Abstract
Porcine enterotoxigenic Escherichia coli (ETEC) continues to result in major morbidity and mortality in the swine industry via postweaning diarrhea. The key virulence factors of ETEC strains, their serotypes, and their fimbrial components have been well studied. However, most studies to date have focused on plasmid-encoded traits related to colonization and toxin production, and the chromosomal backgrounds of these strains have been largely understudied. Here, we generated the genomic sequences of K88-positive and F18-positive porcine ETEC strains and examined the phylogenetic distribution of clinical porcine ETEC strains and their plasmid-associated genetic content. The genomes of porcine ETEC strains UMNK88 and UMNF18 were both found to contain remarkable plasmid complements containing known virulence factors, potential novel virulence factors, and antimicrobial resistance-associated elements. The chromosomes of these strains also possessed several unique genomic islands containing hypothetical genes with similarity to classical virulence factors, although phage-associated genomic islands dominated the accessory genomes of these strains. Phylogenetic analysis of 78 clinical isolates associated with neonatal and porcine diarrhea revealed that a limited subset of porcine ETEC lineages exist that generally contain common toxin and fimbrial profiles, with many of the isolates belonging to the ST10, ST23, and ST169 multilocus sequencing types. These lineages were generally distinct from existing human ETEC database isolates. Overall, most porcine ETEC strains appear to have emerged from a limited subset of E. coli lineages that either have an increased propensity to carry plasmid-encoded virulence factors or have the appropriate ETEC core genome required for virulence.
INTRODUCTION
Diarrhea is a major cause of morbidity and mortality to young pigs, resulting in significant production losses for the swine industry (10). Enterotoxigenic Escherichia coli (ETEC) strains are the most common cause of diarrhea in young pigs, resulting in a disease known as enteric colibacillosis (10). Neonatal and postweaning pigs are most susceptible to ETEC. While neonatal ETEC diarrhea is generally well controlled by using fimbria-based vaccines, postweaning enteric colibacillosis has been more difficult to control. Porcine ETEC strains are characterized by the production of specific adhesins and enterotoxins. The adhesins most common in porcine ETEC are fimbrial adhesins K88 (also called F4), K99 (F5), 987P (F6), F41, and F18, as well as afimbrial adhesins such as AIDA-I. Porcine ETEC also produce one or more enterotoxins, including heat-labile enterotoxin (LT), heat-stable enterotoxins a and b (STa and STb), and EAST1 toxin (33). These and other described porcine ETEC virulence factors have mostly been localized to plasmids. Additionally, F18-positive porcine E. coli strains implicated in edema disease and diarrhea sometimes possess the Shiga toxin-encoding gene (stx2e) within their chromosomes (16).
Analyses of human ETEC strains have revealed that they are polyphyletic. While other E. coli pathotypes group tightly in phylogenetic trees, human ETEC strains appear to be more diverse. A recent study of 1,019 human ETEC isolates and 8 porcine isolates from several countries revealed 42 different multilocus sequence typing (MLST) lineages, with evidence of virulence factor exchange between lineages (49). Human and porcine strains were sometimes found to fall into the same lineages. Based on the diversity and the patterns in lineages, it was concluded that human ETEC likely emerged as a result of at least three separate events. Examination of a fully sequenced prototypical human ETEC strain (H10407) revealed that the chromosome was similar to those of sequenced E. coli commensal strains, indicating that virulence was most likely attributable to the acquisition of plasmid-encoded virulence factors (8). Another recent study examined entire genomes of five ETEC isolates from children in Guinea-Bissau with diarrhea (45). These isolates were selected based on their representation of diverse clonal groups. While no genomic regions were completely conserved across all ETEC strains, there was more genomic content shared between ETEC genomes than between the sequenced ETEC strains and other pathotypes, suggesting the presence of an ETEC genomic core.
O-antigen and fimbrial diversity is more limited among porcine ETEC strains than human ETEC strains, suggesting that a genomic core also exists for ETEC isolates of porcine origin. However, genomic analysis of the chromosomal content of porcine ETEC strains has been largely understudied, and there is a need for better understanding of the chromosomal background of these important pathogens. Additionally, postweaning diarrhea caused by porcine ETEC has been historically difficult to control. While porcine ETEC strains as a group have clearly defined serogroups and fimbrial types, a gap in our knowledge of this pathotype exists regarding their chromosomal content and diversity. Filling this gap may help to explain their more coherent phenotypes in contrast to human ETEC. In this study, we generated the genomic sequences of K88-positive and F18-positive porcine ETEC isolates and examined the phylogenetic distribution of clinical porcine ETEC and their plasmid-associated genetic content in an effort to better determine whether there is a core porcine ETEC genome and to better understand the evolution of K88-positive and F18-positive pathogens.
MATERIALS AND METHODS
Bacterial strain and sequencing.
Two isolates were selected for whole-genome sequencing from a collection of isolates submitted to the University of Minnesota Veterinary Diagnostic Laboratory (Table 1). Strain UMNK88 was isolated in 2007 from a farm in Minnesota and identified via multiplex PCR as possessing the K88, LT, and STb genes, lacking stx2e, and belonging to serogroup O149. Strain UMNF18 was isolated from a nursery farm in Iowa in 2006. This isolate was identified as possessing the F18, STa, and STb genes and the stx2e Shiga toxin gene and belonging to serogroup O147. These isolates were selected because of their associations with outbreaks of postweaning diarrhea in the Midwestern United States. Strains were grown to mid-logarithmic phase in Luria-Bertani broth (BD Diagnostics, Sparks, MD) at 37°C with shaking. Total DNA was isolated from the strains using a DNeasy kit (Qiagen, Valencia, CA). The whole genomes of UMNK88 and UMNF18 were sequenced to a 30× depth using Roche 454 pyrosequencing with GS FLX Titanium chemistry. Two libraries were generated for each genome, a single-end size-fragmented library and an 8-kb mate-pair library. Sequence reads were assembled into scaffolds using the Newbler assembler, resulting in a single chromosomal scaffold for each genome. Remaining gaps in UMNK88 were closed and checked for consistency using PCR across gaps or regions of low coverage. UMNF18 was draft assembled and gaps were closed, but the assembly was not checked for consistency with PCR due to limited resources (52). Comparisons of UMNK88 and UMNF18 were performed against E. coli K-12 strain MG1655 and human ETEC strain E24377A (5, 42).
Table 1.
General characteristics of porcine enterotoxigenic E. coli strains UMNK88 and UMNF18 compared to those of human enterotoxigenic strain E24377A and laboratory strain K-12 MG1655
| Characteristic | UMNK88 | UMNF18 | E24377A | K-12 MG1655 |
|---|---|---|---|---|
| Serogroup | O149 | O147 | O139 | OR |
| Phylogenetic group | A | A | B1 | A |
| Chromosome length (bp) | 5,186,416 | 5,239,207 | 4,979,619 | 4,639,675 |
| Plasmids (bp) | pUMNK88_Hly (65,549), pUMNK88_Ent (IncFIB; 81,475), pUMNK88_K88 (IncFIIA; 81,883), pUMNK88_91 (IncI1; 90,868), pUMNK88_IncA/C (IncA/C; 160,573) | pUMNF18_32 (IncX4; 32,487), pUMNF18_IncI1 (IncI1; 69,065), pUMNF18_87 (IncFIIA; 87,792), pUMNF18_103 (P7-like; 103,130), pUMNF18_IncFV (IncFIIA; 103,624) | pETEC_5 (5,033), pETEC_6 (6,199), pETEC_35 (34,367), pETEC_73 (70,609), pETEC_80 (79,237) | None |
| Chromosome GC content (%) | 50.7 | 50.7 | 50.6 | 50.8 |
| Total chromosome coding sequences | 5,118 | 5,241 | 4,758 | 4,148 |
For multilocus sequence analysis, 78 isolates were selected from those isolated by the University of Minnesota Veterinary Diagnostic Laboratory in 2007 and 2008. These included isolates submitted from different pig farms throughout the United States and identified via PCR as F18+, K88+, or lacking both fimbrial operons. Isolates were verified to have originated from different farms and outbreak cases to eliminate redundancy in the collection. Subsequent gene prevalence studies were performed on a collection of 214 porcine ETEC isolates, including those used for multilocus sequence analysis, identified in a similar manner.
Gene prediction, annotation, and comparative analysis.
Annotation was automated using publicly available tools. Putative coding regions were predicted using GeneMarkS (3), tRNA genes using tRNAscan-SE (26), and rRNA genes using RNAmmer (23). Gene function was assigned using HMMER3 (20) against Pfam-A 24.0 (15), RPS-BLASTp against CDD (30), and BLASTp against all microbial proteins. Genomic islands greater than 3,000 bp were identified by their absence from E. coli strain K-12 MG1655 (5). For UMNK88, manual inspection of genomic islands was performed, while UMNF18 annotation was automated. Comparisons between the coding regions of UMNK88, UMNF18, and sequenced E. coli genomes were performed using BlastP with a similarity cutoff of 90% (38).
Multilocus sequence analysis.
Phylogenetic relationships were determined using the E. coli MLST protocols and scheme described at http://mlst.ucc.ie/mlst/dbs/Ecoli (55). The following genes were included: adk, gyrB, icd, mdh, purA, and recA. Evolutionary history was inferred using the neighbor-joining method. Bootstrapping was performed with 500 replicates. A total of 3,618 nucleotide positions were used in the final data set. Phylogenetic analyses were conducted in MEGA version 4 (50).
Gene prevalence.
The prevalence of 25 genes found on the virulence-associated plasmids of UMNK88 and UMNF18 was sought using multiplex PCR (Table 2). Primers were designed using PrimerSelect (Lasergene, Madison, WI) (see the supplemental material). Template DNA was prepared from bacterial collections using a boiling lysis procedure, as previously described (19). Multiplex PCR was performed in a 25-μl reaction volume using previously described procedures (44). PCR was performed using an initial denaturing step at 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 1 min. The amplified products were electrophoresed in 2% agarose gels at 200 V for 1 h, stained with ethidium bromide, and visualized under UV light. Several products of expected sizes were excised, gel purified, and sequenced to confirm that they were the desired gene sequences. Positive samples were subsequently identified based on the presence of bands of appropriate sizes compared to positive-control strains. The negative-control strain was E. coli DH10B.
Table 2.
Plasmid-associated genes sought via multiplex PCR
| No. | Plasmid | Gene locus | Gene product | Prevalence (%) ina : |
|
|---|---|---|---|---|---|
| ND isolates | PWD isolates | ||||
| 1 | pUMNF18_87 | 79 | Adhesin AIDA-1 | 11.4 | 20.4 |
| 2 | pUMNF18_87 | 12 | F18 fimbrial chaperone FedD | 17.7 | 32.3 |
| 3 | pUMNF18_87 | 10 | F18 fimbrial adhesin FedF | 16.5 | 32.3 |
| 4 | pUMNF18_87 | 83 | Putative AraC-type transcriptional regulator | 12.7 | 17.2 |
| 5 | pUMNF18_87 | 37 | Hemolysin protein HlyA | 74.7 | 92.5 |
| 6 | pUMNK88_Ent | 35 | Heat-labile enterotoxin subunit LT-A | 22.8 | 52.7 |
| 7 | pUMNK88_Ent | 36 | Heat-labile enterotoxin subunit LT-B | 29.1 | 53.8 |
| 8 | pUMNK88_Ent | 18 | Putative MarR-family transcriptional regulator | 22.8 | 53.8 |
| 9 | pUMNK88_Ent | 30 | Heat-stable enterotoxin STb | 26.6 | 73.1 |
| 10 | pUMNK88_Hly | 15 | Putative invasin | 22.8 | 36.6 |
| 11 | pUMNK88_Hly | 37 | DNA primase SogL | 27.8 | 40.9 |
| 12 | pUMNK88_Hly | 11 | Putative mobilization protein MobA | 67.1 | 68.8 |
| 13 | pUMNK88_Hly | 50 | Hemolysin protein HlyA | 59.5 | 76.3 |
| 14 | pUMNK88_Hly | 52 | Hemolysin protein HlyD | 58.2 | 46.2 |
| 15 | pUMNF18_IncFV | 54 | Colicin B activity protein Cba | 20.3 | 35.5 |
| 16 | pUMNF18_IncFV | 3 | IncFV replication protein RepA1 | 67.1 | 91.4 |
| 17 | pUMNF18_IncFV | 85 | IncFV conjugal transfer protein | 54.4 | 68.8 |
| 18 | pUMNK88_K88 | 43 | Glycoporin RafY lacking transposase insertion | 54.4 | 19.4 |
| 19 | pUMNK88_K88 | 43 | Glycoporin RafY core region | 75.9 | 82.8 |
| 20 | pUMNK88_K88 | 25 | Putative malate dehydrogenase | 54.4 | 55.9 |
| 21 | pUMNK88_K88 | 26 | Putative choloylglycine hydrolase | 25.3 | 63.4 |
| 22 | pUMNK88_K88 | 12 | K88 minor fimbrial subunit FaeH | 50.6 | 64.5 |
| 23 | pUMNK88_K88 | 9 | K88 fimbrial chaperone FaeE | 55.7 | 68.8 |
| 24 | pUMNK88_K88 | 57 | Putative fimbrial protein | 58.2 | 68.8 |
| 25 | pUMNK88_K88 | 56 | Putative fimbrial subunit | 32.9 | 69.9 |
PWD, postweaning diarrhea; ND, neonatal diarrhea.
Phylogenetic typing and serotyping.
Determination of major E. coli phylogenetic groups (A, B1, B2, and D) was performed using the interpretive approach described by Clermont et al. (6).
Nucleotide sequence accession numbers.
The annotated genome sequences of UMNK88 have been deposited at DDBJ/EMBL/GenBank under the accession numbers CP002729 for the UMNK88 complete chromosome and CP002730, CP002731, CP002732, CP002733, and HQ023862 for the UMNK88 plasmids pUMNK88_K88, pUMNK88_IncI1, pUMNK88_Ent, pUMNK88_Hly, and pUMNK88, respectively. This whole-genome shotgun project of UMNF18 has been deposited at DDBJ/EMBL/GenBank under the accession number AGTD00000000. The version described in this paper is the first version, AGTD01000000.
RESULTS AND DISCUSSION
Structure and general features of the UMNF18 and UMNK88 chromosomes.
Strains UMNF18 and UMNK88 were sequenced using 8-kb mate-pair and shotgun libraries on Roche 454 GS-FLX using Titanium chemistry. PCR-based finishing was used to close the chromosomes and plasmids. Closed sequences were obtained for the chromosomes and plasmids of both strains, with average coverages of 29× for both chromosomes and 17× to 101× for the plasmids. The chromosome lengths for UMNK88 and UMNF18 were 5,186,416 bp and 5,239,207 bp, respectively (Table 1).
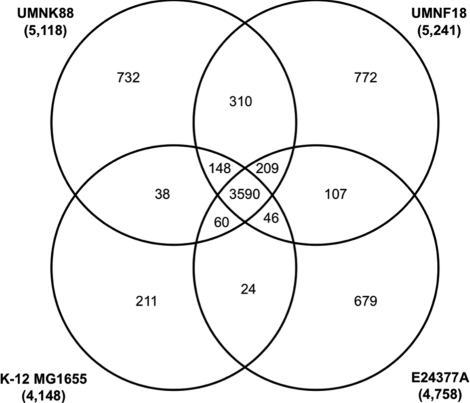
The predicted chromosomal proteins of the UMNK88 and UMNF18 genomes were compared to each other, to those of laboratory E. coli K-12 strain MG1655, and to those of human ETEC strain E24377A using BlastP in RAST (38). Of the 5,118 annotated proteins of UMNK88, 3,590 (70.1%) were shared among the four genomes and 732 (14.3%) were unique to UMNK88 (Fig. 1). Similar results were observed for UMNF18, with 3,590 (68.5%) of 5,241 annotated proteins being shared among all four genomes and 772 (14.7%) being unique to UMNF18. Most of the proteins unique to UMNK88 and UMNF18, and the 310 proteins shared between the two strains but absent from K-12 MG1655, were similar to prophage proteins or proteins of unknown function.
Fig 1.
Venn diagram depicting shared and unique proteins among strains UMNK88, UMNF18, E24377A (human ETEC), and K-12 MG1655. Total numbers of annotated proteins for each chromosome are shown in parentheses.
UMNF18 contained 36 genomic islands, which were defined as chromosomal regions greater than 3 kb that were absent from E. coli K-12 strain MG1655 (Table 3). The UMNF18 chromosome contained a relatively large number of prophage-associated regions, with 21 genomic islands involving prophage-like elements. One prophage-like element on island 10, inserted within the tRNAArg site, possessed the Shiga toxin-encoding gene stx2e and was unique compared to other sequenced E. coli genomes (see Fig. S1 in the supplemental material). This gene is typically present in F18-positive strains implicated in porcine edema disease (31). In addition to prophage-associated genomic islands, several islands containing putative novel virulence and/or regulatory genes were identified. For example, islands 34 and 35 in UMNF18 were adjacent to one another and contained a Tia-like invasion protein (UMNF18_5307), an autotransporter-like protein (UMNF18_5318) also present in Salmonella enterica serovar Heidelberg strain SL476, a phage integrase (UMNF18_5319), an XRE-family transcriptional regulator (UMNF18_5327) also present in S. Heidelberg SL476, an H-NS-like protein (UMNF18_5335) also present in S. Heidelberg SL476, and a putative HTH-type transcriptional regulator (UMNF18_5454) also present in numerous pathogenic E. coli genomes. However, the genetic context in which these genes were located was unique to UMNF18 compared to sequenced E. coli genomes (see Fig. S1). Both UMNF18 and UMNK88 contained genomic islands adjacent to tRNAAsp (island 2 in both genomes) with similarity to a type VI secretion system, present in most other sequenced E. coli genomes, which has been shown to be involved in the translocation of effector molecules across the cell envelope (4). Similarly, both genomes possessed truncated versions of the type III secretion system ETT2 in islands 27 and 26, which are likely nonfunctional (18, 43).
Table 3.
Genomic islands of UMNF18 larger than 3,000 bp
| UMNF18 island | Start | Stop | Size (bp) | Feature(s)a | Associated tRNA(s) |
|---|---|---|---|---|---|
| 1 | 19813 | 23858 | 4,045 | Putative fimbrial chaperone/usher system | |
| 2 | 242548 | 255137 | 12,589 | Type VI secretion system | Asp |
| 3 | 544640 | 569674 | 25,034 | Prophage associated | Arg |
| 4 | 793512 | 837529 | 44,017 | Prophage associated | |
| 5 | 919305 | 936912 | 17,607 | Prophage associated | |
| 6 | 997969 | 1004872 | 6,903 | Prophage associated | |
| 7 | 1088909 | 1147297 | 58,388 | Prophage associated | Thr, Gly, Ser |
| 8 | 1298120 | 1411072 | 112,952 | Prophage associated | Met, Lys, Thr, Gly |
| 9 | 1461047 | 1491867 | 30,820 | Prophage associated | |
| 10 | 1557736 | 1586729 | 28,993 | Prophage associated, containing stx2e | Met, Arg |
| 11 | 1695312 | 1731440 | 36,128 | Prophage associated | |
| 12 | 1934369 | 1940797 | 6,428 | Uncharacterized genomic island | |
| 13 | 2235721 | 2277966 | 42,245 | Prophage associated | |
| 14 | 2356059 | 2376541 | 20,482 | Prophage associated | Gly, Thr, Met, Ser, Asn |
| 15 | 2401215 | 2412982 | 11,767 | Uncharacterized genomic island | Asn |
| 16 | 2420362 | 2425869 | 5,507 | Uncharacterized genomic island | |
| 17 | 2449881 | 2454038 | 4,157 | O147 O-antigen gene cluster | |
| 18 | 2530590 | 2546536 | 15,946 | Prophage associated | |
| 19 | 2557730 | 2561542 | 3,812 | Uncharacterized genomic island | |
| 20 | 2651438 | 2664886 | 13,448 | Prophage associated | Pro |
| 21 | 2814895 | 2851872 | 36,977 | Prophage associated | |
| 22 | 2882510 | 2895186 | 12,676 | Prophage associated | Arg |
| 23 | 3086976 | 3118068 | 31,092 | Prophage associated | |
| 24 | 3181712 | 3218032 | 36,320 | Prophage associated | |
| 25 | 3236446 | 3257467 | 21,021 | Prophage associated | |
| 26 | 3357351 | 3370209 | 12,858 | CRISPR associated | |
| 27 | 3479023 | 3487230 | 8,207 | Truncated ETT2 type 3 secretion system | Gly |
| 28 | 3600025 | 3633904 | 33,879 | Uncharacterized genomic island | Phe |
| 29 | 3635450 | 3643451 | 8,001 | Type 2 general secretory pathway gsp | |
| 30 | 3661507 | 3669899 | 8,392 | Uncharacterized genomic island | |
| 31 | 3754330 | 3762369 | 8,039 | Prophage associated | Met |
| 32 | 4971105 | 4980544 | 9,439 | TRAP transporter system | |
| 33 | 4999864 | 5035394 | 35,530 | Prophage associated | |
| 34 | 5080926 | 5092938 | 12,012 | Genomic island with tia | Leu |
| 35 | 5093380 | 5135899 | 42,519 | Genomic island with putative transcriptional regulators and prophage sequences | |
| 36 | 5178404 | 5190014 | 11,610 | Hydroxyphenolacetic acid (hpa) degradation genes |
CRISPR, clusters of regularly interspaced short palindromic repeats; TRAP, transporter, tripartite ATP-independent periplasmic transporter.
Strain UMNK88 contained 40 genomic islands, totaling 814 kb (15%) in total genome content (Table 4). Of these, 15 islands were prophage-associated loci. A few notable islands contained potential novel virulence and regulatory genes. Island 8 was found adjacent to tRNASer and contained a phage integrase gene, a gene encoding a disrupted protein with similarity to the serum resistance gene traT, urease genes (disrupting traT), and tellurite resistance genes (see Fig. S2 in the supplemental material). This region was similar to the urease-tellurite islands found in E. coli O157:H7 strains, EHEC O26:H11 strain 11368, and EHEC O111:H− strain 11128 (51). Previous studies have identified this island among porcine ETEC O149:K88 strains (39), and portions of this island in E. coli O157:H7 were shown to be involved in colonization to ligated pig intestine (56). Therefore, this island might be important for the colonization of UMNK88 and other K88 strains. UMNK88 island 13 was unique compared to all sequenced E. coli strains and included mostly genes for proteins of unknown function and one with similarity to an XRE-like transcriptional regulator (UMNK88_1920) also found in Salmonella serovar Montevideo strains. Island 37, adjacent to tRNAPhe, was also unique to UMNK88 and displayed synteny and low sequence similarity with an integrative conjugative element (ICE) from the plant-commensal bacterium Pseudomonas fluorescens strain Pf-5 (40) (see Fig. S3 in the supplemental material). This putative ICE was bound by 49-bp direct repeats, typical of ICE elements. While this putative ICE contained no apparent antibiotic resistance genes, it did contain a gene identical to that found in plasmid pUMNK88_Hly of UMNK88 encoding a putative invasin with an immunoglobulin (Ig)-like domain typical of surface proteins such as intimins and invasins, as mentioned below.
Table 4.
Genomic islands of UMNK88 larger than 3,000 bp
| UMNK88 island | Start | Stop | Size (bp) | Feature(s)a | Associated tRNA(s) |
|---|---|---|---|---|---|
| 1 | 19799 | 23844 | 4,045 | Putative fimbrial chaperone/usher system | |
| 2 | 241931 | 271948 | 30,017 | Type IV secretion system and putative toxin/hemolysin | Asp |
| 3 | 295827 | 333555 | 37,728 | Prophage associated | Thr |
| 4 | 351585 | 355290 | 3,705 | eaeH-containing island | |
| 5 | 608025 | 618523 | 10,498 | Uncharacterized genomic island | |
| 6 | 913695 | 949151 | 35,456 | Prophage associated | |
| 7 | 1003314 | 1040757 | 37,443 | Prophage associated | |
| 8 | 1185471 | 1278317 | 92,846 | Urease-tellurite island | Ser |
| 9 | 1364416 | 1406001 | 41,585 | Prophage associated | Met, Lys, Thr, Gly |
| 10 | 1517028 | 1528810 | 11,782 | Prophage associated | |
| 11 | 1616858 | 1672662 | 55,804 | Prophage associated | |
| 12 | 1768109 | 1775102 | 6,993 | Prophage associated | |
| 13 | 1834030 | 1854198 | 20,168 | Uncharacterized genomic island | |
| 14 | 1884155 | 1890440 | 6,285 | Uncharacterized genomic island | |
| 15 | 1905281 | 1927182 | 21901 | Prophage associated | |
| 16 | 1957083 | 1963679 | 6,596 | Uncharacterized genomic island | |
| 17 | 2315225 | 2319891 | 4,666 | Uncharacterized genomic island | |
| 18 | 2353020 | 2380821 | 27,801 | Prophage associated | Thr, Met, Ser |
| 19 | 2405296 | 2430621 | 25,325 | Prophage associated | |
| 20 | 2455693 | 2460757 | 5,064 | O149 O antigen cluster | |
| 21 | 2520153 | 2531275 | 11,122 | Putative sugar utilization/transport system | |
| 22 | 2815712 | 2856228 | 40,516 | Prophage associated | Arg |
| 23 | 3004819 | 3042924 | 38,105 | Prophage associated | Arg |
| 24 | 3167944 | 3204497 | 36,553 | Prophage associated | |
| 25 | 3306702 | 3310865 | 4,163 | CRISPR region check | |
| 26 | 3422930 | 3431136 | 8,206 | Truncated ETT2 type 3 secretion system | Gly |
| 27 | 3544009 | 3582918 | 38,909 | Prophage associated | Phe |
| 28 | 3584464 | 3593465 | 9,001 | Type 2 general secretory pathway gsp | |
| 29 | 3610527 | 3618919 | 8,392 | Uncharacterized genomic island | |
| 30 | 4050914 | 4054357 | 3,443 | Uncharacterized genomic island | |
| 31 | 4285617 | 4290215 | 4,598 | LPS biosynthesis genes | |
| 32 | 4303989 | 4320461 | 16,472 | Prophage associated | |
| 33 | 4518292 | 4526863 | 8,571 | Uncharacterized genomic island | |
| 34 | 4528662 | 4534261 | 5,599 | Uncharacterized genomic island | |
| 35 | 4728127 | 4732015 | 3,888 | Uncharacterized genomic island | Ile, Ala |
| 36 | 4748052 | 4754369 | 6,317 | PTS | |
| 37 | 4888719 | 4951420 | 62,701 | Putative integrative conjugative element | Phe |
| 38 | 4998329 | 5003463 | 5,134 | Uncharacterized genomic island | Gly |
| 39 | 5085662 | 5095934 | 10,272 | Uncharacterized genomic island | Leu |
| 40 | 5106623 | 5113315 | 6,692 | Uncharacterized genomic island |
CRISPR, clusters of regularly interspaced short palindromic repeats; LPS, lipopolysaccharide; PTS, phosphotransferase system.
Two UMNK88 islands contained novel carbohydrate utilization and transport systems. Island 21 in UMNK88 contained a number of genes predicted to be involved in the utilization and transport of ribitol, xylitol, and arabitol sugars and was also present in human ETEC strain H10407 (8). Island 38 of UMNK88 was also present in enteroaggregative E. coli strain 042 and contained an IclR-family transcriptional regulator adjacent to three novel putative carbohydrate transport and metabolism proteins. Overall, the chromosomes of UMNK88 and UMNF18 contained a number of potential virulence factors that remain to be characterized. However, as described below, much of their virulence-associated content was plasmid encoded.
Plasmids of UMNF18.
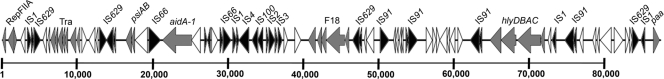
Both UMNK88 and UMNF18 possessed remarkable plasmid complements, each containing both resistance-associated and virulence-associated plasmids (Table 1). UMNF18 possessed five large plasmids, including those containing plasmid replicons similar to incompatibility groups IncFIIA, IncI1, and IncX. Most of the virulence-associated genes were located on a single virulence-associated IncFIIA-like plasmid, pUMNF18_87. Genes of interest in this 87-kb plasmid included those of the F18ac fimbrial operon (fedA to fedF), the aidA adhesin gene, the paa gene, which is capable of inducing attaching and effacing-associated lesions (1, 2), and the hemolysin-encoding hlyDBAC genes (Fig. 2). The AIDA-1 adhesin and F18 fimbriae were previously localized to the same plasmid (29), and AIDA-1 has been associated with Stx2e and the F18 adhesin in porcine strains implicated in postweaning diarrhea and edema disease (34). This plasmid also contained truncated remnants of an F transfer region and a mosaic of mobile genetic elements, suggesting that it is transfer deficient and might be constantly in flux due to insertion sequence-driven genetic recombination. Previous reports have identified F18 and K88 porcine ETEC virulence plasmids on multiple IncF-type plasmid replicons, suggesting that this particular plasmid may represent only one variant of F18 plasmids circulating among swine ETEC isolates (13, 28).
Fig 2.
Linear map of pUMNF18_87, containing the F18 operon. Gray shading indicates genes of known function associated with virulence or plasmid transfer/stability, and black shading indicates insertion sequence-associated genes. The scale is in base pairs.
UMNF18 contained a second IncFIIA-like plasmid, pUMNF18_IncFV, that was 103 kb (see Fig. S4 in the supplemental material). This plasmid is notable for several reasons. First, it contained a number of antimicrobial resistance-associated genes in a single module, including those encoding resistance to mercury (merR to merD), kanamycin (aph), bleomycin (ble), streptomycin (aadA2), trimethoprim (dfrA12), apramycin (aprR), hygromycin (hph), extended-spectrum beta-lactams (blaTEM-1), and silver (silP to silE). Many of these gene acquisitions have apparently involved IS26 elements, as they are embedded throughout the resistance-associated region. Considering that pUMNF18_69, another plasmid of UMNF18 belonging to the IncI1 incompatibility group, also harbored genes encoding resistance to streptomycin (aadA1 and aadA2) and chloramphenicol (cmlA) (22), the plasmid-encoded resistance repertoire in UMNF18 is remarkable. The second interesting aspect of pUMNF18_IncFV is that it contained the genes encoding colicins B (cbi and cba) and M (cmi and cma), which have previously been found only on plasmids of extraintestinal pathogenic E. coli (21, 41, 46). Finally, this plasmid possessed a unique transfer region similar to the F transfer region in terms of organization but with low amino acid sequence similarity to its closest match, the IncFV plasmid pED208 (27). Overall, pUMNF18_IncFV may play an important role in UMNF18's resistance phenotype, the production of colicins, and the propensity to disseminate its plasmids to other bacterial recipients.
As mentioned above, UMNF18 contains pUMNF18_IncI1, an IncI1 plasmid containing multiple antimicrobial resistance genes. IncI1 plasmids were previously found to be highly prevalent among porcine ETEC strains, although many apparently lack genes associated with drug resistance (22). Thus, their dissemination and persistence in these bacterial populations could be attributed to as-yet-unidentified advantages that they provide to the host. IncI1 plasmids have been shown to contribute to the adherence of atypical human enteroaggregative E. coli, suggesting that they could play similar roles in porcine ETEC (12). Finally, UMNF18 possessed two additional plasmids, pUMNF18_32 and pUMNF18_103. pUMNF18_32 is a plasmid belonging to the recently proposed IncX4 group and contains a type IV secretion system and H-NS-like transcriptional regulator typical of this plasmid group (T. J. Johnson et al., presented at the 4th Symposium on Antimicrobial Resistance in Animals and the Environment, Tours, France). pUMNF18_103 was most similar to E. coli bacteriophage P1, which lysogenizes its host as a circular, low-copy-number plasmid (25).
Plasmids of UMNK88.
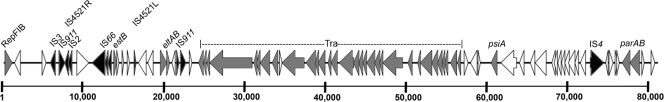
UMNK88 contains three virulence-associated plasmids (pUMNK88_Ent, pUMNK88_K88, and pUMNK88_Hly), an IncA/C multidrug resistance-encoding plasmid, and an IncI1 plasmid. pUMNK88_Ent was an 81-kb plasmid that contains the LT enterotoxin-encoding genes eltAB, surrounded by IS elements with similarity to ISEc18 and IS911 (Fig. 3). pUMNK88_Ent also contains the ST enterotoxin-encoding stb gene, and this region is surrounded by flanking IS4521 left and right junction elements and immediately surrounded within this element by a number of different transposase genes. This plasmid contains the IncFIB replicon and an intact F transfer region. The sequence of pUMNK88_Ent differed substantially from that of pEntH10407, which is an LT/STIa-encoding plasmid containing an IncFIIA replicon with a deficient transfer region (35).
Fig 3.
Linear map of pUMNK88_Ent, containing the eltAB (LT) and estB (STb) genes. Gray shading indicates genes of known function associated with virulence or plasmid transfer/stability, and black shading indicates insertion sequence-associated genes. The scale is in base pairs.
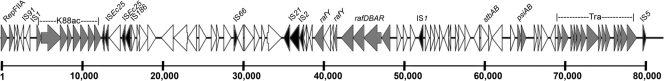
pUMNK88_K88 is an 81-kb plasmid containing the K88ac fimbrial operon and an operon encoding raffinose utilization, as previously described for K88-positive isolates (48) (Fig. 4). This plasmid contains an IncFIIA-like replicon and a truncated F transfer region. Downstream of the K88ac operon is a putative operon encoding an anion permease, amino acid dehydrogenase, and penicillin V acylase. This region and portions of the K88 operon were also found in pSE11-3 from human commensal E. coli strain SE11 (36). pUMNK88_K88 also contains a putative fimbrial operon downstream of the raffinose utilization operon with low amino acid similarity to the F1 capsule fimbrial-usher system of the Yersinia pestis virulence plasmid (47) and the Afa afimbrial adhesin gene clusters of pathogenic E. coli isolates (24). Interestingly, a putative transposon was identified that disrupted the rafY gene, which encodes the porin protein for raffinose utilization (54). However, UMNK88 was still able to utilize raffinose as a sole sugar on MacConkey agar base (data not shown). Overall, pUMNK88_K88 contains previously described virulence factors of porcine ETEC and a number of novel putative virulence factors warranting further study.
Fig 4.
Linear map of pUMNK88_K88, containing the K88ac operon. Gray shading indicates genes of known function associated with virulence or plasmid transfer/stability, and black shading indicates insertion sequence-associated genes. The scale is in base pairs.
pUMNK88_Hly is a 65-kb plasmid containing the hemolysin-encoding hlyCABD genes (see Fig. S5 in the supplemental material). This plasmid is of unknown Inc type and contains an intact transfer region and a mob mobilization region. The transfer region of pUMNK88_Hly most closely resembles that of pADAP, a virulence plasmid from Serratia entomophila associated with amber disease in grass grubs (17). Between the mob and tra regions of this plasmid is a gene product annotated as a putative invasin with an Ig-like domain typical of surface proteins such as intimins and invasins also found on the putative ICE of UMNK88. The role of this protein, if any, in porcine ETEC pathogenesis remains to be determined.
UMNK88 also contains a 161-kb IncA/C plasmid and 90-kb IncI1 plasmid, whose sequences were previously described (14, 22). The IncA/C plasmid (pUMNK88_IncA/C) contains a number of resistance-associated genes, including those encoding resistance to phenicols (floR and cmlA), sulfonamides (sul1 and sul2), aminoglycosides (strAB, aadA, and aadA2), and mercury (mer). IncA/C plasmids have been shown to be highly prevalent among disease-associated porcine ETEC strains, underscoring their importance in the dissemination of multidrug resistance among bacteria of production animals (22). The IncI1 plasmid of UMNK88 (pUMNK88_91) is a ColIa plasmid lacking any resistance-associated genes but containing typical core components of IncI1 plasmids.
Multilocus sequence analysis of K88- and F18-positive porcine ETEC strains.
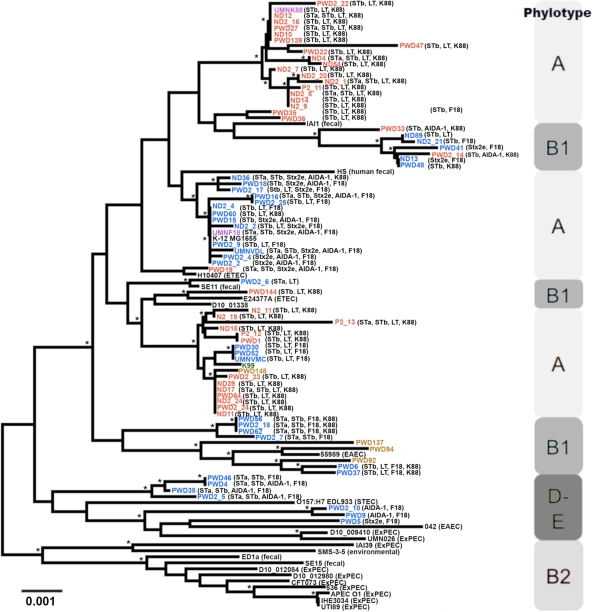
To determine the genetic relatedness of UMNF18 and UMNK88 to one another and additional K88- and F18-positive porcine ETEC isolates, multilocus sequence analysis was performed on these two strains and 76 additional porcine ETEC isolates recently implicated in postweaning and neonatal diarrhea in the midwestern United States. The resulting dendrogram, and the phylogenetic analysis of isolates using the scheme of Clermont et al. (6), revealed several clusters of isolates throughout phylogenetic groups A and B1 (Fig. 5). The K88-positive isolates fell into two primary clusters belonging to phylogenetic group A, whereas the F18-positive isolates fell into seven primary clusters belonging to phylogenetic groups A, B1, and D/E. Postweaning and neonatal diarrhea-associated isolates were mixed throughout these clusters, suggesting that there is no genetic distinction between isolates belonging to these disease groups.
Fig 5.
Multilocus sequence alignment of porcine ETEC isolates. Isolate designations are colored according to type (red, F18-positive isolates; blue, K88-positive isolates; purple, sequenced strains; black, reference strains; olive, non-F18 and non-K88). An asterisk indicates a bootstrap confidence value greater than 50%. Note that “PWD” and “ND” in isolate names do not necessarily reflect the actual age of the animal; they were used simply as designators.
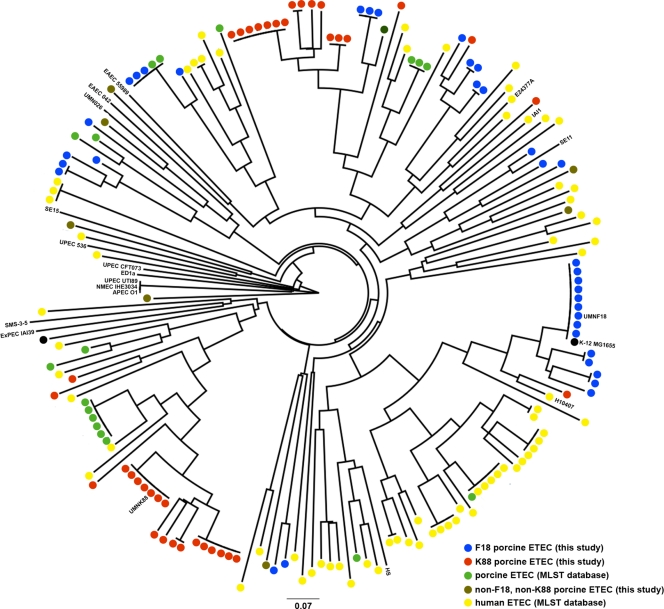
Using the E. coli MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli), allele types from these isolates were used to generate a dendrogram depicting similarities between the isolates in this study and human and porcine ETEC isolates available in the database (Fig. 6). Here, the monophyletic nature of some of the K88- and F18-containing groups was striking, suggesting that a limited number of highly similar chromosomal backgrounds are implicated in K88- and F18-associated porcine diarrhea. This is in contrast to human ETEC strains, which have been shown to arise in a large number of different chromosomal lineages (53). It also appears that the F18-positive ETEC isolates were more diverse than the K88-positive ETEC isolates with respect to chromosomal background. Overall, these results are suggestive of different plausible scenarios. One such scenario is that while the plasmids of porcine ETEC strains play a critical role in the pathogenesis of these organisms, a specific core genome is also required for it to be successful, as has been described for human ETEC strains (45). A second scenario is that the successful transfer and stability of these plasmids is limited to a subset of E. coli lineages. It has been suggested that both human and porcine ETEC likely emerged from similar E. coli lineages, with horizontal gene transfer events driving host adaptation (49). Our results to some extent support this hypothesis, since clusters of isolates are present from an apparent ancestral strain that have diverged into separate ETEC types (Fig. 6). For example, UMNK88 belongs to a cluster of K88-positive isolates that are distinct from other subsets of porcine and human ETEC isolates in the database but likely share an ancestry. Because isolates belonging to the same dendrogram clusters mostly contained similar toxin profiles (Fig. 5), it is likely that toxin acquisition by an E. coli lineage was an initial event in the evolution of ETEC, followed by host-specific fimbria acquisition mediated by horizontal gene transfer. Further dissection of the chromosomal traits required for porcine ETEC pathogenesis and of the nature of porcine ETEC plasmid dissemination and stability in different E. coli lineages is required to address these possibilities.
Fig 6.
Dendrogram based on allele types of porcine ETEC isolates examined in this study and porcine and human ETEC isolates available through the Escherichia coli MLST database (http://mlst.ucc.ie/mlst/dbs/Ecoli). Neighbor joining based on allele types was performed using tools available at http://pubmlst.org/analysis/.
UMNK88 belonged to the ST165 complex, and UMNF18 belonged to the ST10 complex, which also includes E. coli K-12 MG1655 (Fig. 6). Based on our results, these complexes appear to be two primary chromosomal lineages for K88-positive and F18-positive strains, respectively. A third clonal complex, ST23, contained both K88-positive and F18-positive strains. Together, the majority of the porcine ETEC isolates examined belonged to these three clonal complexes. A concern with the association of most porcine ETEC strains examined with the ST10 and ST23 complexes is that these complexes have been shown to be strongly associated with certain resistance-associated elements, such as AmpC-type beta-lactamases, NDM-type carbapenemases, and other extended-spectrum beta lactamases (7, 9, 32, 37). Therefore, many porcine ETEC isolates may have an increased propensity to acquire and maintain these resistance-associated elements, as demonstrated by the genomes of UMNK88 and UMNF18.
Genotyping of porcine ETEC for plasmid-associated genes.
The presence of 25 genes of interest found in the plasmids of UMNK88 and UMNF18 was sought among a collection of 172 postweaning-diarrhea- and neonatal-diarrhea-associated porcine E. coli isolates (Table 2). Genes selected included known porcine ETEC virulence factors, putative virulence factors, and replication or transfer genes serving as markers of particular plasmid types. Hierarchical clustering displaying these data revealed that the isolates clustered into three major groups according to possession of the K88 and F18 fimbrial operons (see Fig. S6 in the supplemental material). Some expected correlations among genes examined were observed, such as between aidA and its associated transcriptional regulator, between fedD and fedF, between the toxin-encoding genes and transcriptional regulator of pUMNK88_Ent, and between genes of pUMNK88_K88. Some other observations were noteworthy, including the presence of aidA among about 50% of the F18-positive strains but none of the K88-positive strains examined; the presence of rafY among many nearly 80% of the isolates examined, with an apparently disrupted rafY in most of the K88-positive isolates examined; the positive correlation between aidA-STb, as previously demonstrated (11); and the negative correlation between aidA and LT. Together, these data confirm that homogeneity exists with regard to plasmid-associated gene content among K88-positive and F18-positive isolates, although these genes might exist on a variety of different plasmid variants and coreside with other virulence factors and plasmid replicons.
Conclusions.
While many of the virulence factors implicated in porcine ETEC pathogenesis have been well studied, this work demonstrates that as-yet-uncharacterized genomic traits exist on the chromosomes and plasmids of porcine ETEC that might play a role in their pathogenesis and may possess potential as future vaccine candidates. Porcine ETEC appear to be limited to a subset of E. coli lineages that either have an increased propensity to carry plasmid-encoded virulence factors or have the appropriate ETEC core genome required for virulence. Future studies based on the sequences and data generated here are needed to address these possibilities.
Supplementary Material
ACKNOWLEDGMENTS
We thank the University of Minnesota Veterinary Diagnostic Laboratory and Simone Oliveira for providing strains for this study. This work was carried out in part using computing resources at the University of Minnesota Supercomputing Institute. This project was funded in part by the Minnesota Pork Board and by Science Foundation of Ireland grant 05/FE1/B882. We gratefully acknowledge the curation of the MLST database by Vimalkumar Velayudhan and serotyping of strains by Chitrita Debroy at the Penn State University E. coli Reference Center.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. An H, Fairbrother JM, Desautels C, Harel J. 1999. Distribution of a novel locus called Paa (porcine attaching and effacing associated) among enteric Escherichia coli. Adv. Exp. Med. Biol. 473: 179–184 [DOI] [PubMed] [Google Scholar]
- 2. Batisson I, et al. 2003. Characterization of the novel factor paa involved in the early steps of the adhesion mechanism of attaching and effacing Escherichia coli. Infect. Immun. 71: 4516–4525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29: 2607–2618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11: 3–8 [DOI] [PubMed] [Google Scholar]
- 5. Blattner F, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462 [DOI] [PubMed] [Google Scholar]
- 6. Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66: 4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cremet L, et al. 2010. Occurrence of ST23 complex phylogroup A Escherichia coli isolates producing extended-spectrum AmpC beta-lactamase in a French hospital. Antimicrob. Agents Chemother. 54: 2216–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crossman LC, et al. 2010. A commensal gone bad: complete genome sequence of the prototypical enterotoxigenic Escherichia coli strain H10407. J. Bacteriol. 192: 5822–5831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Curiao T, et al. 2011. Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-beta-lactamase-producing Escherichia coli clones from humans. Antimicrob. Agents Chemother. 55: 2451–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. DebRoy C, Maddox CW. 2001. Identification of virulence attributes of gastrointestinal Escherichia coli isolates of veterinary significance. Anim. Health Res. Rev. 2: 129–140 [PubMed] [Google Scholar]
- 11. Dubreuil JD. 2010. STb and AIDA-I: the missing link? Crit. Rev. Microbiol. 36: 212–220 [DOI] [PubMed] [Google Scholar]
- 12. Dudley EG, et al. 2006. An IncI1 plasmid contributes to the adherence of the atypical enteroaggregative Escherichia coli strain C1096 to cultured cells and abiotic surfaces. Infect. Immun. 74: 2102–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fekete PZ, Gerardin J, Jacquemin E, Mainil JG, Nagy B. 2002. Replicon typing of F18 fimbriae encoding plasmids of enterotoxigenic and verotoxigenic Escherichia coli strains from porcine postweaning diarrhoea and oedema disease. Vet. Microbiol. 85: 275–284 [DOI] [PubMed] [Google Scholar]
- 14. Fernandez-Alarcon C, Singer RS, Johnson TJ. 2011. Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS One 6: e23415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finn RD, et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38: D211–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helgerson AF, et al. 2006. Edema disease caused by a clone of Escherichia coli O147. J. Clin. Microbiol. 44: 3074–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hurst MR, O'Callaghan M, Glare TR. 2003. Peripheral sequences of the Serratia entomophila pADAP virulence-associated region. Plasmid 50: 213–229 [DOI] [PubMed] [Google Scholar]
- 18. Ideses D, et al. 2005. A degenerate type III secretion system from septicemic Escherichia coli contributes to pathogenesis. J. Bacteriol. 187: 8164–8171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181: 261–272 [DOI] [PubMed] [Google Scholar]
- 20. Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson TJ, Johnson SJ, Nolan LK. 2006. Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids. J. Bacteriol. 188: 5975–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson TJ, Shepard SM, Rivet B, Danzeisen JL, Carattoli A. 2011. Comparative genomics and phylogeny of the IncI1 plasmids, a common plasmid type among porcine enterotoxigenic Escherichia coli. Plasmid. 66: 144–151 [DOI] [PubMed] [Google Scholar]
- 23. Lagesen K, et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: 3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Le Bouguenec C, Bertin Y. 1999. AFA and F17 adhesins produced by pathogenic Escherichia coli strains in domestic animals. Vet. Res. 30: 317–342 [PubMed] [Google Scholar]
- 25. Lobocka MB, et al. 2004. Genome of bacteriophage P1. J. Bacteriol. 186: 7032–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu J, et al. 2002. Analysis and characterization of the IncFV plasmid pED208 transfer region. Plasmid 48: 24–37 [DOI] [PubMed] [Google Scholar]
- 28. Mainil JG, Daube G, Jacquemin E, Pohl P, Kaeckenbeeck A. 1998. Virulence plasmids of enterotoxigenic Escherichia coli isolates from piglets. Vet. Microbiol. 62: 291–301 [DOI] [PubMed] [Google Scholar]
- 29. Mainil JG, Jacquemin E, Pohl P, Kaeckenbeeck A, Benz I. 2002. DNA sequences coding for the F18 fimbriae and AIDA adhesin are localised on the same plasmid in Escherichia coli isolates from piglets. Vet. Microbiol. 86: 303–311 [DOI] [PubMed] [Google Scholar]
- 30. Marchler-Bauer A, et al. 2011. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39: D225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moxley RA. 2000. Edema disease. Vet. Clin. North Am. Food Anim. Pract. 16: 175–185 [DOI] [PubMed] [Google Scholar]
- 32. Mushtaq S, et al. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J. Antimicrob. Chemother. [DOI] [PubMed] [Google Scholar]
- 33. Nagy B, Fekete PZ. 2005. Enterotoxigenic Escherichia coli in veterinary medicine. Int. J. Med. Microbiol. 295: 443–454 [DOI] [PubMed] [Google Scholar]
- 34. Niewerth U, et al. 2001. The AIDA autotransporter system is associated with F18 and stx2e in Escherichia coli isolates from pigs diagnosed with edema disease and postweaning diarrhea. Clin. Diagn. Lab. Immunol. 8: 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ochi S, et al. 2009. Nucleotide sequence analysis of the enterotoxigenic Escherichia coli Ent plasmid. DNA Res. 16: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oshima K, et al. 2008. Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 15: 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Oteo J, et al. 2009. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int. J. Antimicrob. Agents 34: 173–176 [DOI] [PubMed] [Google Scholar]
- 38. Overbeek R, et al. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33: 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parreira VR, Liao JH, Kim SH, Gyles CL. 2008. A homolog of the O157 urease-encoding O island 48 is present in porcine O149:H10 enterotoxigenic Escherichia coli. Vet. Res. 39: 38. [DOI] [PubMed] [Google Scholar]
- 40. Paulsen IT, et al. 2005. Complete genome sequence of the plant commensal Pseudomonas fluorescens Pf-5. Nat. Biotechnol. 23: 873–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Peigne C, et al. 2009. The plasmid of neonatal meningitis Escherichia coli strain S88 (O45:K1:H7) is closely related to avian pathogenic E. coli plasmids and is associated with high level bacteremia in neonatal rat meningitis model. Infect. Immun. 77: 2272–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rasko DA, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190: 6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ren CP, et al. 2004. The ETT2 gene cluster, encoding a second type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186: 3547–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rodriguez-Siek KE, Giddings CW, Doetkott C, Johnson TJ, Nolan LK. 2005. Characterizing the APEC pathotype. Vet. Res. 36: 241–256 [DOI] [PubMed] [Google Scholar]
- 45. Sahl JW, et al. 2011. A comparative genomic analysis of diverse clonal types of enterotoxigenic Escherichia coli reveals pathovar-specific conservation. Infect. Immun. 79: 950–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sasarman A, et al. 1980. Naturally occurring R. ColBM plasmids belonging to the IncFIII incompatibility group. J. Gen. Microbiol. 119: 475–483 [DOI] [PubMed] [Google Scholar]
- 47. Sebbane F, Jarrett C, Gardner D, Long D, Hinnebusch BJ. 2009. The Yersinia pestis caf1M1A1 fimbrial capsule operon promotes transmission by flea bite in a mouse model of bubonic plague. Infect. Immun. 77: 1222–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shipley PL, Gyles CL, Falkow S. 1978. Characterization of plasmids that encode for the K88 colonization antigen. Infect. Immun. 20: 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Steinsland H, Lacher DW, Sommerfelt H, Whittam TS. 2010. Ancestral lineages of human enterotoxigenic Escherichia coli. J. Clin. Microbiol. 48: 2916–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- 51. Taylor DE, et al. 2002. Genomic variability of O islands encoding tellurite resistance in enterohemorrhagic Escherichia coli O157:H7 isolates. J. Bacteriol. 184: 4690–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tettelin H, Radune D, Kasif S, Khouri H, Salzberg SL. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62: 500–507 [DOI] [PubMed] [Google Scholar]
- 53. Turner SM, et al. 2006. Phylogenetic comparisons reveal multiple acquisitions of the toxin genes by enterotoxigenic Escherichia coli strains of different evolutionary lineages. J. Clin. Microbiol. 44: 4528–4536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ulmke C, Lengeler JW, Schmid K. 1997. Identification of a new porin, RafY, encoded by raffinose plasmid pRSD2 of Escherichia coli. J. Bacteriol. 179: 5783–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wirth T, et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60: 1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yin X, et al. 2009. Contributions of O island 48 to adherence of enterohemorrhagic Escherichia coli O157:H7 to epithelial cells in vitro and in ligated pig ileal loops. Appl. Environ. Microbiol. 75: 5779–5786 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.