used as a positive control as previously described (36).
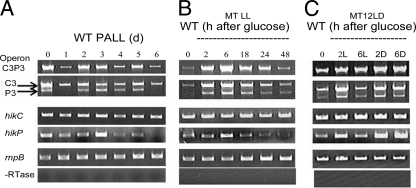
FIG 6.
Transcription of the operon and hik genes of both hik31 operons in the WT under various growth conditions. (A) Expression of the hik31 operon genes in different stages of growth. WT cells were grown under PA LL conditions for 6 days. RNA was extracted just before (t = 0) and after each day (days 1 to 6) of growth for RT-PCR. (B) Expression of the hik31 operon genes after addition of glucose in LL. WT cells were grown for about a day (30 h) under PA LL conditions, and then 5 mM glucose was added and the labeled time points (2, 6, 18, 24, and 48 h) were analyzed in LL. Dotted lines above sample times indicate those taken after the addition of glucose. The 30-h time point represents RNA from cells just before the addition of glucose (t = 0). (C) Expression of the hik31 operon genes after addition of glucose under LD conditions. WT cells were grown for about a day (30 h) under PA LL conditions, 5 mM glucose was added, and samples taken at the labeled time points (2L, 6L, 2D, and 6D) were analyzed in 12 h light, 12 h dark. Dotted lines above sample times indicate samples taken after the addition of glucose. The 30-h time point represents RNA from cells just before the addition of glucose. −RTase, without reverse transcriptase.
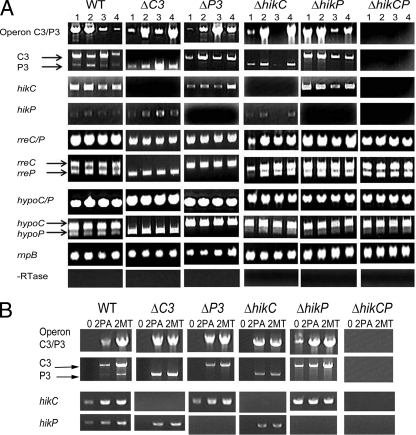
FIG 7.
Transcription of the hik and rre genes of both hik31 operons in the WT and mutants under various growth conditions. (A) Expression of hik31 operon genes in the WT and operon and hik mutants. Cells were grown for about 3 days under PA LL (lanes 1), PA LD (lanes 2), MT LL (lanes 3), and MT LD (lanes 4) conditions. PA, MT, LL (24 h light), and LD (12 h light/12 h dark) conditions. (B) Expression of the operon and hik genes in all of the strains after growth under high-light conditions Cells were grown for 1 day in PA LL and then exposed to HL at 150 μE m−2 s−1 for 2 h in the presence (MT) or absence (PA) of 5 mM glucose. The zero time point was taken after growth for about 1 day (26 h) under PA LL conditions to serve as the control. The RnpB and −RTase (without reverse transcriptase) controls were similar to those in the experiments in panel A (data not shown).
RESULTS AND DISCUSSION
Growth in liquid media—cell doubling and pigmentation.
The mutants described in Fig. 1 were characterized for growth and morphology in liquid medium and on plates. Under LL conditions, the ΔhikC mutant grew somewhat better than the WT, whereas under LD conditions, all 3 hik mutants grew slower (Table 1). The ΔC3P3 mutant was the slowest-growing strain in PA and grew additively worse than both the ΔC3 and ΔP3 mutants. The ΔP3 and ΔC3P3 operon mutant strains grew much slower than the WT under the 6L/18D reduced-light conditions. The ΔC3 mutant was the best-growing strain under both PA LD and PA HL conditions. Under PA HL conditions, all of the mutants except the ΔP3 and ΔC3P3 strains grew fairly well, whereas these two strains were incapable of growth and displayed signs of photoinhibition.
Under MT conditions, the operon mutants grew poorly under different light regimens, whereas all of the hik mutants grew nearly as well as the WT. The ΔC3P3 strain was found to grow less than both individual operon mutants in LL but intermediate between the two individual operon mutants in LD and HL. The ΔP3 strain grew less than half as well as the WT under LD cycles, and this suggests that the ΔP3 strain has defects in dark metabolism involving the processes of sugar catabolism and respiration. The ΔC3 mutant grew poorly in LL and the worst under HL conditions, and the ΔC3P3 mutant also grew slower, resembling the ΔC3 mutant. It was observed that after 4 days of growth, two of the cultures had increased cell concentrations, reaching high cell numbers of 3 × 108 cells/ml (the ΔhikP mutant in MT HL) and 4 × 108 cells/ml (the ΔhikCP mutant in MT 12L/12D), compared to about 2 × 108 cells/ml for the WT. When cultures were grown heterotrophically, both the ΔP3 and ΔC3P3 mutants grew poorly and the ΔhikCP mutant grew the fastest.
The hik mutants demonstrated significant growth defects when grown with 3% CO2 in the presence or absence of glucose. The ΔhikP and ΔhikCP mutants grew poorly in the absence of glucose, and the ΔhikC and ΔhikCP mutants grew poorly in the presence of glucose. The defect exhibited by the ΔhikCP mutant in MT LL and HL became more severe in HC. Furthermore, when grown in high CO2 with glucose, the ΔC3 mutant reached a high cell density of 3.5 × 108 cells/ml and the ΔhikP mutant reached 2.8 × 108 cells/ml, compared to 1.7 × 108 cells/ml for the WT. Thus, the ΔC3 mutant is sensitive to HL conditions but benefits from HC when grown in the presence of glucose. The ΔC3P3 mutant grew better under MT LL than under PA LL conditions and better under PA HC than under PA LL conditions, and intermediately between both operon mutants under MT HC conditions, indicating that both glucose and high CO2 can overcome the growth defect of this strain.
Finally, low-O2 growth was monitored by growth of cultures in PA LL for 2 days and then bubbling with 99.9% N2 and 0.1% CO2 for 2 additional days as previously described (36). The ΔC3, ΔhikC, ΔhikP, and ΔhikCP mutants all showed a mean growth increase in low O2. Moreover, the 3 hik mutants also had a greater growth increase compared to the WT, with the ΔhikCP strain exhibiting the highest level after the first day of low O2. Both the ΔP3 and ΔC3P3 strains grew poorly under low-O2 conditions and did not show an increase in growth like the other strains, suggesting that the plasmid operon, and especially the Rre and/or Hypo on the plasmid, may be important in regulating the growth increase and adapting to low-O2 conditions (data not shown). Based on these results, we concluded that the various mutants had alterations in their growth caused by light or dark, by the presence or absence of carbon sources, and by low O2. Tukey's test was used to measure the significance of the different growth results of the mutants under the various conditions, and those results shown to be statistically significant at α = 0.05 are indicated in Table 1. Based on this analysis, the main growth defects from the growth experiments for each strain are summarized in Table 2. Changes in pigmentation reflected the alterations in growth fairly closely (data not shown, indicated by bold font in Table 1). The main changes in chlorophyll and phycobilisome content were in the ΔP3 and ΔC3P3 mutants, with the strongest influence under glucose, and high-light conditions in the presence and absence of glucose (a 2- to 5-fold reduction in pigment content under most conditions).
TABLE 2.
Growth defects of the mutants compared to the WT
| Strain | Growth defects in liquid mediuma | Cell division defectsb | Metal sensitivityc |
|---|---|---|---|
| ΔC3 mutant | MT LL, MT HL | Yes | NiCl2, ZnCl2, CdCl2 |
| ΔP3 mutant | PA LL, PA LD, PA HL, MT LD, HT DD, low O2 | Yes | CoCl2 |
| ΔC3P3 mutant | PA LL, PA LD, PA HL, MT LL, MT LD, MT HL, HT DD, low O2 | Yes | ZnCl2, CdCl2 |
| ΔhikC mutant | MT HC | No | ZnCl2, CdCl2 |
| ΔhikP mutant | PA LD | No | NiCl2 |
| ΔhikCP mutant | PA LD, MT LL, MT LD, MT HL, PA HC, MT HC | No | NiCl2, ZnCl2, CdCl2 |
Growth on plates.
The mutants were also grown on BG-11 agar plates in order to analyze cultures that might grow well at high density but would manifest defects when diluted. This was an excellent way to differentiate the impact of LD cycles and glucose, either separately or together, and to determine the effect of metals on mutant growth. The main changes in growth patterns seen on plates that were different from liquid growth are shown in Fig. S1 in the supplemental material.
An alternative plating approach consisted of cell growth in MT either for 4 days in LL, followed by 4 days in continuous dark (DD) (38), or for 4 days in DD, followed by 11 days in LL. We also tested the LAHG of all of the cultures on glucose plates with regular BG-11 containing sodium nitrate, as well as ammonium chloride, as previously described (31). The LD treatments were particularly useful in eliciting growth changes in the ΔP3 strain under all of these conditions and for demonstrating that all of the mutants had growth problems if plates were first incubated in complete darkness in the presence of glucose for 4 days or under LAHG conditions (data not shown). The ΔP3 strain grew better under the 4 days of LL, followed by dark, than under continuous cycles of LD. This result also reveals that our glucose-tolerant WT strain is different from other reports in the literature in that it grows heterotrophically without requiring light in liquid and does not grow at all on plates exposed to LAHG. We also tested all of the strains for phenotypes in NaCl and sorbitol stress but did not find significant differences from the WT.
The most interesting plating experiments were those involving metal stress using concentrations that permitted reasonably good growth of the WT (Table 2; also see Fig. S2 in the supplemental material). There are putative transporter genes (efflux pumps) for these metals adjacent to both the chromosomal and plasmid operons. The genes next to C3 may provide tolerance to Ni, Co, and Zn, whereas the genes next to P3 can potentially control Zn and Cd. None of the mutants that were missing hikC could grow on plates with Zn or Cd. These results indicated a role for hikC and C3 in mediating Zn and Cd tolerance. Ni tolerance seems to be dependent on both C3 (specifically, the two genes other than hikC) and hikP. The ΔP3 strain alone has a growth defect in CoCl2. Based on the phenotypes described above, it is possible that there is cross-regulation between the two operons that may control the transporter genes adjacent to the other copy.
Cell morphology and ultrastructure—light and electron microscopy.
All cultures were also carefully analyzed by phase microscopy and electron microscopy. Both of these indicated that the ΔhikC, ΔhikP, and ΔhikCP mutants were similar in overall size and shape to the WT (data not shown), whereas mutants that contained a deletion of the plasmid operon (the ΔP3 and ΔC3P3 mutants) had some morphological alterations under most growth conditions after 3 days of growth for both individual cells and cells that were in the process of doubling. Cells of the ΔC3P3 mutant and, to a lesser extent, the ΔP3 strain demonstrated a slight asymmetry in septum formation even under PA conditions (LL and LD, Fig. 2) and had varied sizes and shapes.
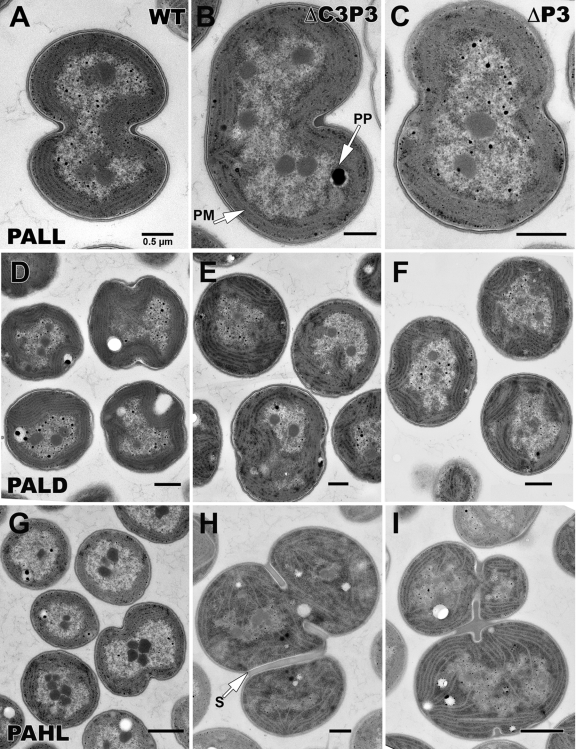
FIG 2.
Transmission electron microscopy under PA conditions. Transmission electron micrographs of WT Synechocystis sp. strain PCC 6803 (A, D, and G), the ΔC3P3 mutant (B, E, and H), and the ΔP3 mutant (C, F, and I) grown under PA conditions under LL (A, B and C), LD (12 h light/12 h dark; D, E, and F), or HL (G, H, and I). Details are described in Materials and Methods. PM, photosynthetic membranes; PP, polyphosphate granules; S, septum. Magnifications: ×28,500 to ×29,400.
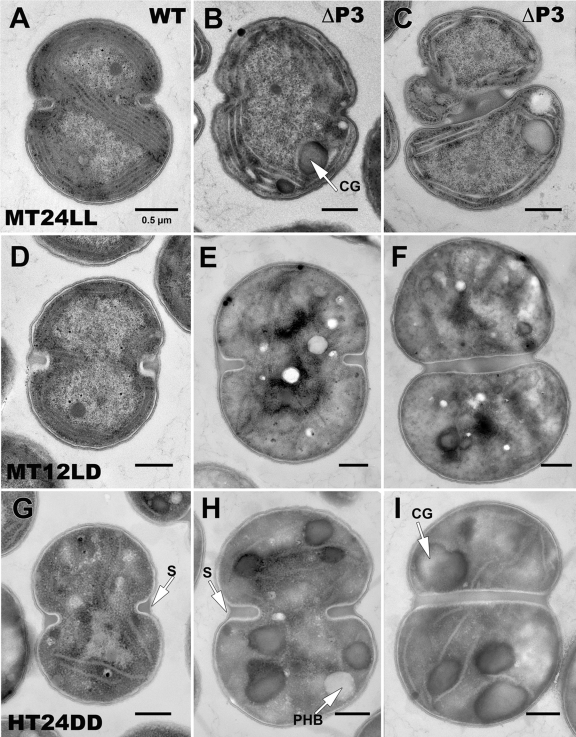
Synechocystis mutants that have changes in genes that are not directly known to have a function in cell division but demonstrate division defects under certain growth conditions have been reported (1, 6, 18). To our knowledge, this is the first report of mutants (ΔP3 and ΔC3P3 mutants) that had division defects under all conditions and among the first published micrographs of Synechocystis under MT LD and HT conditions. Importantly, when the ΔP3 and ΔC3P3 mutants were grown in PA HL, significant changes in cell division were noted (Fig. 2H and I versus G). This inability to divide led to cell clusters, which were the reason for the large sizes of cells in the process of doubling. Kidney bean-shaped, mushroom-shaped, and cloverleaf-shaped tetrad cells of these mutants were seen. Tetrad cells appeared to be formed by two daughter cells dividing even before they completely separated from their twins. Some cells were large at one end and tapered at the other. ΔP3 and ΔC3P3 mutant cells were up to 3 times larger than WT cells under all of the conditions of light, dark, and glucose studied (data not shown) and always had fewer glycogen granules. In the WT, there were many fewer cells with the dividing septa as seen for the mutants (Fig. 3C, F, and I), indicating slower or arrested cell division for the mutants. A summary of cell division defects of each mutant is presented in Table 2.
FIG 3.
Transmission electron microscopy under MT and HT conditions. Transmission electron micrographs of WT Synechocystis sp. strain PCC 6803 (A, D, and G) and the ΔP3 mutant (B, C, E, F, H, and I) grown under MT conditions with 5 mM glucose under LL (A, B, and C) and LD (12 h light/12 h dark; D, E, and F) and under HT conditions in DD (G, H, and I). CG, cyanophycin granules; PHB, polyhydroxybutyrate granules; S, septum. Magnifications, ×20,000 to ×21,000.
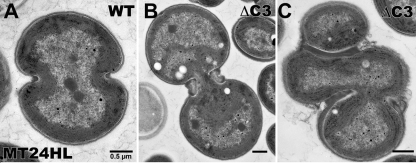
The most intriguing morphology was demonstrated by the ΔP3 and ΔC3P3 strains during growth in MT 12L/12D or HT in the dark. These cells had few, if any, photosynthetic membranes (Fig. 3E and F) and had one or two large storage granules, typically cyanophycin granules (Fig. 3H and I) and to a lesser extent polyhydroxybutyrate (Fig. 3H). In addition, the cytoplasm became opaque and this opacity obscured other typical features. Although these cells doubled at about half the rate of the WT, the cells were clearly altered when grown in glucose. The ΔP3 strain in MT 12L/12D resembled the WT in HT (Fig. 3E versus G), indicating that this mutant is unable to adapt to changing LD conditions and switch its metabolism. Finally, the ΔC3 mutant demonstrated numerous morphological changes when grown in MT HL (Fig. 4). Once again, cell division defects could be seen in the accumulation of excess material at the dividing septum (Fig. 4B) and by clusters of cells that could not separate properly (Fig. 4C). The main difference between the division defects shown by the ΔP3 and ΔC3 strains is that while the ΔP3 strain shows shape and division defects under all conditions, the ΔC3 mutant has these defects only when grown in the presence of glucose and HL. Cells respond to HL stress by reducing the amount of photosynthetic pigments and reaction centers to minimize the oxidative damage. Some of the ΔC3 mutant cells were almost filled with thylakoid membranes, suggesting that the balance between photosynthesis and carbon catabolism is disturbed in these cells in MT HL. All of the operon mutants showed many cells that had thylakoid membranes with branches running across the middle of the cell under conditions of altered cell structure (Table 2). On the other hand, WT cells contained thylakoids arranged in neat rings around the periphery.
FIG 4.
Transmission electron microscopy under MT high-light conditions. Transmission electron micrographs of WT Synechocystis sp. strain PCC 6803 (A) and the ΔC3 mutant (B and C) under MT HL. Magnifications: A, ×18,600; B and C, ×12,000.
Model for the functions of C3 and P3.
Integration of the data on the growth, pigment content, morphology and ultrastructure results of these mutants suggested separate but interacting effects between photosynthetic metabolism (LD) and carbohydrate metabolism (glucose). The ΔC3 and ΔP3 strains grew similarly under PA LL and HC conditions but showed different phenotypes under 10 other growth conditions involving HL or dark and all conditions with glucose and low O2 (Table 1). These results suggested that both operon copies were involved in regulating targets in similar connected pathways in the light but different ones in the dark (Table 2; Fig. 5). Importantly, both operons appear to have a regulatory role in HL. P3 also appears to be responsible for regulating cell division, cell shape, and photosynthesis, and rreP and hypoP are important in regulating growth and adaptation to low O2. The results obtained with the hik mutants differed from those of the corresponding operon mutants when they were grown under HC conditions. The similarity between the growth defects of the ΔhikC and ΔhikCP mutants in high CO2 with glucose and those of the ΔhikP and ΔhikCP mutants in high CO2 without glucose suggests that there is a regulatory switch when glucose is added that is mediated by HikC. This result led us to hypothesize that high CO2 affects the growth of the ΔhikCP mutant more than glucose. In this respect, our results differ from those of Kahlon et al. (11). In our liquid studies, we found only a slight defect in the ΔhikCP mutant with glucose alone. The ΔhikCP mutant displayed growth defects under HC conditions even without glucose, thus differing from the report by Haimovich-Dayan et al. (9). This may be due to slightly different growth conditions and genetic composition of the host strain. The growth conditions we used were 5 mM glucose with 3% CO2, and their conditions were 10 mM glucose and 1 to 5% CO2. The ability of their mutant to grow well in HC may also be due to the unstable phenotypes caused by suppressor mutants. Our deletion mutants were very stable and gave us reproducible phenotypes, and we have taken carefully repeated measurements over a period of 3 years.
FIG 5.
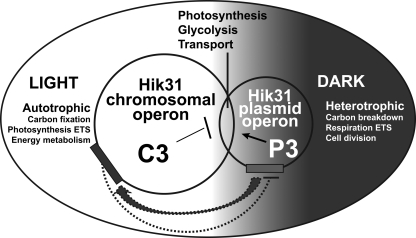
Regulatory functions of the proteins of the chromosomal and plasmid hik31 operons. Regulatory relationship of the hik31 operons as shown by a Venn diagram representing the different and overlapping functions that are regulated by the hik31 operon on the chromosome (C3) and the plasmid (P3). The operons are represented as bars on the chromosome and plasmid. Both operons regulate major metabolic processes in the light and the dark and show both positive (P3 to C3) and negative control (C3 to P3) to their targets and to each other. ETS, electron transport system.
Growth in both liquid and solid media suggested that HikC and RreP and/or HypoP may be dominant and have a hierarchy over their counterparts on the other operon. C3 and HikC seem to be involved in responses to light with or without glucose and high CO2, and P3 and HikP together are involved in responses to L and D with or without glucose and high CO2. Similarly, C3 is involved in MT HL growth and P3 in PA HL growth, whereas HikC is involved in MT HC growth and HikP in PA HC growth. These data indicated that for the ΔC3 and ΔP3 mutant strains, L and D take precedence over glucose, which in turn takes precedence over high CO2. However, in the ΔhikC and ΔhikP mutants, it appears that L and D take precedence over high CO2, which in turn takes precedence over glucose. Such interrelationships are schematically depicted in Fig. 5, with the functional targets of the chromosome and the plasmid operons shown as overlapping circles. We hypothesize that the protein products for both copies regulate major common and separate metabolic processes in the LD. They display both negative (C3) and positive (P3) control. The chromosomal copy C3 is involved in autotrophic growth in light (targets in carbon fixation, photosynthetic electron transport, and energy metabolism). The plasmid copy P3 is involved in heterotrophic growth in the dark (targets in carbon breakdown, respiratory electron transport, and cell division). Both copies control certain shared targets in photosynthesis, glycolysis, and shared transporters, including metal transporters. P3 could be responsible for activating C3, and C3 may, in turn, repress P3.
Growth phase and glucose-dependent expression of hik31 genes.
In order to test the model in Fig. 5, we analyzed the expression of the individual hik31 operons from the WT and mutant strains under the same physiological conditions. We first studied the expression of all 3 genes on both the plasmid and the chromosome in the WT under PA LL (Fig. 6A), MT LL (Fig. 6B), and MT LD (Fig. 6C) conditions. In PA LL, the starter cells at t = 0 showed both operon transcripts. For cells grown from days 1 to 6, we observed a distinct trend in the expression of the individual operons. Both operons were expressed in the active growth phase of cells for 2 to 5 days, corresponding to the log and linear growth phases. However, P3 was downregulated at 1 and 6 days, which correspond to the lag/early log and stationary phases, respectively. P3 was also transcribed less than C3 in a majority of the cases. Both operon copies were present on days 3 and 4, suggesting a high demand that made one copy insufficient. Thus, the C3 operon represented the genes that were primarily expressed, with P3 expressed as a backup under faster growth conditions.
Both operons were strongly induced in MT LL, and their expression levels increased until 6 h and then reduced as cells adapted to the changed conditions. P3 was downregulated at t = 0 but increased gradually along with C3 and was maintained at one-third to two-thirds of the C3 levels. However, in MT LD, both operons were induced and remained high in the dark. The expression of hikC was nearly constant in all of the points in Fig. 6, but hikP increased gradually in LL, reached a peak at 3 days and 6 h, and then declined. In LD, hikP remained high in the dark at both 2D and 6D (Fig. 6C). This indicated that both operons were induced by glucose and that both P3 and hikP were induced by glucose in the dark. There were no changes in the rre and hypo gene expression results in these experiments (data not shown). Both copies of these genes were expressed, and the plasmid rre and hypo genes were found to be of lower abundance, similar to what was found for P3. This could indicate a weaker plasmid operon promoter or be due to structural differences in the plasmid like supercoiling or posttranscriptional changes from antisense RNA (28, 32).
Expression of both operon genes under various growth conditions.
The experiments in Fig. 6 revealed the expression of the hik31 operon genes in the WT under both initial and acclimated growth conditions. We observed that 3 to 4 days of continuous growth in PA LL gave us strong expression of both operons. Hence, we decided to use WT and mutant cells grown for ∼3 days to monitor changes in expression under different growth conditions, especially since the phenotypes recorded in Table 1 were also noted at or after 3 days. The salient findings from Fig. 7A (points i to vii below) and B (points viii to xi below) are as follows. (i) Both copies were cotranscribed as operons under all conditions (Fig. 7A), and C3 was always expressed more than P3 in the WT. (ii) C3 was active in the light, and P3 was active in the dark; C3 was upregulated in PA LL and MT LL in the WT, whereas P3 was upregulated in PA LD. (iii) C3 may downregulate P3 in glucose, and P3 may upregulate C3 in LL; the ΔC3 mutant had upregulated expression of P3 in MT LL and MT LD compared to the WT, suggesting that C3 may normally downregulate P3 under glucose conditions. C3 was downregulated in the ΔP3 strain in PA LL and MT LL compared to the WT, suggesting that P3 may upregulate C3 under these LL conditions. This could mean that RreC represses P3 and RreP activates C3 in the WT. (iv) hikC may be important for the expression of P3; ΔhikC had reduced expression of rreP in PA LL and P3 in MT LL. Thus, hikC could be important for the expression of P3 in MT LL and may have primacy over P3. (v) It is possible that in the absence of hikC, rreC still downregulates P3/hikP, since the ΔhikC and ΔC3 mutants are different in MTLL, with the P3 operon downregulated in ΔhikC. hikP was downregulated, even when rreP and hypoP were transcribed. (vi) The ΔhikP and ΔhikCP mutants had reduced expression of hypoP, suggesting a connection between the two. (vii) Under high-light growth conditions, the expression of both operons was induced after 2 h in all of the strains (Fig. 7B). (viii) Glucose and HL together caused an upregulation of C3 in all of the strains containing C3 (WT and ΔP3 and ΔhikP mutant strains), whereas P3 increased only in the WT and was constant with or without glucose in mutants containing P3 (the ΔC3 and ΔhikC mutants). This may indicate that C3 is needed for the induction of P3 in MT HL. (ix) The ΔhikP mutant may grow better in PA HL and MT HL after 3 days due to the constitutive expression of C3 at all 3 time points. (x) hikC was expressed at all time points in both the WT and ΔP3 mutant strains and constitutively expressed in the ΔhikP mutant, whereas hikP levels varied at t = 0 in the ΔC3 and ΔhikC mutants. Both hik genes were upregulated in HL. (xi) rreP was downregulated only in the ΔC3 mutant at t = 0, whereas rreC and both hypo genes were expressed and unchanged in all of the mutants containing these genes (data not shown).
These data suggest that HL and glucose are sensed by the cell as independent stimuli and had nonoverlapping effects on the transcription of these genes. The expression results in Fig. 6 and 7 demonstrate differential and temporal regulation of both operons in response to the growth phase of the cell, as well as the growth condition. The full-length operon transcripts were downregulated sometimes even as the individual genes are expressed. The different locations of these genes on the chromosome and plasmid could also lead to structural and spatial regulation in the cell, as shown for some other paralogous genes for specificity control (16, 24). We also monitored the expression of these genes after growth under high-CO2 conditions for 2 days but did not observe any significant expression changes. Notably, both of the operons were downregulated, suggesting that high CO2 may be a low-demand condition, as this is the only condition tested where we found no operon transcripts for any of the strains.
Model of the regulatory relationship between C3 and P3.
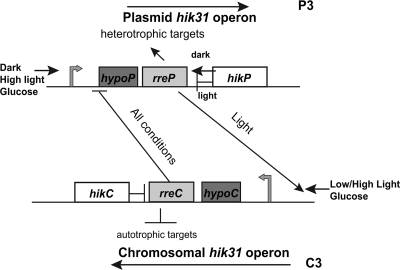
Figure 8 is a working model to explain the relationship between the two hik31 operons and hypotheses about the effects of the Hik proteins on the Rre proteins under different growth conditions. The combination of phenotypic characterization and transcription allowed us to construct a working model of the interrelationships between the two operons that incorporates positive and negative regulation, the role of both operons in the light and the dark, and the effect of glucose, thereby extending the scheme in Fig. 5. The direction of transcription (thin arrows) and the promoter (thick arrows) are marked for both copies. The model suggests that HikP has both negative effects in the light and positive effects in the dark on RreP, which, in turn, has positive effects on heterotrophic targets. RreP also acts to activate C3 in LL. HikC has a negative effect on RreC, which, in turn, has negative effects on its autotrophic targets. RreC may inhibit the transcription of P3 under all conditions, as P3 is always upregulated in the ΔC3 mutant. This model provides an explanation as to why the plasmid operon mutant grows poorly (heterotrophic targets downregulated), but the plasmid hik mutant grows better (HikP cannot inhibit RreP, so targets are upregulated). Similarly, the ΔC3 mutant mostly grows well (RreC cannot downregulate autotrophic targets), but the ΔhikC mutant grows poorly in HC (RreC still inhibits targets).
FIG 8.
Working model of the regulatory relationship between the two hik31 operons. The direction of transcription, expression conditions, and promoters are indicated for each operon, along with the effects of the Hik proteins on the Rre proteins and, in turn, those of the Rre proteins on the targets. The plasmid operon promoter is shown smaller to represent the lower expression results.
We suggest that RreC acts in negative regulation and the C3 promoter is a high-level promoter expressing a low-demand product (41). In contrast, RreP is involved in positive regulation and the P3 promoter is a low-level promoter expressing a high-demand product. The RT-PCR experiments demonstrated that both HL and glucose are high-demand environments inducing the expression of both operons (41). The numerous phenotypic defects displayed by the double operon and hik mutants (Table 2) may be a result of both positive and negative regulation as the cell attempts to fine-tune the levels of both copies.
There are also exceptions, e.g., cultures that grow poorly under many conditions but grow well under certain conditions or vice versa. This behavior could be due to the effects of the other copy. For example, the ΔP3 mutant strain grows well in MT HL, probably due to HikC downregulating RreC so that autotrophic targets are active. The ΔhikC mutant grows better in LL (HikP or some other protein inhibits RreC, so that targets are derepressed). The ΔhikP mutant grows extremely well in MT HL or high CO2 with glucose, and we suggest that this is due to both autotrophic (HikC downregulates RreC) and heterotrophic targets being active (RreP is not downregulated, as HikP is missing). Interestingly, high CO2 with glucose did not affect the ΔC3 mutant but MT HL did, suggesting that autotrophic targets are active in the ΔC3 mutant and this caused photoinhibition in MT HL.
We hypothesize that the Hypo for both copies activates the corresponding Hik protein, which may alter the effect on the Rre protein. Both Hik proteins have ATP lids that closely match the ATP lid of the bifunctional protein EnvZ (2). This would make them bifunctional as well, and both Hik proteins would be able to act as kinases and phosphatases for their partner Rre proteins. However, because the operons maybe a 3CS (e.g., the NRI/NRII system in E. coli), and due to the diversity of signal responses seen, the Hik proteins would switch from being bifunctional to being monofunctional when needed (2). This would allow them to both transduce primary signals, like LD, as well as to integrate secondary signals like HL and glucose as needed. Also, like the EnvZ-OmpR system, there are weaker ribosome binding sites between the Rre and Hik proteins. This leads to lower EnvZ than OmpR protein levels (about 100 EnvZ to 3,500 OmpR molecules per cell of E. coli). The higher OmpR concentrations are considered important for the dual role of activation and repression on the target porins (23). The hik genes have lower transcript levels compared to rre genes in our experiments, suggesting similar behavior that could lead to lower translation of the Hik proteins and degradation of the full-length operon transcripts.
The transcriptional regulation of the well-studied EnvZ-OmpR system is quite complex, with four levels of control and 11 transcription factors that have been shown to regulate expression directly or indirectly (24). It is possible that a similar regulatory control exists in Synechocystis to maintain the two hik31 operon copies and appropriate gene dosages. OmpR-P is thought to undergo a conformational change that relieves self-inhibition of DNA binding. It is possible that the two Rre proteins differ in the ability to relieve self-inhibition and bind DNA on phosphorylation due to the sequence changes between the receiver and output domains. OmpR can both activate and repress its target porins based on its phosphorylation level, and 2 molecules of OmpR-P have been shown to bind to as many as 7 different promoter sites in a hierarchical manner (43). RreC and RreP also have differences in their output domain that may enable them to bind to adjacent regions of the promoters of their common target genes, as well as different regions of different targets, in a hierarchical manner. The model presented in Fig. 8 is a simple one, serving to provide a hypothesis to explain a more complex and unique system combining many previously classified mechanisms of gene fixation after duplication. Nonetheless, it is evident that the right balance between C3 and P3 needs to be maintained in the cell for proper regulation of central metabolic processes.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the help of Tina Summerfield (now at the University of Otago, Dunedin, New Zealand) throughout the initial stages of this project.
This study was funded by DOE grant DE-FG02-99ER20342 and in part by DOE-BER (DE-FC02-07ER64694).
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Akai M, et al. 2011. Plasma membrane aquaporin AqpZ protein is essential for glucose metabolism during photomixotrophic growth of Synechocystis sp. PCC 6803. J. Biol. Chem. 286:25224–25235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alves R, Savageau MA. 2003. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol. Microbiol. 48:25–51 [DOI] [PubMed] [Google Scholar]
- 3. Ashby MK, Houmard J. 2006. Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol. Mol. Biol. Rev. 70:472–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buelow DR, Raivio TL. 2010. Three (and more) component regulatory systems—auxiliary regulators of bacterial histidine kinases. Mol. Microbiol. 75:547–566 [DOI] [PubMed] [Google Scholar]
- 5. Capra EJ, et al. 2010. Systematic dissection and trajectory-scanning mutagenesis of the molecular interface that ensures specificity of two-component signaling pathways. PLoS Genet. 6:e1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferjani A, et al. 2003. Glucosylglycerol, a compatible solute, sustains cell division under salt stress. Plant Physiol. 131:1628–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Galperin MY. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188:4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haimovich-Dayan M, et al. 2011. Cross-talk between photomixotrophic growth and CO2-concentrating mechanism in Synechocystis sp. strain PCC 6803. Environ. Microbiol. 13:1767–1777 [DOI] [PubMed] [Google Scholar]
- 10. Hihara Y, Sonoike K, Kanehisa M, Ikeuchi M. 2003. DNA microarray analysis of redox-responsive genes in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185:1719–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kahlon S, et al. 2006. A putative sensor kinase, Hik31, is involved in the response of Synechocystis sp. strain PCC 6803 to the presence of glucose. Microbiology 152:647–655 [DOI] [PubMed] [Google Scholar]
- 12. Kaneko T, et al. 2003. Structural analysis of four large plasmids harboring in a unicellular cyanobacterium, Synechocystis sp. PCC 6803. DNA Res. 10(5):221–228 [DOI] [PubMed] [Google Scholar]
- 13. Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290:339–348 [DOI] [PubMed] [Google Scholar]
- 14. Kanesaki Y, et al. 2007. Histidine kinases play important roles in the perception and signal transduction of hydrogen peroxide in the cyanobacterium, Synechocystis sp. PCC 6803. Plant J. 49:313–324 [DOI] [PubMed] [Google Scholar]
- 15. Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. 2002. Selection in the evolution of gene duplications. Genome Biol. 3(2):RESEARCH0008. 1–RESEARCH0008.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Laub MT, Goulian M. 2007. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 41:121–145 [DOI] [PubMed] [Google Scholar]
- 17. Li H, Sherman LA. 2000. A redox-responsive regulator of photosynthesis gene expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 182:4268–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liang CW, Zhang XW, Tian L, Qin S. 2008. Functional characterization of sll0659 from Synechocystis sp. PCC 6803. Indian J. Biochem. Biophys. 45:275–277 [PubMed] [Google Scholar]
- 19. Louis EJ. 2007. Evolutionary genetics: making the most of redundancy. Nature 449:673–674 [DOI] [PubMed] [Google Scholar]
- 20. Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155 [DOI] [PubMed] [Google Scholar]
- 21. Marijuán PC, Navarro J, del Moral R. 2010. On prokaryotic intelligence: strategies for sensing the environment. Biosystems 99:94–103 [DOI] [PubMed] [Google Scholar]
- 22. Marin K, et al. 2003. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc. Natl. Acad. Sci. U. S. A. 100:9061–9066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martínez-Hackert E, Stock AM. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301–312 [DOI] [PubMed] [Google Scholar]
- 24. Martínez-Núñez MA, Perez-Rueda E, Gutierrez-Rios RM, Merino E. 2010. New insights into the regulatory networks of paralogous genes in bacteria. Microbiology 156:14–22 [DOI] [PubMed] [Google Scholar]
- 25. Mascher T, Helmann JD, Unden G. 2006. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol. Mol. Biol. Rev. 70:910–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maslov S, Sneppen K, Eriksen KA, Yan KK. 2004. Upstream plasticity and downstream robustness in evolution of molecular networks. BMC Evol. Biol. 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mikami K, Kanesaki Y, Suzuki I, Murata N. 2002. The histidine kinase Hik33 perceives osmotic stress and cold stress in Synechocystis sp PCC 6803. Mol. Microbiol. 46:905–915 [DOI] [PubMed] [Google Scholar]
- 28. Mitschke J, et al. 2011. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. U. S. A. 108:2124–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mizuno T, Kaneko T, Tabata S. 1996. Compilation of all genes encoding bacterial two-component signal transducers in the genome of the cyanobacterium, Synechocystis sp. strain PCC 6803. DNA Res. 3:407–414 [DOI] [PubMed] [Google Scholar]
- 30. Murata N, Suzuki I. 2006. Exploitation of genomic sequences in a systematic analysis to access how cyanobacteria sense environmental stress. J. Exp. Bot. 57:235–247 [DOI] [PubMed] [Google Scholar]
- 31. Osanai T, et al. 2005. Positive regulation of sugar catabolic pathways in the cyanobacterium Synechocystis sp. PCC 6803 by the group 2 sigma factor sigE. J. Biol. Chem. 280:30653–30659 [DOI] [PubMed] [Google Scholar]
- 32. Prakash JS, et al. 2009. DNA supercoiling regulates the stress-inducible expression of genes in the cyanobacterium Synechocystis. Mol. Biosyst. 5:1904–1912 [DOI] [PubMed] [Google Scholar]
- 33. Ramagopal UA, et al. Crystal structure of DUF305 fragment from Deinococcus radiodurans. Research Collaboratory for Structural Bioinformatics Protein Data Bank, Piscataway, NJ. http://pdb201.sdsc.edu/pdb/explore/explore.do?structureId=3BT5. [Google Scholar]
- 34. Singh AK, Sherman LA. 2005. Pleiotropic effect of a histidine kinase on carbohydrate metabolism in Synechocystis sp. strain PCC 6803 and its requirement for heterotrophic growth. J. Bacteriol. 187:2368–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Summerfield TC, Eaton-Rye JJ, Sherman LA. 2007. Global gene expression of a delta PsbO:delta PsbU mutant and a spontaneous revertant in the cyanobacterium Synechocystis sp. strain PCC 6803. Photosynth. Res. 94:265–274 [DOI] [PubMed] [Google Scholar]
- 36. Summerfield TC, Nagarajan S, Sherman LA. 2011. Gene expression under low-oxygen conditions in the cyanobacterium Synechocystis sp. PCC 6803 demonstrates Hik31-dependent and -independent responses. Microbiology 157(Pt. 2):301–312 [DOI] [PubMed] [Google Scholar]
- 37. Summerfield TC, Sherman LA. 2008. Global transcriptional response of the alkali-tolerant cyanobacterium Synechocystis sp. strain PCC 6803 to a pH 10 environment. Appl. Environ. Microbiol. 74:5276–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Summerfield TC, Sherman LA. 2007. Role of sigma factors in controlling global gene expression in light/dark transitions in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 189:7829–7840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki I, et al. 2005. The histidine kinase Hik34 is involved in thermotolerance by regulating the expression of heat shock genes in Synechocystis. Plant Physiol. 138:1409–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suzuki S, Ferjani A, Suzuki I, Murata N. 2004. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 279:13234–13240 [DOI] [PubMed] [Google Scholar]
- 41. Wall ME, Hlavacek WS, Savageau MA. 2004. Design of gene circuits: lessons from bacteria. Nat. Rev. Genet. 5:34–42 [DOI] [PubMed] [Google Scholar]
- 42. Wolanin PM, Thomason PA, Stock JB. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol. 3(10):REVIEWS3013.1–REVIEWS3013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yoshida T, Qin L, Egger LA, Inouye M. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281:17114–17123 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.