Abstract
The phhAB operon encodes a phenylalanine hydroxylase involved in the conversion of l-phenylalanine into l-tyrosine in Pseudomonas putida. The phhAB promoter is transcribed by RNA polymerase sigma-70 and is unusual in that the specific regulator PhhR acts as an enhancer protein that binds to two distant upstream sites (−75 to −92 and −132 to −149). There is an integration host factor (IHF) binding site that overlaps the proximal PhhR box, and, consequently, IHF acts as an inhibitor of transcription. Use of l-phenylalanine is compromised in a crp-deficient background due to reduced expression from the phhAB promoter. Electrophoretic mobility shift assays and DNase I footprinting assays reveal that Crp binds at a site centered at −109 only in the presence of cyclic AMP (cAMP). We show, using circular permutation analysis, that the simultaneous binding of Crp/cAMP and PhhR bends DNA to bring positive regulators and RNA polymerase into close proximity. This nucleoprotein complex promotes transcription from phhA only in response to l-phenylalanine.
INTRODUCTION
The zone of soil surrounding the roots of plants, known as the rhizosphere, is a niche rich in nutrients due to the continuous secretion of root exudates by plants. Recent global transcriptomic studies have revealed that microbial survival in this niche requires a dedicated oxidative stress response and a highly regulated system for the control of food utilization, since compounds in the rhizosphere vary depending on the type of plant, plant growth state, and soil physicochemical parameters (23, 25). In root exudates, proteogenic amino acids are abundant, and Pseudomonas putida KT2440 (Table 1), a model rhizosphere microorganism, uses a number of amino acids, including tyrosine, glutamine, aspartate, proline, and glutamate, as both C and N sources, while phenylalanine is metabolized exclusively as a nitrogen source (8, 29). In P. putida, a hierarchy exists for the use of amino acids that is under the control of the global regulator Crc, which organizes the assimilation of preferred amino acids over nonpreferred ones (2, 30, 33, 34, 41, 42). Crc exerts this control posttranscriptionally by binding to the 5′ end of the target mRNAs, thereby inhibiting translation (31, 32, 33, 34).
Table 1.
Plasmids and strains used in this study
| Strain or plasmid | Genotypea | Reference or source |
|---|---|---|
| Strains | ||
| P. putida | ||
| KT2440 | Cmr | 11 |
| phhR strain | Kmr | 19 |
| crp strain | Cmr Kmrcrp::mini-Tn5 | 8, 9 |
| cya strain | Cmr Kmrcya::mini-Tn5 | 8, 9 |
| E. coli | ||
| BL21(DE3) | F−ompIhsdSB (rB− mB−) galdemmet | Novagen |
| DH5αF′ | F′ hsdR17recA1gyrA | 49 |
| Plasmids | ||
| pMP220 | ′lacZ IncP Tcr | 45 |
| pMCA | PphhA::lacZ Tcr | 19 |
| pET28a (+) | Kmr protein expression vector | Novagen |
| pBend2 | Apr circular permutation vector | 24 |
| pCR2.1 | Apr Kmr | Invitrogen |
| pMCPhhR | KmrphhR | 19 |
| pCRP1 | Apr Kmrcrp | This study |
| pCRP2 | Apr Kmr Crp expression | This study |
| pCRP3 | Apr Kmr PPhhAB | This study |
| pMCCrp | Kmrcrp | This study |
| pCRP3 | Apr | This study |
| pGEMT-phhA-v | Apr, mutant, phhA variant, deleted of the Crp binding site | This study |
| pPhhRBend | Apr | This study |
Apr, Cmr, Kmr, and Tcr, resistance to ampicillin, chloramphenicol, kanamycin, and tetracycline, respectively.
Utilization of certain d- and l-amino acids by P. putida appears to involve the global regulator Crp, since growth with d- and l-lysine, d-alanine, and l-phenylalanine as N sources was compromised in crp mutants (8, 27). In contrast with other microorganisms, it should be noted that the characterization of a P. putida crp-deficient mutant revealed that the profile of C sources used by this mutant is identical to that of the wild-type strain (8, 27). In P. putida, the crp gene product (PP0424) is annotated as a cyclic AMP (cAMP) receptor protein, with high similarity to Crp of Escherichia coli (62% identity) and Vfr of Pseudomonas aeruginosa (82% identity). In E. coli, Crp is a key global regulator of C metabolism that orchestrates glucose-induced catabolite repression control and directly or indirectly controls a number of transporters, catabolic enzymes, and stress response proteins (15), while Vfr in P. aeruginosa seems to be involved in the control of a virulence program, and it is not required for catabolite repression (46). Therefore, it appears that Crp of P. putida has been recruited to control a set of functions involved in the utilization of amino acids as N sources, rather than in catabolism of sugars or virulence.
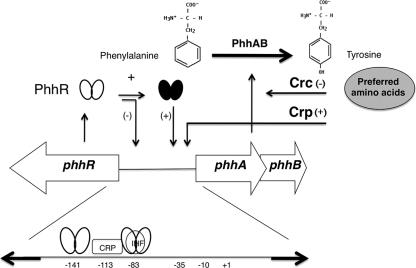
We have previously analyzed the metabolism of l-phenylalanine by P. putida KT2440 (19), and a key pathway for its utilization as an N source was found to involve its initial hydroxylation to yield l-tyrosine in a reaction mediated by the phhAB gene products (Fig. 1). Subsequently, the amino group of tyrosine is transferred to α-ketoglutarate to yield p-hydroxyphenylpyruvate and glutamate, which channels organic nitrogen in the cells. The phhAB genes are transcribed divergently with respect to the phhR gene, which encodes the positive regulator of the phhAB operon. Expression of phhR is constitutive, while that of the phhAB operon occurs only in response to phenylalanine or tyrosine (17). Expression of the phhAB operon is mediated by the PhhR regulator and occurs only in the presence of l-phenylalanine (Fig. 1). Therefore, l-phenylalanine catabolism in P. putida is likely subject to control through the global Crc and Crp regulators and through activation by the specific local regulator PhhR. The phhAB promoter is transcribed by RNA polymerase sigma-70 and is unusual in that for this promoter the PhhR protein functions as an enhancer that binds to two upstream enhancer binding sequences that are centered at positions −83 and −141 from the +1 transcriptional start point with a stoichiometry of one dimer per binding site (19) (Fig. 1). Both target sequences are indispensable for transcription from the phhAB promoter. In addition, overlapping the proximal enhancer site is an integration host factor (IHF) binding site (19). Transcription assays with a PphhA::lacZ fusion, electrophoretic mobility shift assays (EMSAs), and footprint assays showed that IHF acts as a negative regulator and that in a himD mutant background, expression from this promoter in response to l-phenylalanine was maximal (19). Therefore, the phhAB promoter appears to be the target of multiple regulators, which is relatively unusual in prokaryotes (20).
Fig 1.
First step in the catabolic pathway of phenylalanine in Pseudomonas putida and organization of the genes encoding the phenylalanine hydroxylase. l-Phenylalanine is converted into l-tyrosine by the action of phenylalanine hydroxylase, made of the phhA and phhB gene products. The physical organization of the phh genes was established by Herrera and Ramos (19), and gene regulation by PhhR was as established by Herrera et al. (18). Expression of phhR is constitutive, and the ellipses represent PhhR without effector (open); once phenylalanine or tyrosine enters the cell, PhhR with the amino acid (black) is active. Crc and Crp are global regulators that influence negatively (−) or positively (+) the level of expression from the phhAB promoter. The bottom shows the physical sites to which PhhR, IHF, and Crp bind in the intergenic phhA-phhR region.
In this study, we provide insight into the role of Crp/cAMP as a coactivator in the control of l-phenylalanine catabolism, and we show that this global regulator binds to the phhAB promoter at a site located between the two PhhR enhancer sites. Crp/cAMP and PhhR act synergistically and induce DNA bending, which favors the interaction between PhhR and the RNA polymerase. We discuss a model in which a nucleoprotein complex is formed in which Crp, PhhR, and RNA polymerase are involved, but the transcriptional machinery is active only in response to l-phenylalanine. This is the first report that implicates cAMP as an important signaling molecule for amino acid catabolism in P. putida.
MATERIALS AND METHODS
Strain, plasmids, and culture media.
The bacterial strains, cloning vectors, and plasmids used in this study are shown in Table 1. Pseudomonas putida KT2440 and its mutant derivatives were grown in M9 minimal medium supplemented with glucose (0.5% [wt/vol]) as the carbon source (1). When indicated, M8 minimal medium (M9 medium without NH4Cl) was used with either l-phenylalanine or l-tyrosine at a concentration of 5 mM. Cultures were incubated at 30°C and shaken on an orbital platform operating at 200 strokes per minute. When required, antibiotics were used at the following final concentrations (in micrograms per milliliter): ampicillin, 100; chloramphenicol, 30; kanamycin, 50; and tetracycline, 20. Escherichia coli strains were grown at 37°C in LB medium with shaking. E. coli DH5α was used for gene cloning, and E. coli BL21(DE3) was used for protein expression.
β-Galactosidase assays.
The pMCA plasmid bears a fusion of the promoter of the phhAB operon to a promoterless ′lacZ gene in the pMP220 low-copy-number vector (19). The wild-type P. putida KT2440 strain and isogenic mutants were grown overnight in M9 medium with glucose plus tetracycline. The cultures were then diluted 100-fold in the same medium and grown to a turbidity of about 0.8 at 660 nm. Aliquots were prepared and incubated in the absence or presence of 5 mM effector at 30°C for 2 h with shaking. β-Galactosidase activity was assayed in permeabilized whole cells according to Miller's method (28) described elsewhere (13). Assays were run in triplicate and were repeated for at least three independent experiments.
Overexpression and purification of His-tagged Crp.
The crp gene was amplified by PCR to produce pCRP2 (Table 1), which was used to express Crp protein with a 6× histidine tag at its amino-terminal end as described previously (17–19). His6-tagged Crp was purified by nickel affinity chromatography using a Ni+-Sepharose matrix (Amersham-Biosciences) as described by the supplier, and the bound protein was eluted with an imidazole gradient (10 to 500 mM) in 50 mM phosphate buffer (pH 7.6) with 300 mM NaCl. Homogeneous peak fractions were pooled, dialyzed against 50 mM sodium phosphate buffer (pH 7.6), 300 mM NaCl, and stored at −80°C. Protein concentrations were determined spectrophotometrically using molecular extinction coefficients (12).
Electrophoresis mobility shift assays.
A 374-bp DNA fragment containing the phhAB promoter region was amplified by PCR from the P. putida chromosome as described previously (18). A variant of the phhA promoter in which the Crp binding site (5′-CGAATTTTCCGACACTT-3) sequence was replaced by a random sequence (5′-AAGTCAGGTTTTTAATA-3′) was obtained by overlapping PCR mutagenesis as described previously (10). The amplified fragment was cloned in pGEMT, and a clone was used to confirm the sequence of the phhA-variant promoter. This phhA variant was also amplified by PCR for EMSA. To this end, amplified fragments were separated in agarose gels and subsequently extracted from this matrix. The fragments were end labeled with [γ-32P]dATP using the T4 polynucleotide kinase, and about 1 nM (105 cpm) was incubated with increasing amounts of Crp-His6 for 20 min at 25°C in 10 μl of binding buffer containing 20 μg of poly(dI-dC)/ml, 200 μg/ml of bovine serum albumin, 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.1 mM dithiothreitol (DTT), 20 μM cAMP, and 1 mM EDTA (Fig. 2; see Fig. S1 in the supplemental material for the phhA-variant promoter). Reaction mixtures were then electrophoresed in a nondenaturing 4.5% (wt/vol) polyacrylamide gel in 50 mM Tris buffer (pH 7.5), 50 mM borate, and 20 μM cAMP. For the EMSA (Fig. 2), a 10-μl sample containing about 2 nM labeled DNA (1.5 × 104 cpm) was incubated with increasing concentrations of purified Crp and a constant level of PhhR protein for 1 h in 10 μl of binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1 mM DTT, and 20 mM cAMP) containing 20 μg/ml of poly(dI-dC) and 200 μg/ml of bovine serum albumin. The DNA-protein complexes were resolved by electrophoresis in 4% (wt/vol) nondenaturing polyacrylamide gels in TBE (Tris-borate-EDTA) buffer supplemented with cAMP. The results were analyzed with a model GS525 molecular imager (Bio-Rad).
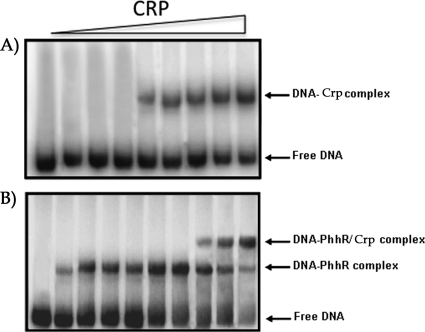
Fig 2.
EMSA of Crp with the phhA operator intergenic region. The whole region was amplified by PCR and end labeled with 32P and 2 nM the phhAB operator was incubated with 1 mM cAMP in the absence or in the presence of increasing concentrations of Crp (from left to right, 0, 2, 4, 6, 8, 10, 15, 20, and 25 μM) and electrophoresed as indicated in Materials and Methods (A). (B) DNA binding assays were carried out using 20 μM purified PhhR protein, 2 nM the phhAB operator region amplified by PCR as described above, and 1 mM cAMP. Then, increasing concentrations of Crp (from left to right starting with the third lane, 0, 2, 4, 6, 8, 10, 15, and 20 μM) were added. The first lane on the left is control DNA with no added protein, and the second lane is DNA with 20 μM Crp.
DNase I footprinting.
The phhAB promoter region generated by PCR (374-bp fragment) was used for footprinting analysis. To this end, we amplified DNA using primers 5′-TGAATTCACCAGCAGGTTGA-3′ (end labeled with [γ-32P]ATP as described above) and 5′-ATCTGCAGATAAAACCATGC-3′. About 5 nM concentrations (104 cpm) of each labeled probe were incubated in 10-μl reaction mixtures of 20 μM PhhR with increasing concentrations of His6-tagged Crp protein (5, 15, and 20 μM). cAMP was added to reach a concentration of 1 mM. Reaction mixtures were incubated for 20 min at 25°C, before being treated with 1.4 × 10−4 U/μl of DNase I diluted in 10 mM Tris-HCl (pH 7) supplemented with 2.5 mM MgCl2, 1 mM CaCl2, 0.1 mM EDTA, and 50 mM KCl. After 4 min at 30°C, reactions were stopped with 2 μl of 0.5 mM EDTA and the sample was extracted with phenol. The DNA was precipitated with 2 volumes of ethanol and resuspended in 5 μl of water and 2.5 μl of loading dye. Equal amounts of DNA (5,000 to 6,000 cpm) were heated at 90°C for 3 min and electrophoresed through a 6.5% (wt/vol) denaturing polyacrylamide gel. Sequencing ladders were generated in each case with the corresponding labeled primer, using a T7 DNA polymerase sequencing kit (USB-Amersham) and the pCRP3 plasmid (Table 1).
Plasmid construction and circular permutation gel retardation assays.
A 257-bp DNA fragment containing the divergent phhR and phhAB promoters was amplified from the P. putida KT2440 chromosome by PCR using primers with XbaI and SalI restriction sites at their 5′ ends. The fragment was isolated and subcloned between the XbaI and SalI restriction sites of DNA bending vector pBend2 to yield pPhhRBend. Purified pPhhRBend plasmid was digested with BglII, NheI, ClaI, XhoI, DraI, SmaI, NcoI, and BamHI restriction enzymes to generate DNA fragments having a circularly permuted phhA-phhR intergenic region. The digested fragments were purified and end labeled with [γ-32P]dATP using T4 polynucleotide kinase. The labeled DNA fragments were mixed with saturated amounts of Crp-His6 and/or PhhR-His6 for 20 min at 25°C in 10 μl of binding buffer containing 20 μg of poly(dI-dC)/ml, 200 μg/ml of bovine serum albumin, 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 0.1 mM DTT, 20 μM cAMP, and 1 mM EDTA. The reaction mixtures were electrophoresed using a nondenaturing 4.5% (wt/vol) polyacrylamide gel in 50 mM Tris buffer (pH 7.5), 50 mM borate, and 20 μM cAMP. The magnitude of the protein-induced bending was calculated using the empirical equation (47) μM/μE = cos(α/2), where μE and μM are the mobility of the protein-DNA complex with the binding site at the ends and at their centers, respectively, and α is the bend angle.
Molecular representation of bending of phhA promoter.
The generation of the three-dimensional (3D) structural bending model of the double-stranded DNA (phhA promoter) was generated from the 3D-Dart server (3DNA-Driven DNA Analysis and Rebuilding Tool) (48), introducing a 120° bending angle on the DNA, as calculated by circular permutation gel retardation assays. The Crp tridimensional structure shown in Fig. 5 corresponds to the crystal structure of DNA binding protein Vfr of Pseudomonas aeruginosa (Protein Data Bank [PDB] accession number 2OZ6), which presents a high identity to the P. putida Crp sequence, and was determined by the Geno3D program Automatic Modeling of Proteins Three-Dimensional Structure (http://geno3d-pbil.ibcp.fr). The representation of RNA polymerase subunits has been constructed by joining of the crystal domain structures of E. coli RNA polymerase (PDB accession number 3LU0, molecular model of the core; PDB accession number 1COO, COOHterminus of the alpha subunit) by using PyMOL software.
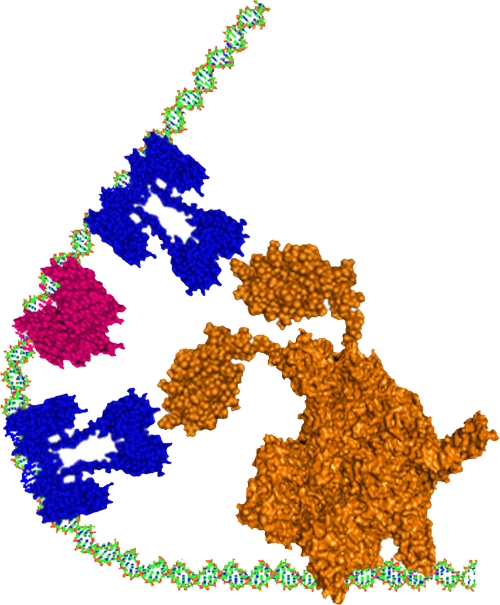
Fig 5.
Schematic representation of the bending of phhA promoter induced by the specific binding of two PhhR dimers and Crp protein. The PhhR dimers are colored in blue and Vfr is in magenta; RNA polymerase is in orange. The PhhR proteins are bound to the distal and the proximal phhR binding boxes, and Crp is bound to its binding site, allowing the PhhR dimers to be in close proximity to RNA polymerase.
The prediction of PhhR tertiary structure was done by homology modeling using the EasyPred 3D server (automated homology modeling program using neural networks) and Swiss-Model server (fully automated protein structure homology-modeling server), accessible via the Expasy Proteomics server.
RESULTS
Maximal expression of the phhAB operon requires Crp and cAMP.
To provide insight into the potential role of the P. putida KT2440 Crp protein in the metabolism of l-phenylalanine, we first determined the growth rate of KT2440 and its isogenic crp mutant in minimal medium with ammonium, l-phenylalanine, or l-tyrosine as the N source. Both the wild type and the mutants grew rapidly with ammonium, exhibiting a 2- to 2.2-h doubling time (Table 2). With phenylalanine the doubling time of the wild type was 8 h, while the crp mutant had a doubling time of over 16 h. We also found that the rate of growth of the crp mutant with l-tyrosine as the sole N source was much slower than that of the parental strain (Table 2). It is known that l-tyrosine (but not l-phenylalanine) is used by P. putida KT2440 as a sole carbon source (19); therefore, we tested the growth of the crp mutant on l-tyrosine as the sole carbon source and ammonium as an N source. We found that the mutant used l-tyrosine as a carbon source as efficiently as the parental strain (i.e., doubling times were in the range of 3.2 to 3.4 h). We therefore concluded that the crp gene product is involved in the utilization of these aromatic amino acids as nitrogen sources. These results are in agreement with those reported by Milanesio et al. (27), which showed that crp mutations do not affect the panel of C compounds that can be used by P. putida as growth substrates and that a crp mutant was impaired in the use of certain dipeptides as an N source.
Table 2.
Growth of KT2440 and its mutants with ammonium, l-phenylalanine, and l-tyrosine as the sole N sourcea
| Genotype | Doubling time (h) |
||
|---|---|---|---|
| NH4+ | l-Phenylalanine | l-Tyrosine | |
| Wild type | 2 ± 0.1 | 8 ± 0.5 | 1.8 ± 0.1 |
| crp | 2.1 ± 0.1 | 16.3 ± 1.5 | 6.5 ± 0.7 |
| cya | 2.2 ± 0.4 | 18 ± 3 | 7.3 ± 3 |
| phhR | 2.1 ± 0.1 | 26.1 ± 2.5 | 3.2 ± 0.2 |
Bacterial cells were grown overnight on M9 minimal medium with glucose. On the following morning, cultures were diluted 100-fold in M8 minimal medium with glucose (0.5% [wt/vol]) and 5 mM the indicated N source. Doubling times were measured in cells growing exponentially. Data shown are the average of 3 independent assays carried out in duplicate.
The only source of cAMP in P. putida is the conversion of ATP into cAMP through the activity of the enzyme adenylate cyclase (Cya) (27). To verify that the Crp protein in P. putida requires cAMP for its regulatory function, we tested growth of a cya-deficient mutant with l-phenylalanine as an N source. We found that this mutant was also impaired in the use of l-phenylalanine and l-tyrosine as an N source (Table 2). Therefore, synthesis of cAMP is also required for optimal transcriptional regulation of the phhAB operon. It should be noted that we have previously shown that a phhR mutant did not grow on l-phenylalanine as an N source. These results suggest a positive effect of PhhR and Crp on the utilization of l-phenylalanine by P. putida KT2440.
To expand our understanding of the role of the Crp/cAMP in the transcriptional control of the phhAB operon, we measured β-galactosidase activity, in exponential phase, using a PphhAB::′lacZ fusion in the wild-type and Crp and Cya mutant backgrounds. Experiments were performed in the presence and in the absence of l-phenylalanine or l-tyrosine in M9 minimal medium with glucose as a C source. We found that the expression levels in cells growing with l-phenylalanine or l-tyrosine were lower in both mutant backgrounds, being about one-third of those measured in the wild type (Table 3). In the PhhR mutant (Table 3) or a double PhhR/Crp mutant, expression from PphhA was practically undetectable. The set of growth results and the results of transcriptional assays using ′lacZ fusion support the idea that PhhR and Crp/cAMP have a positive effect on the activation of expression of the phhAB operon. Expression of phhR was constitutive, and its level was only slightly lower in the crp mutant background than in the parental strain (see Table S1 in the supplemental material).
Table 3.
Expression of PphhA in the parental and in phhR- and cya-deficient backgroundsa
| Strain | PphhA:′lacZ β-galactosidase activity (Miller units) with: |
||
|---|---|---|---|
| Medium alone | Phe | Tyr | |
| Wild type | 50 ± 3 | 2,780 ± 215 | 630 ± 20 |
| crp strain | 70 ± 2 | 832 ± 92 | 234 ± 38 |
| cya strain | 40 ± 3 | 1,109 ± 68 | 272 ± 32 |
| phhR strain | 50 ± 3 | 55 ± 3 | 70 ± 4 |
Pseudomonas putida (wild type) and its isogenic strains (crp deficient, cya deficient, and phhR deficient) were transformed with PphhA::′lacZ in the pMCA plasmid. Bacterial strains bearing pMCA were grown on M9 minimal medium with glucose in the absence (medium alone) or in the presence of 5 mM l-phenylalanine or 5 mM l-tyrosine. β-Galactosidase activity was determined when turbidity of cultures was about 0.8 at 600 nm. Values are the average of three independent experiments carried out in duplicate.
Crp recognizes the target DNA sequence in the phhAB promoter in the presence of cAMP.
Previous work has shown that in E. coli cAMP acts as an allosteric effector, which increases the affinity of the Crp protein for DNA (50). To test if the P. putida Crp protein binds to the phhAB promoter, we carried out electrophoretic mobility shift assays (EMSAs), using a 374-bp phhAB promoter region and purified P. putida Crp protein in the presence and in the absence of cAMP. In the absence of cAMP, no retardation was observed (data not shown), while EMSA revealed that the Crp protein bound to the phhAB promoter when cAMP was present (Fig. 2A). We also carried out EMSAs of the phhAB promoter region in the presence of PhhR at a fixed concentration with increasing levels of purified Crp in the presence of cAMP (Fig. 2B). The results showed that PhhR is able to bind to and retard the migration of the phhA operator region specifically in the absence of Crp (Fig. 2B); however, upon the addition of increasing concentrations of purified Crp in the presence of cAMP, the formation of a second complex, combined with the gradual disappearance of the PhhR-DNA complex, was observed. This suggests a potential cooperativity in binding of these two proteins to target DNA.
To confirm that cAMP/Crp plays a role in the regulation of the phhAB promoter, DNase I footprinting was carried out in the presence or absence of cAMP/Crp. There was no DNA protection by Crp (20 μM) alone. Addition of cAMP/Crp afforded some protection of a region from −103 to −119, which is located between both PhhR boxes (Fig. 3A). In this region, a potential inverted repeat (uppercase), 5′-AAtTtTCcGAcAcTT-3′, was found. Interestingly, the protected region within the phhAB promoter did not match well with the reported consensus for the E. coli cAMP/Crp binding sequence (5′-TGTGA-N6-TCACA-3′). To test if this site is recognized by Crp of P. putida, we carried out EMSA with a 250-bp phhA variant in which the Crp binding site was replaced by a random sequence (see Materials and Methods), but the PhhR binding site was retained. For EMSA we used homogeneous Crp and PhhR protein preparations. We found that while the PhhR protein shifted the phhA variant, Crp was not able to retard the band in either the absence or presence of cAMP (see Fig. S1 in the supplemental material). This result suggests that the mutated site is a specific target of the P. putida Crp protein.
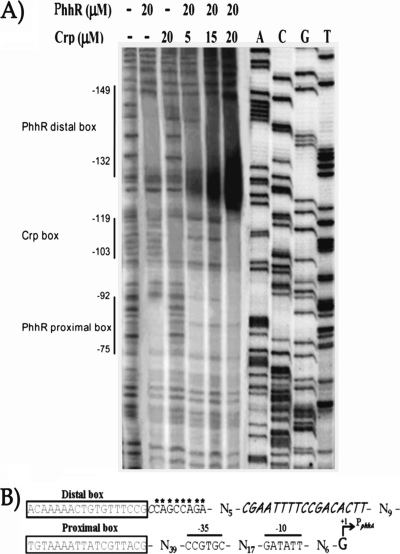
Fig 3.
(A) DNase I footprint of PhhR and Crp in the phhA operator intergenic region. Assays were carried out as described in Materials and Methods. First lane on the left, control in the absence of PhhR and without Crp and 1 mM cAMP; second to sixth lanes, footprint in the presence of PhhR (20 μM); third lane, footprint in the presence of Crp; fourth lane, footprint in the presence of a fixed concentration of PhhR (20 μM), 1 mM cAMP, and increasing concentrations of Crp, as indicated (5, 15, and 20 μM). The regions protected by PhhR and Crp are indicated by vertical lines. (B) Relevant motifs in the intergenic phhA-phhR region. The distal and proximal PhhR boxes are indicated. The Crp binding site is marked in italics, and the bases that exhibit hypersensitivity to DNase I are indicated by asterisks.
We also performed footprint assays with PhhR and Crp/cAMP simultaneously, leading to an expansion of the protected area, which covered both PhhR boxes and a more pronounced protection of the Crp site. We also observed, for Crp/cAMP and PhhR, that the DNase I hypersensitivity of the sequence between the Crp and the distal PhhR site increased with increasing concentration of Crp/cAMP (indicated by asterisks in Fig. 3B). This suggests that Crp/cAMP along with PhhR forms a nucleoprotein complex and that the bending induced by the combination of Crp and PhhR creates a distortion in the DNA helix that widens the DNA minor grove, making it more sensitive to DNase I.
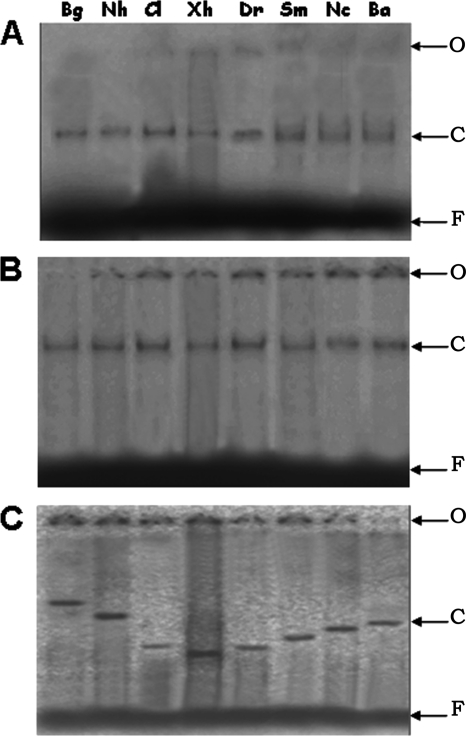
To further elucidate the structural effect of PhhR/Crp-cAMP binding to the phhAB promoter, we used circular permutation analysis (35, 50) to measure the curvature of DNA when PhhR or Crp/cAMP, or both, was added to target DNA (Fig. 4). We found that neither PhhR nor Crp/cAMP alone provoked DNA curvature; however, the simultaneous presence of both regulators induced a marked curvature in the DNA with an angle of 120° (Fig. 5).
Fig 4.
Electrophoresis mobility of circularly permuted DNA fragments complexed to the PhhR and Crp proteins. The fragments were generated from the digestion of the PhhRBend construct with restriction enzymes BglII (Bg), NheI (Nh), ClaI (Cl), XhoI (Xh), DraI (Dr), SmaI (Sm), NcoI (Nc), and BamHI (Ba). O, origin of the wells; F, unbound DNA; C, the PhhR-DNA complex (A), the cAMP/Crp-DNA complex (B), and the PhhR-cAMP/Crp-DNA complex (C). Bending angle (α) was calculated by the equation described by Thompson and Landy (47): μM/μE = cos(α/2), where μE and μM are the mobility of the protein-DNA complex with the binding site at the ends and at their center, respectively.
DISCUSSION
Transcriptional synergy involving multiple activators is a common occurrence in gene regulation in eukaryotes, and a number of examples of synergy have been reported in prokaryotic organisms, although much less frequently (6, 7, 16, 26, 38). The phhAB promoter of P. putida appears to be the target of at least two global regulators, IHF and Crp, and a specific regulator, PhhR. The positive action of PhhR occurs after binding to effector molecules so that the protein, like other positive transcriptional activators, acquires a modified confirmation and binds with higher affinity to its target sites (14, 16, 20). The global regulators IHF and Crp can enhance or inhibit transcription of a number of promoters (3, 5, 43). Our previous studies (19) and the present study suggest that at the phhAB promoter, IHF and Crp have opposite roles in the maximal PhhR-mediated activation of transcription. From previous studies it is known that PhhR binds to the phhAB promoter at two similar, yet nonidentical distal and proximal enhancer sites that are centered at positions −83 and −141 upstream from the +1 position, respectively (17–19). The regulatory function of these boxes appears to be obligatory to the function of the phhA promoter, since elimination of either of them results in a silent promoter. Overlapping the proximal PhhR box is an IHF binding site, and in an IHF-deficient background, expression from the phhA promoter was maximal (18). This then suggests that IHF competes with PhhR and that IHF binding introduces a bend that prevents contact with RNA polymerase and limits local PhhR concentration. Here, we have shown that maximal expression from phhA requires Crp/cAMP. Synergistic coactivation by Crp and another regulator has been described with the LuxR homolog SmcR for activation of the vvpE promoter in Vibrio (21, 22, 24) and, in the case of MalT, in the activation transcription from malEp and malKp promoters in E. coli (39).
It is surprising that the cAMP-Crp complex is a transcriptional activator of the phhAB promoter for l-phenylalanine utilization as a nitrogen source, given the fact that Crp is a generally recognized global regulator of gene expression of carbon sources in E. coli and other microbes (4–6); however, our data clearly show that Crp/cAMP recognizes the phhA operator and that it recognizes a specific binding site in the phhAB promoter located between the proximal and distal PhhR boxes centered at position −109. From several studies, it has been firmly established that both Crp and IHF induce DNA bending near their respective target sites (36, 40, 44, 49, 50). Presumably, protein-induced bending allows protein-protein and/or protein-DNA interactions to occur from a distance, thereby optimizing the appropriate transcriptional response (37). Our results show that the simultaneous binding of Crp/cAMP and PhhR provokes a strong DNA distortion that results in the hypersensitivity of the sequence between the distal site and the crp site to DNase I. The induced curvature likely promotes expression by bringing the activators into close proximity with RNA polymerase.
Taken together, our data suggest a model in which the formation of a higher-order complex containing the two dimers of PhhR is the key feature in the transcriptional activation of phhA. We hypothesized that cAMP/Crp, together with PhhR, induces the bending of DNA to increase the local concentration of PhhR dimers within the vicinity of RNA polymerase. The possibility of direct interaction between cAMP/Crp and the PhhR has not yet been addressed. It is not clear whether PhhR and Crp physically interact to achieve optimal promoter expression, but our results show that the simultaneous occupancy of DNA binding sites is a critical feature. DNase I footprinting experiments in the presence of the two proteins PhhR and Crp indicate cooperative effects in DNA distortion. We have previously shown that the α-CTD (carboxy-terminal domain) arms of RNA polymerase were involved in interactions with PhhR, and in the model proposed in Fig. 5, we suggest that each of the α-CTD arms interacts with a PhhR dimer. The binding of the two regulators and RNA polymerase results in a scaffold that remains in a transcriptionally closed state until l-phenylalanine or l-tyrosine is recognized by the PhhR regulator.
The PhhR protein of P. putida is known to regulate the expression of a number of genes involved in the biosynthesis and transport of aromatic amino acids (17). In the hmg and PP2827 promoters, PhhR binds to a single binding sequence located at −9 and −66, respectively, and, in contrast with phhA, expression is maximal under an IHF-proficient background. Our inspection of the hmg and PP2827 promoters did not reveal the existence of any Crp sites, and, accordingly, we found that expression of hmg and PP2827 promoters was unchanged under a Δcrp background. Therefore, the mechanism of PhhR regulation seems to vary with specific promoters, and the degree of activation of the promoters is critically related to the location of the PhhR enhancer site(s) and the formation of specific DNA-protein interactions that lead to the creation of active transcriptional scaffolds.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by FEDER and grants from the Ministry of Science and Innovation (BIO2010-17227), Consolider-C BIO2006-05668, and Consolider Ingenio 2010 CSD2007-0005. We received FEDER support from the Junta de Andalucía Programme of excellence CVI-3010 and group CVI-191. We also received a grant from the ERANET Pathogenomics program (BIO2008-04419-E/).
We thank M. M. Fandila for secretarial assistance and Ben Pakuts for critical reading of the manuscript.
Footnotes
Published ahead of print 11 November 2011
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Abril MA, Michán C, Timmis KN, Ramos JL. 1989. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171:6782–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aranda-Olmedo I, Ramos JL, Marqués S. 2005. Integration of signals through Crc and PtsN in catabolite repression of Pseudomonas putida TOL plasmid pWW0. Appl. Environ. Microbiol. 71:4191–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bai Q, Somerville RL. 1998. Integration host factor and cyclic AMP receptor protein are required for TyrR-mediated activation of tpl in Citrobacter freundii. J. Bacteriol. 180:6173–6186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bettenbrock K, et al. 2007. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J. Bacteriol. 189:6891–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Botsford JL, Harman JG. 1992. Cyclic AMP in prokaryotes. Microbiol. Rev. 56:100–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Busby S, Kolb A. 1996. The CAP modulon, p 255–270 In Lin ECC, Lynch AS. (ed), Regulation of gene expression in Escherichia coli. Chapman & Hall, Austin, TX [Google Scholar]
- 7. Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199–213 [DOI] [PubMed] [Google Scholar]
- 8. Daniels C, et al. 2010. Global regulation of food supply by Pseudomonas putida DOT-T1E. J. Bacteriol. 192:2169–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duque E, et al. 2007. Towards a genome-wide mutant library of Pseudomonas putida strains KT2440, p 227–254 In Ramos JL. (ed), Pseudomonas, vol V. Kluwer, London, United Kingdom [Google Scholar]
- 10. Fillet S, et al. 2009. TtgV represses two different promoters by recognizing different sequences. J. Bacteriol. 191:1901–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Franklin FCH, Bagdasarian M, Bagdasarian MM, Timmis KN. 1981. Molecular and functional analysis of the TOL plasmid pWW0 from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc. Natl. Acad. Sci. U. S. A. 78:7458–7462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gill SC, von Hippel PH. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319–326 [DOI] [PubMed] [Google Scholar]
- 13. González-Pérez MM, Ramos JL, Gallegos MT, Marqués S. 1999. Critical nucleotides in the upstream region of the XylS-dependent TOL meta-cleavage pathway operon promoter as deduced from analysis of mutants. J. Biol. Chem. 274:2286–2290 [DOI] [PubMed] [Google Scholar]
- 14. Gosh T, Bose D, Zhang X. 2010. Mechanisms for activating bacterial RNA polymerase. FEMS Microbiol. Rev. 34:611–622 [DOI] [PubMed] [Google Scholar]
- 15. Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MJ. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J. Bacteriol. 186:3516–3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gralla JD, Collado-Vides J. 1996. Organization and function of transcription regulatory elements, p 1232–1245 In Neidhardt FC, et al. (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC [Google Scholar]
- 17. Herrera MC, Duque E, Rodríguez-Herva JJ, Ramos JL. 2010. PhhR controls a regulon including two phenylalanine degradation pathways and the MexEF/OprN system in Pseudomonas putida. Environ. Microbiol. 12:1427–1438 [DOI] [PubMed] [Google Scholar]
- 18. Herrera MC, Krell T, Zhang X, Ramos JL. 2009. PhhR binds to target sequences at different distances with respect to RNA polymerase in order to activate transcription. J. Mol. Biol. 394:576–586 [DOI] [PubMed] [Google Scholar]
- 19. Herrera MC, Ramos JL. 2007. Catabolism of phenylalanine by Pseudomonas putida: the NtrC-family PhhR regulator binds to two sites upstream from the phhA gene and stimulates transcription with σ70. J. Mol. Biol. 366:1374–1386 [DOI] [PubMed] [Google Scholar]
- 20. Ishihama A. 2010. Prokaryotic genome regulation: multifactor promoters, multitarget regulators and hierarchic networks. FEMS Microbiol. Rev. 34:628–645 [DOI] [PubMed] [Google Scholar]
- 21. Jeong HS, Kim SM, Lim MS, Kim KS, Choi SH. 2010. Direct interaction between quorum-sensing regulator SmcR and RNA polymerase is mediated by integration host factor to activate vvpE encoding elastase in Vibrio vulnificus. J. Biol. Chem. 285:9357–9366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeong HS, Lee MH, Lee KH, Park SJ, Choi SH. 2003. SmcR and cyclic AMP receptor protein coactivate Vibrio vulnificus vvpE encoding elastase through the RpoS-dependent promoter in a synergistic manner. J. Biol. Chem. 278:45072–45081 [DOI] [PubMed] [Google Scholar]
- 23. Kennedy N, Brodie E, Connolly J, Clipson N. 2004. Impact of lime, nitrogen and plant species on bacterial community structure in grassland microcosm. Environ. Microbiol. 6:1070–1080 [DOI] [PubMed] [Google Scholar]
- 24. Kim CM, Shim SH. 2010. Regulation of the Vibrio vulnificus vvpE expression by cyclic AMP-receptor protein and quorum-sensing regulator SmcR. Microb. Pathog. 49:348–353 [DOI] [PubMed] [Google Scholar]
- 25. Matilla MA, Espinosa-Urgel M, Rodríguez-Herva JJ, Ramos JL, Ramos-González MA. 2007. Genomic analysis reveals the major driving forces of bacterial life in the rhizosphere. Genome Biol. 8:R179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McLeod SM, Xu J, Johnson RC. 2000. Coactivation of the RpoS-dependent proP P2 promoter by Fis and cyclic AMP receptor protein. J. Bacteriol. 182:4180–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Milanesio P, et al. 2011. Regulatory exaptation of the catabolite repression protein (Crp)-cAMP system in Pseudomonas putida. Environ. Microbiol. 13:324–339 [DOI] [PubMed] [Google Scholar]
- 28. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 29. Molina-Henares AM, et al. 2009. Functional analysis of aromatic biosynthetic pathways in Pseudomonas putida KT2440. Microb. Biotechnol. 2:91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morales G, et al. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186:1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morales G, Ugidos A, Rojo F. 2006. Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 8:1764–1774 [DOI] [PubMed] [Google Scholar]
- 32. Moreno R, Fonseca P, Rojo F. 2010. The Crc global regulator inhibits the Pseudomonas putida pWW0 toluene/xylene assimilation pathway by repressing the translation of regulatory and structural genes. J. Biol. Chem. 285:24412–24419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moreno R, Martinez-Gomariz M, Yuste L, Gil C, Rojo F. 2009. The Pseudomonas putida Crc global regulator controls the hierarchical assimilation of amino acids in a complete medium: evidence from proteomic and genomic analyses. Proteomics 9:2910–2928 [DOI] [PubMed] [Google Scholar]
- 34. Moreno R, Marzi S, Romby P, Rojo F. 2009. The Crc global regulator binds to an unpaired A-rich motif at the Pseudomonas putida alkS mRNA coding sequence and inhibits translation. Nucleic Acids Res. 37:7678–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nagaich AK, Appella E, Harrington RE. 1997. DNA bending is essential for the site-specific recognition of DNA response elements by the DNA binding domain of the tumor suppressor protein p53. J. Biol. Chem. 272:14842–14849 [DOI] [PubMed] [Google Scholar]
- 36. Parkinson G, et al. 1996. Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface. J. Mol. Biol. 260:395–408 [DOI] [PubMed] [Google Scholar]
- 37. Pérez-Martín J, de Lorenzo V. 1997. Clues and consequences of DNA bending in transcription. Annu. Rev. Microbiol. 51:593–628 [DOI] [PubMed] [Google Scholar]
- 38. Rhodius VA, Busby SJ. 1998. Positive activation of gene expression. Curr. Opin. Microbiol. 1:152–159 [DOI] [PubMed] [Google Scholar]
- 39. Richet E, Vidal-Ingigliardi D, Raibaud O. 1991. A new mechanism for coaction of transcription: repositioning of an activator triggered by the binding of a second activator. Cell 66:1185–1195 [DOI] [PubMed] [Google Scholar]
- 40. Robertson CA, Nash H. 1988. Bending of the bacteriophage X attachment site by Escherichia coli integration host factor. J. Biol. Chem. 263:3554–3557 [PubMed] [Google Scholar]
- 41. Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34:658–684 [DOI] [PubMed] [Google Scholar]
- 42. Rojo F, Dinamarca A. 2004. Catabolic repression and physiological control, p 365–387 In Ramos JL. (ed), Pseudomonas. Kluwer Academic/Plenum Publishers, London, United Kingdom [Google Scholar]
- 43. Saier MH., Jr 1989. Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Microbiol. Rev. 53:109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schultz SC, Shields GC, Steitz TA. 1991. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science 235:1001–1007 [DOI] [PubMed] [Google Scholar]
- 45. Spaink HP, Okker JH, Wijffelman CSA, Pees E, Lugtemberg BJJ. 1987. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmids pRL1JL. Plant Mol. Biol. 9:27–39. [DOI] [PubMed] [Google Scholar]
- 46. Suh SJ, Runyen-Janecky LJ, et al. 2002. Effect of vfr mutation on global gene expression and catabolite repression control of Pseudomonas aeruginosa. Microbiology 148:1561–1569 [DOI] [PubMed] [Google Scholar]
- 47. Thompson JF, Landy S. 1988. Empirical estimation of protein-induced DNA bending angles: application to lambda site-specific recombination complexes. Nucleic Acids Res. 16:9687–9705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Dijk M, Bonvin AM. 2009. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 37:W235–W239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu H-M, Crothers DM. 1984. The locus of sequence-directed and protein-induced DNA bending. Nature 308:509–513 [DOI] [PubMed] [Google Scholar]
- 50. Young H, Koh J, Roberts GP. 2008. Two-state allosteric modeling suggests protein equilibrium as an integral component for cyclic AMP (cAMP) specificity in the cAMP receptor protein of Escherichia coli. J. Bacteriol. 190:4532–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.