Abstract
Miniature inverted terminal repeat elements (MITEs) are nonautonomous mobile elements that have a significant impact on bacterial evolution. Here we characterize E622, a 611-bp virulence-associated MITE from Pseudomonas syringae, which contains no coding region but has almost perfect 168-bp inverted repeats. Using an antibiotic coupling assay, we show that E622 is transposable and can mobilize an antibiotic resistance gene contained between its borders. Its predicted parent element, designated TnE622, has a typical transposon structure with a three-gene operon, consisting of resolvase, integrase, and exeA-like genes, which is bounded by the same terminal inverted repeats as E622. A broader genome level survey of the E622/TnE622 inverted repeats identified homologs in Pseudomonas, Salmonella, Shewanella, Erwinia, Pantoea, and the cyanobacteria Nostoc and Cyanothece, many of which appear to encompass known virulence genes, including genes encoding toxins, enzymes, and type III secreted effectors. Its association with niche-specific genetic determinants, along with its persistence and evolutionary diversification, indicates that this mobile element family has played a prominent role in the evolution of many agriculturally and clinically relevant pathogenic bacteria.
INTRODUCTION
Transposons and other mobile genetic elements are prevalent within bacterial genomes. These selfish genes promote rearrangements and facilitate assimilation of new genetic information, thereby providing bacteria with enormous evolutionary potential (27, 51). One of the smallest and simplest autonomous prokaryotic mobile elements is the insertion sequence (IS), which contains a single transposase gene bordered by inverted repeats (IRs) (42, 43). The transposase enzyme binds the inverted repeats and catalyzes excision and subsequent integration of the IS element, in either a replicative or nonreplicative manner (30). Transposases recognize a specific target sequence, with integration resulting in target sequence duplication ranging from 2 bp to 10 bp (14). The incorporation of functional gene cassettes within these IS element borders results in a transposon, which may confer specific capabilities on its host, including those related to resistance, virulence, and catabolism (27, 51).
More recently, even smaller genetic repeat elements have been identified in bacteria, which are analogous to the miniature inverted terminal repeat elements (MITEs) found in eukaryotic genomes (72). Like eukaryotic MITEs, bacterial MITEs are nonautonomous elements that are usually under 200 bp and lack their own transposase but retain IRs and the ability to be mobilized in trans by coresident transposases (75). Integration of MITEs, much like that of IS elements, is often sequence specific and results in a target site duplication (19). It has been suggested that MITEs fall into two types, either type I or type II, based on their predicted origin (4). Type I MITEs retain their IRs but lack an internal transposase gene and are proposed to have originated from an IS element whose transposase has undergone deletion (4, 23). Transposition of these elements is catalyzed by the transposase enzyme of the coresident parent IS element, which shares homologous IRs. Type II MITEs have IRs that share some similarity to the IRs of existing IS elements due to random convergence and are likely to be transactivated by transposases that recognize similar IR sequences (4). Although few MITEs have been identified in bacteria, some have been well studied, including the enterobacterial repetitive intergenic consensus (ERIC) sequences found in Yersinia spp. (18), the Neisseria miniature insertion sequences (NEMIS) in Neisseria, the repeat unit of pneumococcus (RUP) element found in Streptococcus (49), and the bcr1 element found in Bacillus (50). Other bacterial MITEs include a putative type I MITE in Porphyromonas (48) and a type II MITE in Deinococcus radiodurans (47). MITEs have also been identified in the cyanobacterial Microcystis (74), Nostoc, and Anabaena (34), as well as in the archaeal species Sulfolobus solfataricus (54) and Methanococcus jannaschii (8).
The presence of MITEs in a genome has broad implications for niche-specific adaptation. Mobile MITEs have resulted in the disruption of virulence genes, such as the Ralstonia solanacearum type III secreted effector (T3SE) gene, avrA (57), thereby functioning to modulate virulence in natural populations. Similarly, MITEs have been implicated in the disruption of microcystin toxin biosynthetic genes of Anabaena, contributing to the formation of non-toxin-producing populations in natural cyanobacterial communities (24). It has also been suggested that MITEs contribute to the movement of genes both within a chromosome and between the chromosome and coresident plasmids, driving genome rearrangements and increasing genetic diversity. In some cases, MITEs, like transposons, may carry passenger genes, which can provide a selective, niche-specific advantage to their hosts. The mobile insertion cassette MIC231 from Bacillus carries a fosfomycin resistance gene within its borders (11, 21), while a MITE in Pseudomonas putida carries the PheBA operon of Pseudomonas putida, which encodes phenol-degrading capabilities (53). These miniature elements, however, also have the capacity to influence the posttranslational processing of neighboring genes, as with the NEMIS/Correia elements of Neisseria (7, 13, 17, 41, 45, 59), or promote large-scale genomic rearrangements, as with the MITEs of Sulfolobus (5). Despite their importance in bacterial evolution, many MITEs have gone unnoticed.
In this study, we characterize a new MITE-like element from the plant pathogen Pseudomonas syringae pv. maculicola ES4326 (PmaES4326), originally identified during a comparative genomic analysis of its native plasmids (62). The 611-bp sequence, named E622, has perfect IRs, lacks an obvious transposase enzyme, and is found in different genomic contexts in three of the five plasmids within this strain (62). The pPMA4326D and pPMA4326E plasmids were identical in genetic content and arrangement, except for a difference of 627 bp, 617 bases of which are accounted for by the presence E622 (62). On the pPMA4326B plasmid, the IRs were found flanking the virulence-associated T3SE gene hopW1-1PmaES4326 (formerly hopPmaA) (40) and a three-gene operon consisting of resolvase, integrase, and exeA-like (RIE) genes; it was previously suggested that this element resembles the Tn3 family of transposons (62). In this study, we determined that E622 is a mobile type I MITE, likely derived from and mobilized by the larger RIE gene-containing transposon-like element, which we designated TnE622. We show that the E622 element and its predicted TnE622 parent have diversified into three distinct subgroups and are frequently associated with virulence-associated genes in both agriculturally and clinically relevant bacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Pseudomonas syringae was grown overnight with continuous shaking in 5 ml King's medium B at 30°C. Escherichia coli bacteria were grown in 5 ml Luria-Bertani medium at 37°C. Antibiotics were supplemented in the following concentrations: streptomycin, 100 μg ml−1; kanamycin, 50 μg ml−1; ampicillin, 150 μg ml−1; tetracycline, 30 μg ml−1.
Construction of ampicillin-labeled E622.
The E622 element was amplified from the pPMA4326C plasmid of ES4326 using primers E622+ (5′-CAAGGGTTGCCAACTTTGCACG-3′) and E622− (5′-AGGTGCACAGATCGCGTAGCAAG-3′) and cloned into the XbaI site of the integration vector pCKTR (28). The resulting construct, pCKTR-E, was then linearized with MluI, which cuts once at base 210 of E622. The bla gene was amplified from Bluescript using bla+ (5′-AATGTGCGCGGAACCCCTATTTG-3′) and bla− (5′-CGAAAACTCACGTTAAGGGATTTTGG-3′) and ligated into pCKTR-E to yield pCKTR-Ea. All constructs were created and maintained in DH5a (Table 1).
Table 1.
Bacterial strains and constructs
| Strain or plasmid | Relevant characteristicsa | Reference(s) or source |
|---|---|---|
| Strains | ||
| E. coli DH5a | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169recAlendAIhsdR17 (rK− mK+) supE44 λ−thi-1gyrA96relAl | |
| P. syringae pv. maculicola ES4326 | Wild type; Smr | 16 |
| P. syringae pv. maculicola YM7930 | Wild type | MAFF |
| P. syringae pv. thea K93001 | Wild type | MAFF |
| Plasmids | ||
| pCKTR | pBluescript derivative with recA internal fragment; Kmr | 28 |
| pCKTR-E | pCKTR derivative containing E622; Kmr | This study |
| pCKTR-Ea | pCKTR-E derivative with E622::bla; Kmr Apr | This study |
| pLAFR | Broad host range cosmid; Tcr | 15, 70 |
| pLAFR-Ea | pLAFR derivative containing transposed E622::Apr | This study |
| pUCP20Tk | Kmr derivative of pUCP20T (73) | This study |
| pUCP20Tk-Ea | pUCP20Tk containing pLAFR-Ea fragment; Apr Kmr | This study |
Abbreviations: Sm, streptomycin; Tc, tetracycline; Ap, ampicillin; Km, kanamycin.
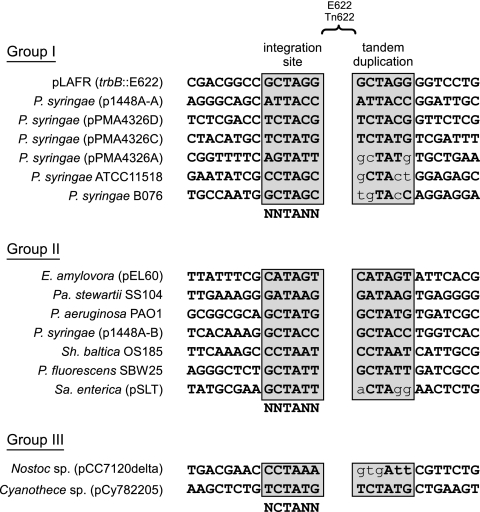
To establish experimentally whether E622 was transpositionally active, we designed and performed an antibiotic coupling mobility assay, which involved tagging E622 with an antibiotic marker and determining whether it could transpose into a plasmid tagged with a second resistance gene. We first cloned E622 into a suicide vector and inserted the beta-lactamase gene (bla) conferring ampicillin resistance into the center of E622. The resulting construct was integrated into the genome of a close relative of PmaES4326, P. syringae pv. maculicola YM7930. Because this strain carries only the pPMA4326B-like plasmid, which has the RIE element, there were far fewer copies of E622 that could complicate integration. The self-replicating cosmid pLAFR, carrying tetracycline resistance, was then introduced into the same line, and if E622 was mobile, it would occasionally integrate into pLAFR, coupling ampicillin (E622) and tetracycline (pLAFR) resistance. Three transformants were recovered, and following restriction mapping, cloning, and sequencing, E622 integration into pLAFR was confirmed in two of the three isolates. Sequence analysis of the E622 integration site suggested that excision of E622 from the chromosome occurred perfectly at the IRs, with subsequent integration in the trbB gene on pLAFR. The putative integration site was determined to be CGCTAGG, the last 6 bases of which (GCTAGG) were duplicated on the 3′ end (Fig. 2). The two recovered isolates had identical integration sites, suggesting they represented the same clone.
Fig 2.
Alignment of the flanking regions of E622/TnE622 for each of the major groups. The integration sites and tandem duplications, which are separated by a gap representing the location of E622 or TnE622, are boxed and highlighted in gray. The integration consensus sequence for each group is shown beneath all the integration sequences. Mismatches between the integration sequence and the tandem duplication are shown in lowercase. The first sequence in group I shows the integration point and tandem duplication for the antibiotic coupling assay. All accession numbers are available in Table S1 in the supplemental material.
Antibiotic coupling mobility assay.
The mobility assay was conducted in a close relative of PmaES4326, P. syringae pv. maculicola YM7930 (PmaYM7930), because fewer plasmids and thus fewer E622 elements could serve as competing target sites for integration. PmaYM7930 has only the pPMA4326B-like plasmid, which carries TnE622, which is almost identical to that of PmaES4326. The pCKTR-Ea construct was electroporated into PmaYM7930 using a Bio-Rad Electropulser at 2.5 kV, 600 Ω, and 25 μF, with 0.2-cm cuvettes. Transformants were selected on kanamycin and ampicillin, although integration into the recA locus could not be confirmed. The cosmid pLAFR, conferring tetracycline resistance (15, 70), was introduced into one of the recombinants via electroporation using the same conditions as above, with transformant selection being achieved on kanamycin, ampicillin, and tetracycline. The resulting line was grown overnight in a standard 5-ml culture with antibiotics, and plasmid extraction was performed from 1.5 ml of culture using the TENS method (76). Plasmid extracts were transformed into chemically competent DH5a, with selection of transformants on tetracycline and ampicillin. Three colonies were obtained, and following incubation for 1 to 2 days, transformants were cultured and grown overnight. Plasmids were extracted from transformants using the TENS method (76), and integration of E622-amp was confirmed by PCR using internal primers. The approximate location of E622 integration was determined through restriction digestion with BsaAI. The region surrounding the E622 integration site was identified through partial digestion with HincII, followed by shotgun cloning of the fragments into pUCP20Tk (Kmr), with subsequent selection on kanamycin-ampicillin. Sequencing outward from E622-amp to the pUCP20Tk backbone led to the identification of the integration point in pLAFR. pLAFR-specific primers pLAFR+10625 (5′-TGGTCAGACGGAACGGAACAGC-3′) and pLAFR−10844 (5′-CGGCTCGCCATGCTTATCAG-3′) were designed to cross the integration site of the E622-amp cassette. Two of the three colonies were shown to have the same site of integration into pLAFR; the third was a false positive.
E622 island PCR and sequencing.
E622 islands were PCR amplified in 50-μl reaction mixtures containing 0.2 μM (each) E622F (5′-TTGCGTCTATCCGAGCCAAC-3′) and E622R (5′-CCTGCAATCGAAATGGCAC-3′) primers, 2.5 U of High Fidelity PCR enzyme mix (Fermentas, Burlington, Canada), and approximately 100 ng of plasmid DNA using the following cycling conditions: 1 cycle of 94°C for 3 min; 10 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 8 min; 25 cycles of 95°C for 30 s, 55°C for 30 s, and 68°C for 8 min plus 10 s/cycle); and 1 cycle of 68°C for 10 min. Gel purification of E622 islands was achieved with the QIAquick PCR purification kit (Qiagen Inc., Valencia, CA) from 1% Tris-borate-EDTA (TBE)–agarose gels run at 8 V/cm. Gel-purified amplicons were enzymatically digested with HincII, BsaAI, and MspAI (New England BioLabs, Beverly, MA), and, following purification with the QIAquick PCR purification kit, fragments were shotgun cloned into Bluescript. Colonies were picked with toothpicks and dipped into a 25-μl PCR mixture containing 0.1 μM M13F (5′-GTAAAACGACGGCCAGT-3′) and 0.1 μM M13R (5′-CAGGAAACAGCTATGACCATG-3′). Cycling conditions were as follows: 1 cycle of 94°C for 5 min; 40 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2.5 min; and 1 cycle of 72°C for 5 min. Fragments larger than 600 bp were cleaned enzymatically with 0.2 μl calf intestinal phosphatase and 0.2 μl exonuclease (New England BioLabs, Beverly, MA) and incubated at 37°C for 30 min, followed by 85°C for 15 min. Sequencing was performed with CEQ Dye Terminator cycle sequencing (DTCS) Quick Start kit (Beckman Coulter Canada, Inc., Mississauga, ON, Canada) on a Beckman CEQ2000 XL sequencer. A 10-μl cycle sequencing reaction mixture contained 2.5 μl PCR product, 2.5 μl Beckman DTCS mix, 1.5 μl 1× PCR buffer with 1.5 mM MgCl2, 3 μl water, and either 0.5 μl (0.5 μM) T3 primer (5′-GCCAAGCTCAGAATTAACCCTCACTAAAGG-3′) or 0.5 μl (0.5 μM) T7 primer (5′-CGACGGCCAGTGAATTGTAATACGACTC-3′). Cycling conditions were 96°C for 20 s, 55°C for 20 s, and 60°C for 4 min, repeated 55 times. Following ethanol precipitation, products were resuspended in Beckman sample loading solution. Chromatogram sequence data were viewed and edited using BioEdit (version 7.0.5) and were subsequently imported into the contig assembly program Sequencher (Gene Codes Corp., Ann Arbor, MI). Contigs were connected by primer amplification from contig ends. A minimum of 3-fold coverage was obtained for all regions.
Comparative genomics and phylogenetic techniques.
E622 from pPMA4326C was compared against the nr database using BLASTN, and approximately 3 kb flanking both sides of each hit was extracted. Each fragment was subsequently BLAST searched and reannotated to include both open reading frames (ORFs) and pseudogenes. More-divergent borders of E622 were identified through a discontinuous MEGABLAST search of the P. syringae and Pantoea stewartii/Erwinia amylovora homologs. To initiate phylogenetic analyses, both incomplete and complete IRs were first aligned using ClustalX 1.83 (67) and neighbor-joining and maximum-likelihood trees were made in MEGA 4 (39) using Maximum Composite Likelihood, with 1,000 bootstrap replicates. Inverted repeats of each E622 element were determined with EINVERTED from EMBOSS (55).
Nucleotide sequence accession number.
E622 island sequences have been deposited under GenBank accession number JN581999.
RESULTS
Comparative genomics and antibiotic coupling assay indicate E622 mobility.
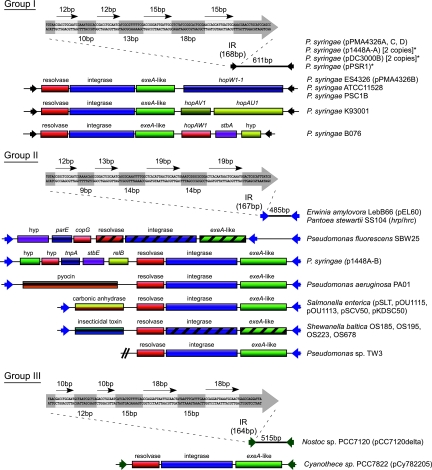
E622 resembles an IS element, having almost perfect IRs of 168 bp, each of which contains at least two sets of direct repeats (Fig. 1). The IRs have no significant sequence identity to those of any described IS element family, and unlike IS elements, the central region of E622 contains no open reading frame, although a small region exhibits weak similarity (E = 0.019) to the coding sequence for 50 amino acids in the C-terminal end of the TnpA family of site-specific resolvases. Because of these features, and the fact that it was found in a distinct genomic context on each of the plasmids of ES4326, we looked for evidence that E622 may have been recently mobilized. We took a comparative approach and examined the plasmids of the close relative Pseudomonas syringae pv. cilantro 0788-9 (Pci0788-9) and focused on the plasmids in this strain that were homologous to the E622-containing pPMA4326A and pPMA4326C plasmids of PmaES4326. We found that E622 was present in the pPMA4326A-like plasmid of Pci0788-9 but was absent from the corresponding region in the pPMA4326C-like plasmid. The amplicon generated for the pPMA4326C-like plasmid of Pci0788-9 was approximately 600 bp shorter than that obtained for pPMA4326C, with subsequent sequencing revealing that the homologous regions on the two plasmids differed by an insertion/deletion (indel) of exactly 617 bp. The 617-bp indel began and ended with the E622 IRs (611 bp), with an additional 6 bases at the 3′ end that corresponded to a duplication of the predicted integration sequence. This suggested that E622 could be mobile.
Fig 1.
Genetic organization of the three major groups of E622/TnE622s. For all three groups, the E622 MITE is shown at the top, represented as a line with two arrowheads. The first 120 bases of one of the inverted repeats (IRs) are shown in a thick arrow; the four direct repeats are indicated by four thin arrows, and their nucleotide sequences are highlighted (light gray shading). The genetic content and organization of the related TnE622 islands are shown beneath each MITE. The group I E622/TnE622 elements contain the prototypical E622 element (611 bp), which appears to be restricted to P. syringae and is found mostly on plasmids. Some E622 elements identified are truncated and disrupted (asterisks). The group I TnE622 islands possess the resolvase-integrase-exeA-like (RIE) gene cassette between their IRs (black arrowheads), along with various genes, including type III effector genes (hopW1-1, hopAV1, hopAU1, and hopAW1) that are localized toward the 3′ end of the exeA-like gene. The group II E622 element, shown as a line with blue arrowheads, is 485 bp and is found in the phytopathogens Pantoea stewartii and Erwinia amylovora. The related group II TnE622 islands are represented in P. syringae, as well as in the human-pathogenic Pseudomonas aeruginosa and Salmonella enterica and the environmental species Shewanella baltica. Genes involved in virulence (pyocin, carbonic anhydrase, and insecticidal toxin genes) are passenger genes on the group II elements and are localized toward the 5′ end of the resolvase gene. Only a partial sequence for the Pseudomonas sp. TW3 TnE622 island is available, with the missing portion being shown as a double slash. The group III elements are represented only in the cyanobacteria, with the 515-bp E622 MITE being present in Nostoc sp. PCC7120 (shown as a line with green arrowheads). The only group III TnE622 island is present in Cyanothece spp., and it is the only element that lacks any genes other than the RIE cassette between its borders. Hatching indicates pseudogenes. All accession numbers are available in Table S1 in the supplemental material. hyp, hypothetical protein.
E622 is prevalent among P. syringae strains and is associated with virulence genes and RIE.
A nucleotide BLAST search for E622 against the three completed P. syringae genomes and all completed P. syringae plasmids revealed numerous homologs and remnants. E622 and its borders were identified in both P. syringae pv. tomato DC3000 (6) and P. syringae pv. phaseolicola 1448A (37), but not in the plasmid-less strain P. syringae pv. syringae B728A (22). Of the 11 fully sequenced and completed plasmid genomes, E622 was absent only from pFKN (58) and pPMA4326E (62). An analysis of the genome context of E622 and its IRs in all completed plasmids and chromosomes revealed frequent linkage with various T3SE genes, including avrB4, avrRps4, hopD1, schF-hopF1, hopO1-1, hopQ1-2, hopQ1-1, hopT1-1, hopW1-1, hopW1-2, hopZ1, hopAB1, hopAB3, hopAJ1, hopAQ1, hopAV1, and hopAW1 (40) (Fig. 1; see Fig. S1 in the supplemental material). Several E622 homologs are also located adjacent to genes involved in coronatine biosynthesis, and almost all E622 homologs are flanked by a diversity of full-length and truncated IS elements (see Fig. S1 in the supplemental material). One plasmid-carried region within P. syringae pv. phaseolicola 1449B and P. syringae pv. phaseolicola 1448A contains IRs oriented toward two distinct T3SE genes, with a third T3SE gene being located in between (see Fig. S1 in the supplemental material).
In several instances, T3SE genes are found adjacent to pseudogenes of the RIE operon. A pseudogene of the exeA-like gene is adjacent to the hopAB1 T3SE gene in P. syringae pv. syringae B728a, although no IR is present (see Fig. S1 in the supplemental material). This arrangement is similar to that found on the P. syringae plasmids pAV511, p1448A-A, and pAV520, with the IR being adjacent to pseudogenes of both the integrase and exeA-like genes, which in turn are adjacent to the hopAB3 T3SE gene (35, 68). Numerous IRs are located adjacent to pseudogenes of the exeA-like gene in the genome of PtoDC3000, and all of these are near or adjacent to virulence genes (see Fig. S1 in the supplemental material).
A BLAST search of the P. syringae and Pseudomonas savastanoi draft genomes revealed the presence of E622 borders in P. syringae pv. tabaci ATCC 11528 (Pta11528), P. syringae pv. glycinea B076 (PgyB076), P. syringae pv. tomato T1 (PtoT1), P. syringae pv. oryzae I6 (PorI6), and P. savastanoi pv. savastanoi NCBPP3335 (Psv3335); however, for the last three genomes, the specific genomic context of the E622 could not be determined due to the current state of genome assembly. The IRs in Pta11528 and PgyB076, however, flank an RIE cassette and a T3SE gene, forming the characteristic TnE622 transposon structure. The Pta11528 TnE622 is identical in organization and genetic composition to the pPMA4326B plasmid, which carries the T3SE gene hopW1-1, while the PgyB076 TnE622 carries the hopAW1 T3SE gene, along with a plasmid stability determinant and a hypothetical gene (Fig. 1). We also performed a BLAST search of all incomplete genomes using just the RIE sequence as the query and found the intact operon in P. syringae pv. lachrymans M302278PT, P. syringae pv. actinidae M302091, and P. syringae pv. morsprunorum M302280PT; however, because these genomes are incomplete, the flanking sequences were unavailable, making it unclear if they have the E622 IRs and if these encompass any T3SE genes or other virulence genes.
The prevalence of E622 on the plasmids of PmaES4326 and its frequent association with virulence genes in P. syringae prompted a large-scale PCR survey of plasmid extracts from 136 P. syringae strains using E622-specific primers. The majority of strains appear to carry the approximately 600-bp E622 element, with most isolates producing additional amplicons of various sizes and up to 8 kb (data not shown). Partial sequencing of several random amplicons revealed random gene cassettes between borders, including several conserved hypothetical genes and an IS51 element (data not shown). PCRs of plasmids from several isolates, including P. syringae pv. broussonetiae KOZ8101, P. syringae pv. glycinea KN127, P. syringae pv. glycinea MOC601, P. syringae pv. syringae Ps9220 (PsyPs9220), P. syringae pv. syringae PSC1B (PsyPSC1B), and P. syringae pv. thea K93001 (PthK93001) each produced a 6- to 8-kb amplicon. To determine the composition of these larger amplicons, we sequenced the approximately 6.5-kb island from PthK93001 in its entirety and identified the T3SE gene hopAU1 and a pseudogene of hopAV1, adjacent to the three-gene RIE cassette originally identified on pPMA4326B (Fig. 1). The T3SE genes are arranged in an operon, with a single promoter hrp box, GGAACCCTCCTGTGATTTTCGAACACTCA (conserved motifs are underlined). The hopAV1PthK93001 T3SE gene is truncated relative to the original allele identified (9, 71) and has several frameshifts after the coding sequence for the first 44 amino acids. The overall genetic structure of the PthK93001 island closely resembles that found on pPMA4326B, including the placement and orientation of the T3SE genes relative to the RIE operon. Partial sequencing suggested that TnE622s from PsyPs9220 and PthK93001 had similar genetic contents, while PsyPSC1B contains a TnE622 element similar to that found on pPMA4326B. Given its conservation and prevalence among multiple P. syringae strains, the shared IRs with the mobile E622 MITE, the conservation of its structure, and organizational similarity to the Tn3 and Tn7 transposons, TnE622 is likely the E622 parent.
TnE622 is represented in clinically relevant bacteria and the cyanobacteria.
We attempted to determine whether E622 and its predicted parent were restricted to P. syringae. Using BLAST search parameters that were optimized for identifying more-divergent E622 homologs, we identified E622 and its characteristic IRs within the chromosomally carried hrp-hrc pathogenicity island in the plant pathogen Pantoea stewartii pv. stewartii (485 bp) (26) and on plasmid pEL60 of the plant pathogen Erwinia amylovora (485 bp) (25) (Fig. 1). A subsequent discontinuous MEGABLAST search with these IRs as the query identified additional E622 IRs in P. syringae, as well as in Pseudomonas sp. TW3 (36), Pseudomonas aeruginosa PAO1 (66), Salmonella enterica (12, 29, 32, 46, 56), Shewanella baltica (GenBank accession numbers NC_009665, NC_009997, CP001252, and CP002383), and Yersinia pestis (20). All of the E622 IRs identified in these organisms encompassed the same three-gene RIE operon, along with virulence-associated passenger genes. The S. baltica TnE622 contains an insecticidal toxin gene, the P. aeruginosa PAO1 TnE622 contains a virulence-associated S-type pyocin biosynthetic gene, and the S. enterica TnE622 contains a virulence-associated carbonic anhydrase gene (mig-5) (69). The TnE622 of P. syringae plasmid p1448A-B carries multiple genes, including a toxin-antitoxin plasmid addiction system and two hypothetical genes, but these have not been shown to be involved in virulence. Likewise, the plant growth-promoting Pseudomonas fluorescens SBW25 TnE622 carries multiple genes, including a transcriptional regulator gene (copG) as well as a gene encoding a hypothetical protein (Fig. 1). More-divergent E622 homologs were also identified on plasmid pCC7120delta in the cyanobacterium Nostoc (515 bp) (38) and pCy782205 in the cyanobacterium Cyanothece (TnE622) (GenBank accession number CP002203). The Nostoc IRs appear to be part of an E622 element, while the IRs of the Cyanothece element flank an RIE cassette, with no additional genes being present within the IRs (Fig. 1). When the sequences of the IRs in the cyanobacteria were used as the query, no additional E622/TnE622 homologs could be identified.
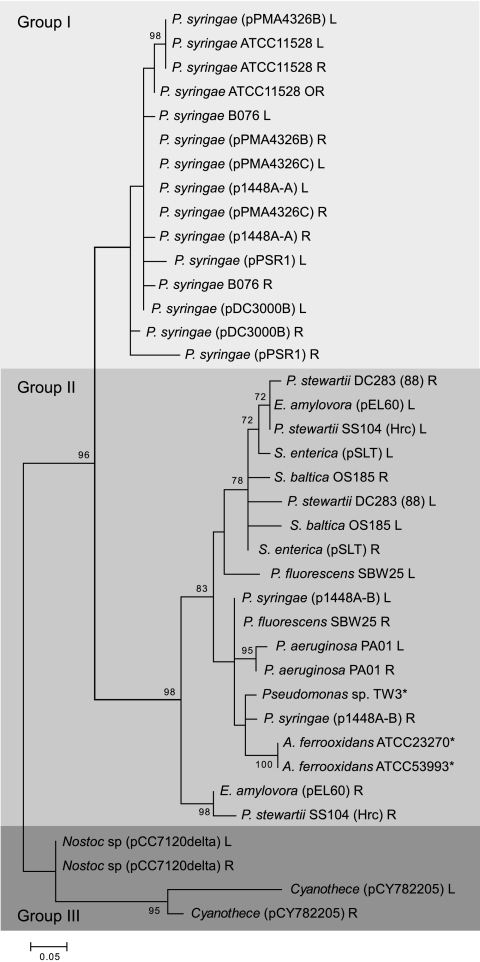
The presence of three seemingly distinct groups of E622 prompted a phylogenetic analysis of all E622 and TnE622 IRs, which established the diversification of this family into three evolutionary groups (Fig. 3). Group I is composed of E622/TnE622 elements found only in P. syringae and includes those that were originally identified in PmaES4326. Group II comprises homologs in a broader set of bacteria, including S. enterica, S. baltica, E. amylovora, P. stewartii, and P. syringae, while the group III IRs are found only in cyanobacterial Nostoc and Cyanothece spp. The phylogenetic groups are consistent with the distinctive TnE622 genetic structures, with TnE622 from group I having gene insertions toward the 5′ end of the resolvase gene and the group II elements having insertions toward the 3′ end of the exeA-like gene. There is only one representative of TnE622 in group III, and it has no gene insertions. The phylogenetic analysis also shows that the left and right IRs from the same E622/TnE622 element always fall within the same group and are generally closely related. In some cases, however, as with E. amylovora and P. stewartii, one of the IRs is more divergent.
Fig 3.
Neighbor-joining tree of the E622/TnE622 inverted repeats. Three distinct phylogenetic groups are represented, with group I consisting solely of elements from P. syringae, group II consisting of elements from several different pathogens in addition to P. syringae, and group III consisting of E622/TnE622 elements from the cyanobacteria. Species marked with asterisks have only one IR represented in the phylogeny. All accession numbers are available in Table S1 in the supplemental material.
Despite the phylogenetic distinctiveness of the three established groups, the integration site consensus for each group remains conserved. The group I and group II element integration site is predicted to be NNTANN, which differs from NCTANN of group III only slightly (Fig. 2); however, the one base difference is likely due to underrepresentation of group III elements. The insertion of E622/TnE622 from all groups appears to generate a tandem duplication, and the “TA” motif appears to be an important part of the recognition sequence. The tandem duplication is conserved for most E622/TnE622 elements identified, with some containing point mutations (Fig. 2).
DISCUSSION
E622 is a new mobile MITE, and at 611 bp, it is substantially longer than most characterized prokaryotic MITEs, which are typically only several hundred bases long (19). We demonstrated that E622 is mobile using an antibiotic coupling assay, which resulted in the E622 element carrying an ampicillin resistance gene being integrated into the trbB gene of pLAFR. E622 was integrated beginning from its left IR to its right IR, with a tandem duplication of the integration site (Fig. 2). The IRs of E622, which are highly conserved and range from 164 to 168 bp, are significantly longer than the 10- to 40-bp IRs found in bacterial IS elements, transposons, or MITEs (19, 42, 43). IRs typically contain multiple domains, including the transposase binding site, and the domain for cleavage and strand transfer, with the direct repeats within the IRs functioning to facilitate transposase recognition and positioning (42, 43). Despite the longer repeats, we observed a relatively low transposition rate for E622, which is likely due to the electroporation efficiency of the cosmid used in our antibiotic assay, along with the fact that E622 is trans-activated.
E622 lacks any ORF that could encode a transposase enzyme, suggesting that it is trans-activated like other MITEs. Trans transposition has been shown to contribute to the mobility of inactivated IS elements in P. syringae, as illustrated by the 10 copies of the IS801 element, each with the same nonsense mutation in the transposase enzyme coding sequence (37). The IRs of E622 are clearly homologous to those of TnE622, indicating that these two elements are related and that the TnE622 enzymes are likely responsible for mobilizing E622. The parent element of the RUP MITE was proposed to be IS630-Spn1, based on the high sequence identity of their IRs (49). TnE622, which contains the RIE operon bordered by the E622 IRs, contains both a resolvase gene and an integrase gene, two genes that characterize the Tn3 family of transposons (42). In Tn3, however, these two genes are divergently transcribed, with the center region containing a cointegrate resolution (res) site that facilitates completion of replicative transposition (31). We were unable to identify an analogous intergenic region that might serve as a res site; however, the resolvase gene homolog in Salmonella enterica (rlgA) has been shown to regulate its expression by binding a region immediately upstream of the resolvase, which may also serve as a res site (44). An analysis of the resolvase gene using the ISFinder database (61) reveals that this gene belongs to the Tn3 family, but a similar analysis of the integrase gene places this element in the IS481 family, which has a consensus integration site that closely matches that of E622/TnE622 (65). Although TnE622 has properties of both the Tn3 and IS481 family, it also carries an exeA-like gene, which encodes a predicted ATPase belonging to the protein family “ATPases associated with diverse cellular activities” (Interpro IPR003593) (33). The conservation of this predicted ATPase and its coregulation with the resolvase and integrase suggest that this locus is not merely a passenger gene but likely plays a role in transposition. An ATPase has been shown to be involved in regulation of transposition for Tn7 (52), as well as for Mu phage, with the latter requiring the MuB ATPase for target selection and delivery of the target to the transposase (60). Thus, the E622/TnE622 family appears to have properties of IS481 (homologous integrase), Tn3 (presence of integrase and resolvase), and Tn7 (presence of ATPase), as well as bacteriophage (ATPase and long inverted terminal repeats), making it sufficiently distinct from defined transposon families.
Transposition of E622/TnE622 results in a 6-bp tandem duplication, suggesting that the integrase is likely a site-specific recombinase. In almost all integration sites and corresponding tandem duplication sequences, the “TA” dinucleotide motif appears to be a key part of the target site sequence. For the IS481 family, NCTANN, NCTAGN, or NNTANN appears to flank many insertions, with the first two occurring at greater frequency (65). Insertion into target sites containing TA has also been documented for the RUP element (19, 49), a member of the IS630 family that shows similarity to the Tc1/Tc3/Mariner family of eukaryotic mobile elements (42, 43). The fact that the duplicated tandem repeats of many of the E622/TnE622 elements have remained conserved could indicate a more recent integration for these transposons, whereas the integration of E622 of Nostoc sp. PCC7120 (pCC7120delta) and the TnE622s of S. enterica and P. syringae (pPMA4326B) may be older since the accumulation of mutations and/or genomic rearrangements have resulted in the tandem duplication no longer matching the target integration site.
Many passenger genes of TnE622 have been implicated in virulence for many clinical pathogens. The S-type pyocin gene in P. aeruginosa (Fig. 1) has been directly implicated in bacterial virulence and toxigenicity (2, 3, 10), while the carbonic anhydrase gene (mig-5) within TnE622 on the virulence plasmid pKDSC50 of Salmonella has been shown to be essential for virulence and bacterial survival in the host (69). The SepC/Tcc insecticidal toxin gene in TnE622 of S. baltica (Fig. 1) is homologous to that found adjacent to an IR in Yersinia pestis (see Fig. S1 in the supplemental material), both of which are homologous to the gene shown to be involved in the insecticidal activity of Yersinia enterocolitica W22703 against Manduca sexta larvae (3). In P. syringae, TnE622 is frequently found to be plasmid localized, and its passenger genes include the virulence-associated T3SE genes hopW1-1, hopAV1, hopAU1, and hopAW1. TnE622 may therefore have played a central role in the dissemination and transfer of T3SE genes across strains, also providing support for previous reports of conserved sequences flanking T3SE genes (1).
Approximately 33% of the T3SE genes located proximally to the IRs appear to encode either chimeras or orphan effector termini (ORPHETs), which are truncated versions of full-length T3SEs (63). This could indicate that TnE622/E622 may contribute to the evolution of new T3SEs through terminal reassortment, a process by which new protein coding domains are fused to preexisting T3SE secretion signals (63, 64). hopAV1, identified on TnE622 from PthK93001, is a new ORPHET coding sequence, since it constitutes the 5′ end (encoding the N-terminal region) of hopAT1Pph1448A and the longer hopAV1Pph1448A. This ORPHET may have served as the precursor for the formation of these T3SEs through terminal reassortment. In addition, the arrangement and structure of hopAU1 and hopAV1 on this island explain previous observations that these two effector genes share homologous upstream regions (63). The presence of these two T3SE genes separately on the Pph1448A chromosome, although they have the same promoter region, suggests that the linkage between hopAU1 and hopAV1 on TnE622 likely represents the ancestral state and that past deletions of the hopAU1PthK93001 and hopAV1PthK93001 T3SE genes independently have resulted in each T3SE gene having the same promoter. Interestingly, E622 was also previously reported to contain a sequence that resembles a type III promoter (62).
The IRs of E622/TnE622 fall into three evolutionarily distinct groups, as established by phylogenetic analysis. Group I and group II E622/TnE622 elements are represented in P. syringae, with the group I E622/TnE622 being exclusive to P. syringae. The relatively broad representation of this transposon family across very diverse P. syringae isolates suggests that it is an ancestral element. The variation in the distribution of E622 versus TnE622 is also distinctive, with the group I E622 element being exclusive to P. syringae (Fig. 3; see Fig. S1 in the supplemental material). Intraspecific dissemination via plasmid transfer appears to be a contributing factor to the broad distribution within P. syringae, since E622 appears to be tightly associated with plasmids. In contrast, TnE622 is somewhat more broadly distributed, possibly due to transfer of TnE622 from P. syringae into related pseudomonads, such as P. aeruginosa, P. fluorescens, and Pseudomonas sp. TW3, and subsequent dissemination from these organisms to other clinically important bacteria.
The noncoding nature of the E622 MITE makes it highly amenable to assimilating gene cassettes, and it is therefore more versatile than an IS element, whose transposase is more likely to be disrupted by the insertion of a gene cassette. The IRs of E622/TnE622 are scattered throughout the genomes of P. syringae and other bacteria, suggesting that shuffling mediated through genomic indels and rearrangements would have resulted in IRs being reorganized to encompass various gene cassettes, as demonstrated by the relatively high diversity of amplicon size that resulted from the PCR-based screen for E622 elements (data not shown). Our antibiotic coupling assay that demonstrated that an ampicillin resistance gene can be mobilized when contained between the E622 IRs is consistent with the idea that any gene cassette between intact IRs is likely mobilizable if the necessary transposases are present. While this structure is clearly efficient, the dependency of E622 on a trans-acting transposase limits its distribution and ability to spread beyond the range of its predicted parent. Nonetheless, E622 and its predicted parent, TnE622, appear to be functioning as efficient vessels for shuttling genes between plasmids and chromosomes and may ultimately provide substantial adaptive potential for clinically and agriculturally relevant bacterial pathogens.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Douglas Rawlings for helpful discussions and advice on the mobility assay and Linda Tan, Afzal Hassan, and Irene Chair for their valuable assistance and contribution to this work.
D.S.G. is supported by grants from the Canada Research Chairs Program and the Natural Science and Engineering Council of Canada. J.S. is supported by grants from the Natural Science and Engineering Council of Canada and the Canada Foundation for Innovation.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Arnold DL, et al. 2001. Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi. Microbiology 147:1171–1182 [DOI] [PubMed] [Google Scholar]
- 2. Bowen D, et al. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129–2132 [DOI] [PubMed] [Google Scholar]
- 3. Bresolin G, Morgan JAW, Ilgen D, Scherer S, Fuchs TM. 2006. Low temperature-induced insecticidal activity of Yersinia enterocolitica. Mol. Microbiol. 59:503–512 [DOI] [PubMed] [Google Scholar]
- 4. Brugger K, et al. 2002. Mobile elements in archaeal genomes. FEMS Microbiol. Lett. 206:131–141 [DOI] [PubMed] [Google Scholar]
- 5. Brugger K, Torarinsson E, Redder P, Chen L, Garrett RA. 2004. Shuffling of Sulfolobus genomes by autonomous and non-autonomous mobile elements. Biochem. Soc. Trans. 32:179–183 [DOI] [PubMed] [Google Scholar]
- 6. Buell CR, et al. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000.Proc. Natl. Acad. Sci. U. S. A. 100:10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buisine N, Tang CM, Chalmers R. 2002. Transposon-like Correia elements: structure, distribution and genetic exchange between pathogenic Neisseria sp. FEBS Lett. 522:52–58 [DOI] [PubMed] [Google Scholar]
- 8. Bult CJ, et al. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058–1073 [DOI] [PubMed] [Google Scholar]
- 9. Chang JH, et al. 2005. A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc. Natl. Acad. Sci. U. S. A. 102:2549–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chang W, Small DA, Toghrol F, Bentley WE. 2005. Microarray analysis of Pseudomonas aeruginosa reveals induction of pyocin genes in response to hydrogen peroxide. BMC Genomics 6:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen YH, Braathen P, Leonard C, Mahillon J. 1999. MIC231, a naturally occurring mobile insertion cassette from Bacillus cereus. Mol. Microbiol. 32:657–668 [DOI] [PubMed] [Google Scholar]
- 12. Chu CS, et al. 2008. Evolution of genes on the Salmonella virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics 92:339–343 [DOI] [PubMed] [Google Scholar]
- 13. Correia FF, Inouye S, Inouye M. 1988. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263:12194–12198 [PubMed] [Google Scholar]
- 14. Craig NL. 1997. Target site selection in transposition. Annu. Rev. Biochem. 66:437–474 [DOI] [PubMed] [Google Scholar]
- 15. Daniels MJ, et al. 1984. Cloning of genes involved in pathogenicity of Xanthomonas campestris pv campestris using the broad host range cosmid pLAFR1. EMBO J. 3:3323–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis KR, Schott E, Ausubel FM. 1991. Virulence of selected phytopathogenic pseudomonads in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 4:477–488 [Google Scholar]
- 17. De Gregorio E, Abrescia C, Carlomagno MS, Di Nocera PP. 2003. Asymmetrical distribution of Neisseria miniature insertion sequence DNA repeats among pathogenic and nonpathogenic Neisseria strains. Infect. Immun. 71:4217–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Gregorio E, Silvestro G, Petrillo M, Carlomagno MS, Di Nocera PP. 2005. Enterobacterial repetitive intergenic consensus sequence repeats in yersiniae: genomic organization and functional properties. J. Bacteriol. 187:7945–7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delihas N. 2008. Small mobile sequences in bacteria display diverse structure/function motifs. Mol. Microbiol. 67:475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Deng W, et al. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Palmenaer D, Vermeiren C, Mahillon J. 2004. IS231-MIC231 elements from Bacillus cereus sensu lato are modular. Mol. Microbiol. 53:457–467 [DOI] [PubMed] [Google Scholar]
- 22. Feil H, et al. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feschotte C, Jiang N, Wessler SR. 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3:329–341 [DOI] [PubMed] [Google Scholar]
- 24. Fewer DP, et al. 2011. Non-autonomous transposable elements associated with inactivation of microcystin gene clusters in strains of the genus Anabaena isolated from the Baltic Sea. Environ. Microbiol. Rep 3:189–194 [DOI] [PubMed] [Google Scholar]
- 25. Foster GC, McGhee GC, Jones AL, Sundin GW. 2004. Nucleotide sequences, genetic organization, and distribution of pEU30 and pEL60 from Erwinia amylovora. Appl. Environ. Microbiol. 70:7539–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Frederick RD, et al. 2001. Genetic organization of the Pantoea stewartii subsp stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol. Plant-Microbe Interact. 14:1213–1222 [DOI] [PubMed] [Google Scholar]
- 27. Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732 [DOI] [PubMed] [Google Scholar]
- 28. Guttman DS, Greenberg JT. 2001. Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant-Microbe Interact. 14:145–155 [DOI] [PubMed] [Google Scholar]
- 29. Haneda T, Okada N, Nakazawa N, Kawakami T, Danbara H. 2001. Complete DNA sequence and comparative analysis of the 50-kilobase virulence plasmid of Salmonella enterica serovar choleraesuis. Infect. Immun. 69:2612–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haren L, Ton-Hoang B, Chandler M. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245–281 [DOI] [PubMed] [Google Scholar]
- 31. Heffron F, McCarthy BJ, Ohtsubo H, Ohtsubo E. 1979. DNA-sequence analysis of the transposon Tn3-3 genes and 3 sites involved in transposition of Tn3. Cell 18:1153–1163 [DOI] [PubMed] [Google Scholar]
- 32. Hong SF, Chiu CH, Chu CS, Feng Y, Ou JT. 2008. Complete nucleotide sequence of a virulence plasmid of Salmonella enterica serovar Dublin and its phylogenetic relationship to the virulence plasmids of serovars Choleraesuis, Enteritidis and Typhimurium. FEMS Microbiol. Lett. 282:39–43 [DOI] [PubMed] [Google Scholar]
- 33. Hunter S, et al. 2009. InterPro: the integrative protein signature database. Nucleic Acids Res. 37:D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ilyina TS. 2010. Miniature repetitive mobile elements of bacteria: structural organization and properties. Mol. Genet. Microbiol. Virol. 25:139–147 [Google Scholar]
- 35. Jackson RW, et al. 1999. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. U. S. A. 96:10875–10880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. James KD, Williams PA. 1998. ntn genes determining the early steps in the divergent catabolism of 4-nitrotoluene and toluene in Pseudomonas sp. strain TW3. J. Bacteriol. 180:2043–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joardar V, et al. 2005. Whole-genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals divergence among pathovars in genes involved in virulence and transposition. J. Bacteriol. 187:6488–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaneko T, et al. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp strain PCC 7120. DNA Res. 8:205–213 [DOI] [PubMed] [Google Scholar]
- 39. Kumar S, Tamura K, Nei M. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinformatics 5:150–163 [DOI] [PubMed] [Google Scholar]
- 40. Lindeberg M, et al. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275–282 [DOI] [PubMed] [Google Scholar]
- 41. Liu SV, Saunders NJ, Jeffries A, Rest RF. 2002. Genome analysis and strain comparison of Correia repeats and Correia repeat-enclosed elements in pathogenic Neisseria. J. Bacteriol. 184:6163–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mahillon J, Leonard C, Chandler M. 1999. IS elements as constituents of bacterial genomes. Res. Microbiol. 150:675–687 [DOI] [PubMed] [Google Scholar]
- 44. Massey RC, Bowe F, Sheehan BJ, Dougan G, Dorman CJ. 2000. The virulence plasmid of Salmonella typhimurium contains an autoregulated gene, rlgA, that codes for a resolvase-like DNA binding protein. Plasmid 44:24–33 [DOI] [PubMed] [Google Scholar]
- 45. Mazzone M, et al. 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 278:211–222 [DOI] [PubMed] [Google Scholar]
- 46. McClelland M, et al. 2001. Complete genome sequence of Salmonella enterica serovar typhimurium LT2. Nature 413:852–856 [DOI] [PubMed] [Google Scholar]
- 47. Mennecier S, Servant P, Coste G, Bailone A, Sommer S. 2006. Mutagenesis via IS transposition in Deinococcus radiodurans. Mol. Microbiol. 59:317–325 [DOI] [PubMed] [Google Scholar]
- 48. Nelson KE, et al. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 185:5591–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Oggioni MR, Claverys JP. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145:2647–2653 [DOI] [PubMed] [Google Scholar]
- 50. Okstad OA, et al. 2004. The bcr1 DNA repeat element is specific to the Bacillus cereus group and exhibits mobile element characteristics. J. Bacteriol. 186:7714–7725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Osborn AM, Boltner D. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48:202–212 [DOI] [PubMed] [Google Scholar]
- 52. Peters JE, Craig NL. 2001. Tn7: smarter than we thought. Nat. Rev. Mol. Cell Biol. 2:806–814 [DOI] [PubMed] [Google Scholar]
- 53. Peters M, Tomikas A, Nurk A. 2004. Organization of the horizontally transferred pheBA operon and its adjacent genes in the genomes of eight indigenous Pseudomonas strains. Plasmid 52:230–236 [DOI] [PubMed] [Google Scholar]
- 54. Redder P, She QX, Garrett RA. 2001. Non-autonomous mobile elements in the crenarchaeon Sulfolobus solfataricus. J. Mol. Biol. 306:1–6 [DOI] [PubMed] [Google Scholar]
- 55. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 56. Richardson EJ, et al. 2011. Genome sequences of Salmonella enterica serovar Typhimurium, choleraesuis, dublin, and gallinarum strains of well-defined virulence in food-producing animals. J. Bacteriol. 193:3162–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Robertson AE, Wechter WP, Denny TP, Fortnum BA, Kluepfel DA. 2004. Relationship between avirulence gene (avrA) diversity in Ralstonia solanacearum and bacterial Wilt incidence. Mol. Plant-Microbe Interact. 17:1376–1384 [DOI] [PubMed] [Google Scholar]
- 58. Rohmer L, Kjemtrup S, Marchesini P, Dangl JL. 2003. Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol. Microbiol. 47:1545–1562 [DOI] [PubMed] [Google Scholar]
- 59. Rouquette-Loughlin CE, Balthazar JT, Hill SA, Shafer WM. 2004. Modulation of the mtrCDE-encoded efflux pump gene complex of Neisseria meningitidis due to a Correia element insertion sequence. Mol. Microbiol. 54:731–741 [DOI] [PubMed] [Google Scholar]
- 60. Schweidenback CTH, Baker TA. 2008. Dissecting the roles of MuB in Mu transposition: ATP regulation of DNA binding is not essential for target delivery. Proc. Natl. Acad. Sci. U. S. A. 105:12101–12107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stavrinides J, Guttman DS. 2004. Nucleotide sequence and evolution of the five-plasmid complement of the phytopathogen Pseudomonas syringae pv. maculicola ES4326. J. Bacteriol. 186:5101–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stavrinides J, Ma W, Guttman DS. 2006. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog. 2:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stavrinides J, McCann HC, Guttman DS. 2008. Host-pathogen interplay and the evolution of bacterial effectors. Cell. Microbiol. 10:285–292 [DOI] [PubMed] [Google Scholar]
- 65. Stibitz S. 1998. IS481 and IS1002 of Bordetella pertussis create a 6-base-pair duplication upon insertion at a consensus target site. J. Bacteriol. 180:4963–4966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stover CK, et al. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959–964 [DOI] [PubMed] [Google Scholar]
- 67. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tsiamis G, et al. 2000. Cultivar-specific avirulence and virulence functions assigned to avrPphF in Pseudomonas syringae pv. phaseolicola, the cause of bean halo-blight disease. EMBO J. 19:3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Valdivia RH, Falkow S. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007–2011 [DOI] [PubMed] [Google Scholar]
- 70. Vanbleu E, Marchal K, Vanderleyden J. 2004. Genetic and physical map of the pLAFR1 vector. DNA Seq. 15:225–227 [DOI] [PubMed] [Google Scholar]
- 71. Vencato M, et al. 2006. Bioinformatics-enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol. Plant-Microbe Interact. 19:1193–1206 [DOI] [PubMed] [Google Scholar]
- 72. Wessler SR, Bureau TE, White SE. 1995. LTR-retrotransposons and MITEs—important players in the evolution of plant genomes. Curr. Opin. Genet. Dev. 5:814–821 [DOI] [PubMed] [Google Scholar]
- 73. West SEH, Schweizer HP, Dall C, Sample AK, Runyenjanecky LJ. 1994. Construction of improved Escherichia-Pseudomonas shuttle vectors derived from pUC18/19 and sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 148:81–86 [DOI] [PubMed] [Google Scholar]
- 74. Xu M, Kong RQ, Xu XD. 2006. A miniature insertion element transposable in Microcystis sp FACHB 854. Prog. Nat. Sci. 16:486–491 [Google Scholar]
- 75. Yang GJ, Hall TC. 2003. MAK, a computational tool kit for automated MITE analysis. Nucleic Acids Res. 31:3659–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou C, Yang YJ, Jong AY. 1990. Mini-prep in ten minutes. Biotechniques 8:172–173 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.