Abstract
Campylobacter jejuni, a microaerophilic bacterium, is the most frequent cause of human bacterial gastroenteritis. C. jejuni is exposed to harmful reactive oxygen species (ROS) produced during its own normal metabolic processes and during infection from the host immune system and from host intestinal microbiota. These ROS will damage DNA and proteins and cause peroxidation of lipids. Consequently, identifying ROS defense mechanisms is important for understanding how Campylobacter survives this environmental stress during infection. Construction of a ΔCj1386 isogenic deletion mutant and phenotypic assays led to its discovery as a novel oxidative stress defense gene. The ΔCj1386 mutant has an increased sensitivity toward hydrogen peroxide. The Cj1386 gene is located directly downstream from katA (catalase) in the C. jejuni genome. A ΔkatAΔ Cj1386 double deletion mutant was constructed and exhibited a sensitivity to hydrogen peroxide similar to that seen in the ΔCj1386 and ΔkatA single deletion mutants. This observation suggests that Cj1386 may be involved in the same detoxification pathway as catalase. Despite identical KatA abundances, catalase activity assays showed that the ΔCj1386 mutant had a reduced catalase activity relative to that of wild-type C. jejuni. Heme quantification of KatA protein from the ΔCj1386 mutant revealed a significant decrease in heme concentration. This indicates an important role for Cj1386 in heme trafficking to KatA within C. jejuni. Interestingly, the ΔCj1386 mutant had a reduced ability to colonize the ceca of chicks and was outcompeted by the wild-type strain for colonization of the gastrointestinal tract of neonate piglets. These results indicate an important role for Cj1386 in Campylobacter colonization and pathogenesis.
INTRODUCTION
Campylobacter jejuni is the most common cause of human food-borne bacterial gastroenteritis in both industrial and nonindustrial countries (13). Morphologically, C. jejuni is a Gram-negative, curved, rod-shaped bacterium that grows best under microaerophilic conditions at temperatures ranging from 37 to 42°C (1). Infection caused by C. jejuni can occur from the ingestion of quantities of as few as 800 organisms (12), with colonization occurring in the jejunum, ileum, and colon (16). Typically, the symptoms of gastroenteritis include diarrhea, abdominal pain, nausea, fever, and fatigue which can last up to 10 days (26). In addition, C. jejuni infection has also been associated with the development of a rare neuromuscular disease, Guillain-Barré syndrome, with approximately 1 in 1,000 C. jejuni infection cases leading to the development of this disease (30).
While unable to grow at high concentrations of oxygen, C. jejuni still requires the presence of free oxygen for growth because of the use of an oxygen-dependent ribonucleotide reductase (41). Additionally, C. jejuni lacks an anaerobic-type ribonucleotide reductase (34, 41). Ribonucleotide reductase is essential for DNA synthesis, resulting in C. jejuni's dependence on oxygen for bacterial growth (41). C. jejuni also utilizes oxygen as a terminal electron acceptor in its respiratory chain (43), and a consequence of oxidative phosphorylation is the inadvertent production of reactive oxygen species (ROS), specifically, the superoxide radical, O2−, and hydrogen peroxide, H2O2 (28). These intracellular ROS are predominantly formed due to O2 oxidizing the redox enzymes (dehydrogenases) of the respiratory chain, which generates ROS and prevents these enzymes from being oxidized by their intended substrates (28). Additionally, a third ROS, the hydroxyl radical (OH), is produced within the cell via Fenton chemistry when endogenous ferrous ion is oxidized by H2O2 (27). In combination with the production of ROS from C. jejuni's normal metabolic processes, the host immune system and intestinal microflora also expose C. jejuni to ROS (2, 40). The three sources of ROS listed above are particularly detrimental to bacteria as they damage DNA and proteins and cause lipid peroxidation. Therefore, bacteria have evolved elaborate, inducible defense mechanisms to protect themselves from the harmful effects of these various sources of oxidative stress (18).
C. jejuni senses oxidative stresses through the peroxide-sensing regulator PerR (45). Recent transcriptome profiling experiments suggested that Cj1386 might be part of the PerR regulon (32). Cj1386 is located directly downstream from katA in the C. jejuni NCTC11168 genome (34). KatA, the encoded catalase, is an enzyme responsible for the decomposition of H2O2 into O2 and H2O and is an important enzyme in protecting C. jejuni against the oxidative damage induced by H2O2 (19). The genomic location of Cj1386 downstream from katA, in conjunction with its regulatory pattern, suggests that Cj1386 may have an important role in oxidative stress defense in C. jejuni. The aim of this study was to investigate the function of Cj1386 in response to oxidative stress and to characterize its role in the colonization and pathogenesis of animal models. Our work demonstrates that Cj1386 contributes to bacterial catalase activity by trafficking heme to the KatA enzyme and that Cj1386 is required for the colonization of the gastrointestinal tract of chicks and piglets.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli DH5α, K-12, and BL21 strains were grown in Luria-Bertani (LB) broth or on LB agar plates at 37°C under aerobic conditions. LB plates and broth were supplemented with 100 μg/ml ampicillin, 50 μg/ml kanamycin, and/or 10 μg/ml chloramphenicol as required. Campylobacter jejuni NCTC11168 (wild type [WT]) was cultured on Mueller-Hinton (MH) agar plates or in biphasic flasks under microaerophilic conditions (83% N2, 4%H2, 8% O2, and 5% CO2) at 37°C in a MACS-VA500 workstation (Don Whitley, West Yorkshire, England). MH agar plates were supplemented with 10 μg/ml kanamycin and/or 20 μg/ml chloramphenicol as required for strains containing antibiotic resistance cassettes. Minimal essential medium alpha (MEMα; Invitrogen) was supplemented with 20 mM sodium pyruvate to enhance Campylobacter growth. The bacterial strains used in this study are listed in Table 1.
Table 1.
Bacterial strains used in this study
| Strain or plasmid | Genotypea | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| K-12 | endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 lacF′ [proA+B+lacIqZΔM15::Tn10(Tetr)] | Clontech |
| DH5α | endA1 hsdR17 (rK− mK−) supE44 thi-1 recA1 gyrA relA1Δ(lacZYA-argF)U169 deoR[ϕ80dlacΔ(lacZΔM15)] | Invitrogen |
| BL21(DE3) | F−ompT gal dcm lon hsdSB(rB− mB−) λ(DE3) | Novagen |
| AS1082 | E. coli BL21(DE3) + (pGST+KatA) Ampr | This study |
| C. jejuni | ||
| AS144 | C. jejuni NCTC11168 | National Collection of Type Cultures |
| AS433 | AS144 ΔkatA::Camr | Palyada et al. (32) |
| AS942 | AS144 ΔCj1386::Camr | This study |
| AS1029 | AS144 ΔkatA ΔCj1386::Camr | This study |
| AS978 | AS433 +katA::Camr Kanr | This study |
| AS1028 | AS942 +Cj1386::Camr Kanr | This study |
| AS1031 | AS1029 +(katA+Cj1386)::Camr Kanr | This study |
| Plasmids | ||
| pRY111 | Camr resistance gene | Yao et al. (50) |
| pRRK | Cloning vector used for complementation of mutants, Kanr | Reid et al. (37) |
| pUC19 | Cloning vector, Ampr | Biolabs |
| pGST | Protein expression vector with GST tag and IPTG-inducible promoter | Sheffield et al. (42) |
Camr, chloramphenicol resistance gene; Kanr, kanamycin resistance gene; Ampr, ampicillin resistance gene.
Construction of isogenic deletion mutants.
Construction of the ΔCj1386 and ΔkatA ΔCj1386 mutants was performed using the In-Fusion Dry-Down PCR cloning kit (Clontech), which uses homologous recombination to clone gene products into the cloning vector in the correct orientation. The primers used in this study are listed in Table 2. Briefly, the gene of interest and the flanking regions were amplified using Taq polymerase (Invitrogen) and subsequently cloned into pUC19 using the cloning kit. Inverse PCR was performed using primers designed to introduce a deleted region into the gene of interest, yielding the amplification of pUC19 with the gene regions flanking the end of the vector. A chloramphenicol resistance cassette was subsequently cloned into the vector to disrupt the gene, and the constructed vectors were sequenced to confirm the absence of mutations. The final constructions were naturally transformed into C. jejuni NCTC11168 as previously described (48). MH agar plates containing 20 μg/ml chloramphenicol were used to select for transformed mutants. The deletion mutants were confirmed by PCR amplification of the corresponding chromosomal regions followed by DNA sequencing.
Table 2.
Primers used in this study
| Primer name | Primer sequence (5′–3′)a |
|---|---|
| Cj1386-sense | CGGTACCCGGGGATCCAAGGCGTAGCACA |
| AAGATATGC | |
| Cj1386-antisense | CGACTCTAGAGGATCCAAAAAGAAGAAATTGCTGAAAAGC |
| Cj1386-SE-inverse | GAACTAAAGGGCGCATTCTTATGAAAGCGCTAAAATGC |
| Cj1386-AS-inverse | GAACACCGCCGAGCAGCAAGCATAAGCAAGCTATCG |
| katA + Cj1386-SE | CGGTACCCGGGGATCCCGATTTTGGAAACATTATAGCTGA |
| katA + Cj1386-AS | CGACTCTAGAGGATCCTAAAAGGGGCGGTTCCTATC |
| katA + Cj1386-SE-inv | GAACTAAAGGGCGCAGGCGATAGCTTGCTTATGCT |
| katA + Cj1386-AS-inv | GAACACCGCCGAGCACGGATGAAGAATGTCGGAGT |
| Cj1386-SE-comp | GATTTAGATGTCTAGCGCTTCTATGGAAGGAGTTGA |
| Cj1386-AS-comp | GGGGAAGCTTTCTAGAAAAAGAAGAAATTGCTGAAAAGC |
| katA + Cj1386-SE-comp | GATTTAGATGTCTAGTTACGTGCATCCCAGTGTTC |
| katA + Cj1386-AS-comp | GGGGAAGCTTTCTAGTAAAAGGGGCGGTTCCTATC |
| Cat-SE | TGCTCGGCGGTGTTCCTTTCCAAG |
| Cat-AS | TGCGCCCTTTAGTTCCTAAAGGGT |
| AR56-AS | CATCCTCTTCGTCTTGGTAGC |
| ak233-SE | GCAAGAGTTTTGCTTATGTTAGCAG |
| ak234-SE | GAAATGGGCAGAGTGTATTCTCCG |
| ak235-SE | GTGCGGATAATGTTGTTTCTG |
| katA-RT-SE | AAGTGGAGCTTATGGCGAAA |
| katA-RT-AS | ACTTCGCTTTTTGCACGATT |
| katA-RTint-SE | AGCTGCCGAGCTTATAGCAA |
| Cj1386-RT-SE | GAGCTTTGCAAAATGGCTTT |
| Cj1386-RT-AS | CCACATCTTTGCGTCCAAAC |
| Cj1386-RTint-AS | TAATGGGGTTTGTCCACGAT |
| Cj1384c-RT-SE | TGGGATGCACGTAATGAAAA |
| Cj1387c-RT-AS | TTGATTTTCCAACAAGAGCCTTA |
| katA-SE-qPCR | GCGATGTGAGAGGTTTTGCT |
| katA-AS-qPCR | CAAAAATCCCAAGCAGCATT |
| Cj1386-SE-qPCR | GGCGATAGCTTGCTTATGCT |
| Cj1386-AS-qPCR | TAATGGGGTTTGTCCACGAT |
| metC_SE | CTAAACTTATTCATTGTGGCAGAGG |
| metC_AS | CTCTGTATTTTTCCAAGTTGCGTG |
| KatA_NcoI | GCCATGGCTATGAAAAAATTGACTAACGA |
| KatA_NotI | GCGGCCGCTTAGTTTGCCACCAAAAGTGG |
Restriction sites are shown in bold.
Construction of complemented strains.
Construction of the complemented mutant strains was performed as previously described (37). The katA, Cj1386, and katA + Cj1386 gene regions were amplified from the C. jejuni NCTC11168 genome using either Pfx (Invitrogen) or Pwo (Roche) high-fidelity polymerase. The amplified PCR product was cloned into the pRRK vector (37) using the In-Fusion Dry-Down PCR cloning kit (Clontech). The resulting construct was sequenced to confirm the absence of PCR-induced errors in the insert. The C. jejuni NCTC11168 deletion mutants were subsequently naturally transformed with the final construct as described above. Positive colonies were selected for on MH agar plates supplemented with 20 μg/ml chloramphenicol and 10 μg/ml kanamycin. The insertion of the gene into the chromosomal rRNA locus was confirmed by PCR and DNA sequencing as previously described (37).
Disk inhibition assay.
Wild-type C. jejuni, ΔCj1386 mutant, and complemented strains were grown on MH agar plates for 3 days. Several colonies from each strain were subcultured in biphasic flasks and grown overnight to mid-log phase. Next, the bacteria were pelleted by centrifugation and resuspended in MH broth to an optical density at 600 nm (OD600) of 1.0. For each strain, 4 ml of this bacterial suspension was added to 100 ml of melted MH agar (cooled to 45°C), and 25 ml was poured into petri dishes. After solidification of the agar, three 6-mm disks were placed on the surface and 10 μl of 3% H2O2, 3% cumene hydroperoxide (CHP) in dimethyl sulfoxide (DMSO), or 90 mM menadione sodium bisulfite was added to the disks as previously described (32). Plates were incubated under microaerophilic conditions for 28 h, and cleared zones of growth inhibition were measured in mm for each strain. All experiments were performed in quadruplicate for each strain. Averages for each zone of inhibition were calculated, and the average of the average was statistically analyzed with analysis of variance (ANOVA). P values of <0.05 were considered statistically significant.
Total cell extract and cell fractionation preparation.
Whole-cell extracts of C. jejuni wild-type, ΔCj1386 mutant, and complemented strains were prepared using the Peripreps Periplasting kit (Epicentre Technologies, Madison, WI), which utilizes osmotic shock to disrupt the outer cell membrane followed by the use of lysozyme to digest the cell wall. Briefly, C. jejuni strains were grown to mid-log phase in MEMα followed by centrifugation and resuspension to an OD600 of 2.0 in fresh MEMα. Subsequently, 1 ml of this culture was centrifuged, resuspended in 50 μl Peripreps Periplasting buffer, and incubated for 5 min at room temperature. This was followed by addition of 50 μl Peripreps lysis buffer with an additional 5-min incubation at room temperature. Cell debris was pelleted by centrifugation, and the supernatant containing the total cellular protein fraction was transferred to a clean tube and stored at −20°C. As required, mid-log cultures of C. jejuni were exposed to 1 mM H2O2 for 15 min prior to protein extraction (described above). It has been previously shown that exposure of C. jejuni (grown in MEMα) to 1 mM H2O2 does not affect cell viability (32).
For cytoplasmic and periplasmic protein preparations, the same procedure was employed as described above with a slight modification to isolate the two different fractions. Bacterial cells were resuspended in 50 μl Peripreps Periplasting buffer followed by a 5-min incubation at room temperature as described above. Next, 50 μl of chilled, purified water was added and this suspension was incubated on ice for 5 min. The culture was centrifuged, and the supernatant, constituting the soluble periplasmic protein fraction, was transferred to a clean Eppendorf tube. The remaining insoluble fraction (containing the spheroplasts) was resuspended in 100 μl of Peripreps lysis buffer and incubated at room temperature for 5 min. Cell debris was pelleted by centrifugation, and the supernatant containing the cytoplasmic cellular protein fraction was transferred to a clean tube and stored at −20°C.
Catalase activity gel.
The catalase activities of the wild-type C. jejuni, ΔCj1386 mutant, ΔkatA mutant, and complemented strains were assessed by the use of the negative gel staining protocol as described by Woodbury et al. (49). Whole-cell extracts were run on an 8% nondenaturing polyacrylamide gel at 25 mA and 4°C. The gel was washed 3 times in H2O for 5 min followed by exposure to 0.003% hydrogen peroxide for 15 min. The gel was quickly rinsed in H2O before staining with a solution of 2% (wt/vol) potassium ferricyanide (Sigma-Aldrich) and 2% (wt/vol) ferric chloride (Fisher Scientific) until bands became visible. The gel was washed 2 more times in H2O for 10 min and photographed using a multi-image light cabinet (Alpha Innotech Corporation).
Catalase activity assay.
Catalase activities were quantified using the method developed by Beers and Sizer (7). Briefly, 30 μl of cytoplasmic or periplasmic protein preparation (130 ng/μl) was added to 220 μl KH2PO4 and 250 μl 20 mM hydrogen peroxide in a 500-μl-volume quartz cuvette. The cuvette was quickly inverted several times to ensure uniform mixing and placed in a Beckman DU 640B spectrophotometer. Decomposition of the 10 mM hydrogen peroxide was measured at 240 nm with absorbance readings taken at 15-s time intervals for a total time of 3.5 min. The unit of activity of each sample is expressed as μmol hydrogen peroxide decomposed per min (U) and per mg of protein (μmol · min−1 · mg−1). Each sample was tested in quadruplicate for each strain assayed. Statistical analysis was performed using the Student t test with P values < 0.05 considered statistically significant.
RNA isolation.
RNA extractions were performed as described previously (33). Briefly, strains were grown in 8 ml MH broth in biphasic flasks or in MEMα to mid-log phase (OD600 of 0.7 or 0.2) at 37°C under microaerophilic conditions for the reverse transcriptase PCR (RT-PCR) or quantitative RT-PCR (qRT-PCR) assays, respectively. Cells were harvested following the addition of 800 μl of cold RNA degradation stop solution (10% buffer-saturated phenol [pH 4.3] in ethanol) to prevent RNA turnover (10). The cultures were centrifuged and resuspended in Tris-EDTA (TE) buffer, and total RNA was extracted using the hot phenol-chloroform method (44). The RNA was subsequently precipitated, resuspended in diethyl pyrocarbonate (DEPC)-treated water, and treated with DNase I (Invitrogen) to remove contaminating DNA. Finally, the RNA samples were further purified using the RNeasy kit (Qiagen). PCR was performed to ensure the absence of genomic DNA. RNA integrity and quantity were determined using the Experion RNA STDsens analysis kit (Bio-Rad Laboratories).
RT-PCR assay.
Reverse transcriptase PCR experiments were performed using the Qiagen One Step RT-PCR kit (Qiagen). All primers used in the RT-PCR experiments are listed in Table 2. Primers were designed to amplify transcripts specific for katA, Cj1386, and the intragenic region between katA and Cj1386. The reverse transcription was performed at 50°C for 30 min followed by an initial PCR activation step at 95°C for 15 min. The PCR cycling steps consisted of denaturing at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 1 min for a total of 30 cycles, followed by a final extension step at 72°C for 10 min. A positive control using C. jejuni genomic DNA was included to confirm that the primers were able to anneal and amplify the intended target regions. Additionally, a negative control without reverse transcriptase was included to ensure the absence of contaminating genomic DNA.
qRT-PCR.
The relative expression levels of the Cj1386 and katA transcripts in the ΔperR and ΔCj1386 mutants, respectively, were determined using the QuantiTect Sybr green RT-PCR kit (Qiagen) and a 7300 real-time PCR system (Applied Biosystems) as described previously (33). The primers used for the qRT-PCR experiment are listed in Table 2. The relative expression levels of Cj1386 and katA were normalized to metC (putative cystathionine beta-lyase). metC expression levels remained unchanged in the wild-type C. jejuni, ΔperR, and ΔCj1386 strains, ensuring that metC was a suitable choice of gene to use as a reference for the quantification of the Cj1386 and katA transcripts (37). The threshold cycle (ΔΔCT) method was used to calculate the relative fold change of the katA transcript, and specific RT-PCR products were confirmed by dissociation curve analysis according to the manufacturer's recommendations (Applied Biosystems). Experiments were done in biological duplicate with technical triplicates for each gene.
Purification of C. jejuni KatA and anti-KatA antiserum production.
Overexpression of KatA was performed in E. coli BL21 cells using the protein expression vector pGST as described previously (42). Briefly, the C. jejuni katA gene was PCR amplified using Pfx (Invitrogen) high-fidelity polymerase and the KatA_NcoI and KatA_NotI primers listed in Table 2. The amplified gene was cloned into the protein overexpression vector pGST using NcoI and NotI restriction sites, followed by transformation of the final construct into E. coli BL21 cells. Sequencing was performed to confirm the absence of polymerase-introduced mutations in the katA gene. The strain containing the pGST+KatA construct was grown in 400 ml of LB broth supplemented with 100 μg/ml of ampicillin to an OD600 of 0.6 at 37°C with continual shaking. IPTG (isopropyl-β-d-thiogalactopyranoside; 500 μM) was added to the broth, and the bacterial culture was incubated for an additional 3 h at 37°C. The cells were then pelleted and resuspended in phosphate-buffered saline (PBS) containing protease inhibitor (Roche), and the cell membranes were disrupted by sonication. Cell membranes and debris were removed by centrifugation at 13,000 rpm for 15 min. The cell lysate containing the glutathione S-transferase (GST)–KatA fusion protein was then affinity purified using glutathione Sepharose 4B resin according to the manufacturer's specifications (GE Healthcare). Cleavage of the GST tag from KatA was performed on the resin by addition of tobacco etch virus (TEV) protease (42) and gentle shaking overnight at 4°C. The KatA protein was washed from the resin the following day using 6 washes of 200 μl of PBS buffer. Purified protein was frozen and stored at −20°C until use.
Approximately 2.5 mg of KatA protein was used for antibody production by Immuno-Precise Antibodies Limited using 2 rabbits (Victoria, BC, Canada). For each rabbit, a preimmune bleed was performed prior to the primary immunization with the KatA antigen (0.5 mg) using complete Freund's antigen. Over the course of the project, each rabbit received 3 additional boosts with KatA antigen (0.5 mg) using incomplete Freund's antigen followed by a terminal bleed and serum collection. Anti-KatA antiserum was stored at −20°C until use.
Western blotting.
Protein lysates from C. jejuni strains were separated by SDS-PAGE on a 12% denaturing gel and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore) using a wet-electroblotting transfer cell (Bio-Rad). Membranes were blocked overnight in 5% (wt/vol) skim milk-0.1% Tween 20 in PBS followed by incubation with 0.1 μg/ml anti-KatA antiserum in PBS for 1 h at room temperature. The membranes were washed 6 times with PBS followed by incubation with a 1:3,000 dilution of anti-rabbit horseradish peroxidase (HRP)-conjugated antibody (Invitrogen) for 1 h at room temperature. The immunoblot membrane was developed with a 1:1 mixture of luminol-peroxide solution (Thermo Scientific) for 1 min, and chemiluminescence was detected by X-ray film (Thermo Scientific). Densitometry of immunoblot results was performed with the use of Adobe Photoshop software (version 10.0).
Immunoprecipitation of KatA.
Immunoprecipitation experiments using wild-type C. jejuni, ΔCj1386, ΔkatA, and ΔCj1386+Cj1386 strains were performed using protein A-conjugated Dynabeads (Invitrogen). Strains were grown to mid-log phase (OD600 of 0.2) in MEMα under microaerophilic conditions at 37°C prior to harvesting total soluble proteins. Bacterial strains were spun at 6,000 rpm for 10 min, resuspended in 1 ml PBS containing a bacterial protease inhibitor cocktail (Sigma) and 10 mg/ml lysozyme, and incubated on ice for 15 min. Cells were briefly sonicated on ice (five 5-s pulses), followed by centrifugation at 13,000 rpm for 5 min at 4°C to remove membranes and cellular debris. Two hundred fifty micrograms of anti-KatA antiserum diluted in 200 μl PBS-0.02% Tween 20 was incubated with 50 μl of Dynabeads for 1 h with end-over-end rotation at room temperature. The anti-KatA-bound Dynabeads were washed once with 500 μl PBS-0.02% Tween 20 before addition of 5 mg of protein lysate. The bead-lysate mixture was incubated overnight at 4°C with end-over-end rotation. The beads were washed 3 times with 200 μl of ice-chilled PBS, and KatA was eluted from the beads twice in 100 μl soft-elution buffer (50 mM Tris, pH 8.0, 0.2% SDS, 0.1% Tween 20) (3) with end-over-end rotation for 7 min at room temperature. Immunoprecipitation of KatA from each strain was visualized by SDS-PAGE run on a 10% denaturing gel followed by Coomassie blue staining. KatA protein concentration was determined for each immunoprecipitated sample by densitometry of the SDS-PAGE gel using Adobe Photoshop software (version 10.0). KatA protein content was normalized, and equal amounts of KatA (250 ng) from the wild-type NCTC11168, ΔCj1386, and ΔCj1386+Cj1386 strains were assayed for catalase activity using the method described above. Immunoprecipitate from the ΔkatA strain was used as a negative control for the catalase activity assay. Catalase activity assays were performed in quadruplicate, and statistical significance was determined using the Student t test. P values of <0.05 were considered significant.
Hemin quantification assays.
Heme content of KatA protein isolated from wild-type C. jejuni NCTC11168, ΔCj1386, ΔkatA, and ΔCj1386+Cj1386 strains was quantified using the hemin assay kit (Biovision, Mountain View, CA), which utilizes the peroxidase activity of hemin to produce a colored substrate which can be assayed spectrophotometrically. Briefly, KatA was immunoprecipitated from wild-type C. jejuni NCTC11168, ΔkatA, ΔCj1386, and ΔCj1386+Cj1386 strains using the same method as described above with slight modification to the elution step. Elution was carried out by addition of 20 μl of 50 mM glycine, pH 2.8, to the Dynabeads to release protein complexes and to extract heme from the KatA protein. Samples were incubated at room temperature for 2 min with end-over-end rotation and subsequently transferred to a clean tube. The samples were brought to neutral pH by the addition of 1 M Tris, pH 7.4. Samples were visualized by SDS-PAGE run on a 10% denaturing gel followed by Coomassie blue staining. KatA protein content was normalized using densitometry (Adobe Photoshop software [version 10.0]). Total hemin concentration was assayed from 40 ng of KatA protein prepared from each immunoprecipitated sample, and the assay was performed according to the manufacturer's instructions (hemin assay kit; Biovision, Mountain View, CA). Background absorbance of IgG (determined from the ΔkatA strain immunoprecipitated sample) was subtracted from each of the strains assayed. Experiments were performed in biological quadruplicate. Statistical significance was determined by the Student t test with P values of <0.05 considered significant.
Chick colonization model.
The chick colonization model for C. jejuni was used as described previously (33). One-day-old specific-pathogen-free broiler chicks were checked upon arrival to ensure the absence of Campylobacter contamination by culturing the fecal contents on Karmali plates (Oxoid). Chicks were provided with water and commercial chicken starter diet ad libitum. Wild-type C. jejuni, ΔCj1386 mutant, and ΔCj1386+Cj1386 strains were grown to mid-log phase in MH broth under microaerophilic conditions at 37°C. The strains were recovered by centrifugation, washed in PBS, and resuspended in fresh MH broth at a concentration of 2 × 104 cells per ml. Food and water were withheld from the chicks for 2 h prior to inoculation. Each 3-day-old chick was inoculated with 0.5 ml of the bacterial suspension (containing approximately 104 viable C. jejuni bacteria). Each bacterial suspension was serially diluted and plated on MH agar plates to confirm the inoculum. Six days later, the chicks were euthanized and necropsy was performed. The ceca were collected and weighed, and the cecal contents were extracted and homogenized in MH broth. Cecal contents were serially diluted and plated onto Karmali agar plates supplemented with either 20 μg/ml chloramphenicol or 10 μg/ml kanamycin for the mutant and complemented strains, respectively. The plates were incubated for 48 h at 42°C under microaerophilic conditions, and the resulting colonies were counted and expressed as CFU per gram of cecum. Statistical analysis was done using the nonparametric Mann-Whitney rank sum test with P values of <0.05 being considered statistically significant.
Neonate piglet infectious model.
The procedure used for the neonate piglet infectious model has been previously described (31). Newborn piglets were housed under sterile conditions at room temperature provided with additional warmth from heat lamps. Newborn piglets were fed a milk replacer five times daily. On the first day of feeding, piglets were syringe fed 60 to 120 ml of the milk replacer. Wild-type, ΔkatA, and ΔCj1386 strains were grown up to mid-log phase overnight under microaerophilic conditions at 37°C in MH broth. Piglets were inoculated orally by the use of a syringe 1 day after they were received. Each piglet received approximately 10 ml of a 1:1 mixture of wild-type and ΔCj1386 mutant or wild-type and ΔkatA bacterial suspension (approximately 108 viable C. jejuni bacteria). Serial dilutions of the inoculum were spread onto MH agar plates and MH agar plates supplemented with 20 μg/ml chloramphenicol to confirm the ratio of ΔCj1386 mutant to wild type and ΔkatA mutant to wild type. The piglets were euthanized 2 days following inoculation, and necropsy was performed immediately after. Two- to three-inch segments of the duodenum, jejunum, ileum, colon, and cecum were removed and weighed, and the mucus layer from each segment was collected and homogenized. Serial dilutions were made for each homogenate of intestine, of which 100 μl was spread in triplicate onto Karmali agar plates (Campylobacter selective supplement; Oxoid) with chloramphenicol added to half the plates to select for the ΔCj1386 and ΔkatA mutants. Karmali agar plates were held under microaerophilic conditions at 37°C for 2 days. The resulting Campylobacter colonies were counted. The wild-type strain and mutant strains were tested in at least 3 piglets each. The colonization levels of the mutant strains were directly obtained by counting the colonies present on the Karmali agar plates supplemented with chloramphenicol. The wild-type C. jejuni titer was obtained by subtracting the number of mutant colonies from the total number of colonies recovered on the Karmali agar plates. The competitive index was calculated by dividing the in vivo ratio of mutant to wild-type strain recovered (output ratio) by the in vitro ratio of the mutant to wild-type strain inoculated (input ratio). The data were analyzed for statistical significance using a nonparametric Mann-Whitney rank sum test. P values of <0.05 were considered statistically significant.
RESULTS
Cj1386 plays an important role in H2O2 detoxification.
Analysis of the katA genetic region of C. jejuni NCTC11168 revealed the presence of a downstream gene, Cj1386, encoding an ankyrin repeat-containing protein of unknown function. Transcriptome studies have demonstrated that Cj1386 transcript level increases in response to iron starvation (24, 33) and suggested that Cj1386 might be a member of the PerR regulon (32). Indeed, quantitative reverse transcriptase PCR confirmed the regulation of Cj1386 expression by PerR with up to a 72- ± 24-fold increase in Cj1386 transcript abundance in the ΔperR mutant strain compared to the wild type. Interestingly, orthologues for Cj1386 are conserved among Campylobacter species, and in Pseudomonas aeruginosa a gene encoding an ankyrin repeat-containing protein (AnkB) has been also identified downstream of the katB gene and was found to enhance Pseudomonas catalase activity (25). To assess the potential role for Cj1386 in defense against ROS, an isogenic deletion mutant was constructed by allelic exchange as described in Materials and Methods. Additionally, a complemented strain was constructed by introducing Cj1386 into one of the three rRNA clusters in the ΔCj1386 mutant background. Next, the ΔCj1386 mutant was tested for its sensitivity toward three different oxidants: hydrogen peroxide (H2O2), cumene hydroperoxide (CHP), and menadione sodium bisulfite. As shown in Table 3, the ΔCj1386 mutant exhibited increased sensitivity toward 3% H2O2 relative to the wild-type C. jejuni (P < 0.0005), and the phenotype was fully restored by complementation with the Cj1386 gene (strain ΔC1386+Cj1386). The ΔCj1386 mutant was not, however, affected in its sensitivity toward either cumene hydroperoxide or menadione relative to the wild-type strain. These results indicate that Cj1386 plays an important function in resistance to hydrogen peroxide in C. jejuni and that this function is oxidant specific.
Table 3.
Sensitivity of wild-type C. jejuni, ΔkatA and ΔCj1386 mutants, and corresponding complemented strains to three oxidantsa
| Strain | Diam (mm) of zone of inhibition after exposure to oxidant: | ||
|---|---|---|---|
| H2O2 | CHP | MND | |
| C. jejuni NCTC11168 | 19.70 ± 0.38 | 22.90 ± 0.52 | 34.8 ± 0.79 |
| ΔkatA mutant | 25.30 ± 0.07* | 24.58 ± 0.19 | 29.85 ± 1.39 |
| ΔkatA+ katA strain | 10.83 ± 1.24 | 23.63 ± 0.47 | 31.00 ± 1.20 |
| ΔCj1386 mutant | 25.21 ± 0.28* | 25.67 ± 0.76 | 35.04 ± 1.36 |
| ΔCj1386 + Cj1386 strain | 19.90 ± 0.57 | 24.20 ± 0.51 | 37.30 ± 0.76 |
| ΔkatAΔCj1386 mutant | 25.80 ± 0.39* | 22.90 ± 0.71 | 35.30 ± 1.12 |
| ΔkatAΔCj1386 + (katA + Cj1386) strain | 10.70 ± 0.85 | 23.90 ± 0.40 | 37.50 ± 0.60 |
The diameter of the zone of inhibition is represented as the mean clear zone ± standard error for each strain (in mm) after exposure to 10 μl of 3% H2O2 (hydrogen peroxide), 3% cumene hydroperoxide (CHP), or 90 mM menadione (MND) bisulfite. Each experiment was repeated in quadruplicate. Values were considered significant (*) at P < 0.05 using ANOVA statistical analysis.
As indicated above, Cj1386 is located directly downstream from katA in the NCTC11168 genome (34). The gene katA encodes the only catalase of C. jejuni, which is the main enzyme responsible for the detoxification of hydrogen peroxide into oxygen and water. KatA has been previously demonstrated, both in vitro and in vivo, to have an important role in hydrogen peroxide defense in multiple strains of C. jejuni (17). Additionally, a ΔkatA mutant also displayed an increased sensitivity toward 3% H2O2 relative to the wild-type C. jejuni NCTC11168 (32) (also Table 3). Because of the similar sensitivities of the ΔkatA and ΔCj1386 mutants toward hydrogen peroxide and because of the genomic location of Cj1386 directly downstream from katA, we postulated that Cj1386 may be involved in the same hydrogen peroxide detoxification pathway as KatA. To test this hypothesis, a double katA and Cj1386 deletion mutant, the ΔkatA ΔCj1386 mutant, was constructed and assessed for H2O2 sensitivity. In the case of two independent H2O2 detoxification pathways encoded by katA and Cj1386, it would be expected that the loss of function of both KatA and Cj1386 would lead to an overall increase in sensitivity toward H2O2 relative to that of the single mutants. However, this phenotype was not observed (Table 3). Indeed, as shown in Table 3, the ΔkatA, ΔCj1386, and ΔkatA ΔCj1386 mutants exhibited equal susceptibilities to H2O2 (with a growth inhibition zone of approximately 25 mm compared to 19.7 mm for the wild-type strain). ANOVA statistical analysis confirmed that the three mutant strains were not significantly different from each other (P > 0.05). Complementation in trans of the ΔkatA, ΔCj1386, and ΔkatA ΔCj1386 mutants with the corresponding wild-type genes restored their sensitivity toward H2O2 at levels comparable to the parental strain. The increased resistance to H2O2 in the ΔkatA+katA and ΔkatA ΔCj1386+(katA+Cj1386) complemented strains relative to the wild type is likely due to the loss of iron- and H2O2-mediated regulation of the katA gene expression (32, 45) (as it is expressed from the kanamycin promoter in our construction). Altogether, these results indicate that Cj1386 is involved in the same detoxification pathway or mechanism as KatA.
The katA and Cj1386 genes are independently transcribed.
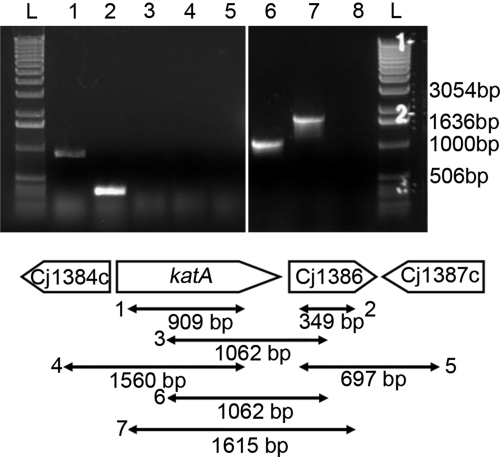
The genetic proximity of katA and Cj1386 genes suggests that these two genes might be cotranscribed. Therefore, to determine if these two genes belong to the same operon, we mapped the potential transcripts by RT-PCR using primers that anneal within and across these two genes. As shown Fig. 1, amplicons of the expected sizes were obtained for the katA (Fig. 1, lane 1) and Cj1386 (Fig. 1, lane 2) transcripts. However, RT-PCR of the katA-Cj1386 region using one primer annealing within katA in conjunction with a Cj1386-specific primer failed to yield any product (Fig. 1, lane 3). Of note, RT-PCR using the same primers but with genomic DNA as a template was successful (Fig. 1, lane 6). These results indicate that katA and Cj1386 are not cotranscribed.
Fig 1.
The katA and Cj1386 genes are independently transcribed. RT-PCR products from RNA extracted from C. jejuni NCTC11168 were analyzed by agarose gel electrophoresis for operon identification analysis. Lanes 1 to 5, RT-PCR from RNA extracted from C. jejuni NCTC11168; lanes 6 and 7, RT-PCR from genomic DNA extracted from C. jejuni NCTC11168. Lanes: L, 1-kb ladder; 1, katA-RT-SE and katA-RT-AS (909-bp band visible, demonstrating presence of katA transcript); 2, Cj1386-RT-SE and Cj1386-RT-AS (349-bp band visible, demonstrating presence of Cj1386 transcript); 3, katA-RTint-SE and Cj1386-RTint-AS (absence of predicted 1,062-bp band, indicating that katA and Cj1386 are not cotranscribed); 4, Cj1384c-RT-SE and katA-RT-AS (absence of predicted 1,560-bp band, indicating that Cj1384c and katA are not cotranscribed); 5, Cj1386-RT-SE and Cj1387-RT-AS (absence of predicted 697-bp band, indicating that Cj1386 and Cj1387c are not cotranscribed); 6, positive-control katA-RTint-SE and Cj1386-RTint-AS (1,062-bp band present); 7, positive-control katA-RT-SE and Cj1386-RT-AS (1,615-bp band present); 8, negative control (no RT added to reaction mixture).
Zymographic analysis of C. jejuni catalase activity detected a single catalase.
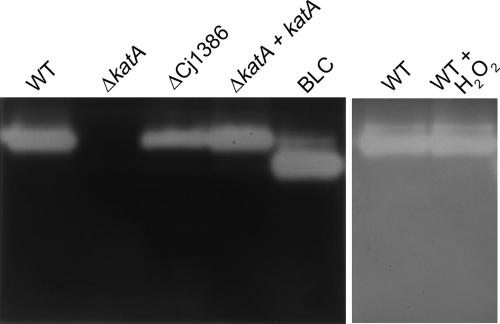
To further test the hypothesis that Cj1386 is involved in the same H2O2 detoxification pathway as KatA, we assessed the catalase activity of the wild-type strain and the ΔkatA and ΔCj1386 mutants by zymographic analysis (Fig. 2). The mutants and the wild-type strain were grown in iron-limited MEMα in order to ensure high expression of katA and Cj1386 genes (24, 33). As shown in Fig. 2, the wild-type C. jejuni strain contained a single enzyme with catalase activity (lane 1), the ΔkatA mutant did not produce detectable catalase activity (lane 2), and the ΔCj1386 mutant showed catalase activity, yet at a reduced level relative to that of the wild-type strain (lane 3). The catalase activity was fully restored in the ΔkatA+katA complemented strain to levels of enzymatic activity comparable to those of the wild-type C. jejuni. Bovine liver catalase (Sigma) was used as a positive control for catalase activity (lane 5). Finally, the addition of 1 mM H2O2 to wild-type C. jejuni did not induce expression of additional enzymes with detectable catalase activity (lane 7). These results indicate that C. jejuni possesses a single enzyme with catalase activity (KatA). Furthermore, similar levels of catalase activity were observed in C. jejuni in the presence and in the absence of H2O2 exposure. This observation is in agreement with the relief of PerR repression under iron-restricted conditions (used in this assay), enabling high levels of katA expression (32). Overall, the reduced catalase activity observed in the ΔCj1386 mutant suggests that Cj1386 contributes to KatA enzymatic function.
Fig 2.
The C. jejuni strain contains a single enzyme with catalase activity, and the ΔCj1386 mutant exhibits a reduced catalase activity. All bacterial strains (C. jejuni NCTC11168 and ΔkatA and ΔCj1386 mutants) were grown in MEMα to mid-log phase (OD600 of 0.2) under microaerophilic conditions at 37°C prior to cell extract preparation. When required, wild-type C. jejuni was exposed to 1 mM H2O2 for 10 min prior to total protein isolation. Fifty micrograms of whole-cell extract was loaded onto an 8% nondenaturing gel run at 25 mA at 4°C and assayed for catalase activity. BLC, bovine liver catalase (Sigma-Aldrich).
Cj1386 plays a role in enhancing KatA catalase activity.
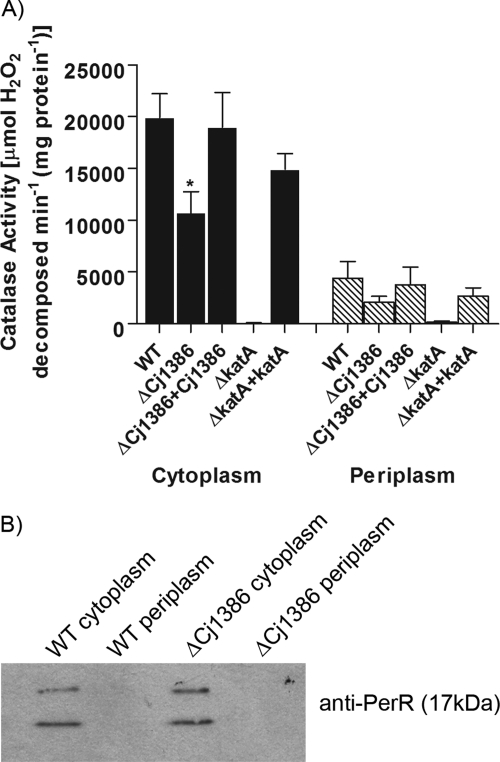
According to our zymographic analysis, the absence of Cj1386 decreased catalase activity. To confirm this finding, the catalase activities of the wild-type strain and the ΔkatA and ΔCj1386 mutants were quantified and compared according to the method developed by Beers and Sizer (7). Unlike many other bacteria, which have two catalases, one cytoplasmic and one periplasmic, C. jejuni harbors a single catalase of as-yet-undefined location. A previous study by van Vliet et al. has shown KatA to be present within the periplasmic space in C. jejuni (45). However, it remains unknown how KatA is transported into the periplasmic space as the translocation process appears to be TAT (twin-arginine translocase) and Sec independent due to the absence of a TAT (36) or Sec leader sequence (SignalP) (8) in the KatA protein sequence. Therefore, to confirm the localization of C. jejuni catalase, the enzymatic activity was tested in both cytoplasmic and periplasmic protein preparations from C. jejuni grown to mid-log phase (OD600 of 0.2) in MEMα. Cytoplasmic and periplasmic protein fractions were prepared by using the Peripreps Periplasting kit (Epicentre Technologies, Madison, WI), which utilizes a combination of osmotic shock and lysozyme treatment. We have previously used and validated this kit to isolate periplasmic fractions from C. jejuni (31). Nevertheless, we further confirmed the absence of significant contamination of our periplasmic protein fractions with cytoplasmic proteins by Western blot analysis using antibodies against the transcriptional regulator PerR, which should be localized exclusively in the cytoplasm (Fig. 3B). Figure 3 shows the catalase activities of the wild-type C. jejuni, the ΔkatA and ΔCj1386 mutants, and their complemented strains for both cytoplasmic and periplasmic protein fractions. The ΔCj1386 mutant displayed an overall decrease in KatA activity in both the cytoplasm and periplasm. The ΔCj1386 mutant has significantly reduced catalase activity (1.1 × 104 ± 2.0 × 103 U/mg protein) relative to the wild-type C. jejuni (2.0 × 104 ± 2.3 × 103 U/mg protein) in the cytoplasmic preparation (P < 0.05). The ΔCj1386 mutant also had reduced catalase activity compared to the wild-type strain in the periplasmic fraction, but this reduction was not statistically significant. The complemented ΔCj1386 strain exhibited catalase activity equal to that of the wild type in both the periplasmic and cytoplasmic fractions. As expected, no appreciable catalase activity could be detected in the ΔkatA mutant in either the cytoplasm or periplasmic fractions, while catalase activity was restored in the ΔkatA+katA complemented strain. Altogether, these results indicate that that Cj1386 is important in promoting proper catalase activity in C. jejuni and that C. jejuni catalase is located in both the periplasmic and cytoplasmic fractions. Catalase enzymes have also been shown to be present within both the cytoplasmic and periplasmic spaces of other bacterial species, including KatA in Helicobacter pylori (20), Vibrio rumoiensis (51), and Pseudomonas aeruginosa (14); CatF in Pseudomonas syringae (29); and a catalase/peroxidase in Caulobacter crescentus (38).
Fig 3.
Cj1386 enhances KatA catalase activity. (A) Catalase activities of cytoplasmic and periplasmic cellular protein fractions for wild-type C. jejuni and ΔCj1386, ΔCj1386+Cj1386, ΔkatA, and ΔkatA+katA strains are expressed as μmol of hydrogen peroxide decomposed per minute per mg of protein. Error bars indicate standard errors of the means (n = 4). An asterisk indicates a P value of <0.05 using the Student t test. (B) Cell fractionation control assay using Western blot analysis. Five micrograms of wild-type C. jejuni and ΔCj1386 mutant cytoplasmic and periplasmic protein extracts were loaded into each well and subsequently assessed for cytoplasmic protein contamination by immunoblotting using an anti-PerR antibody.
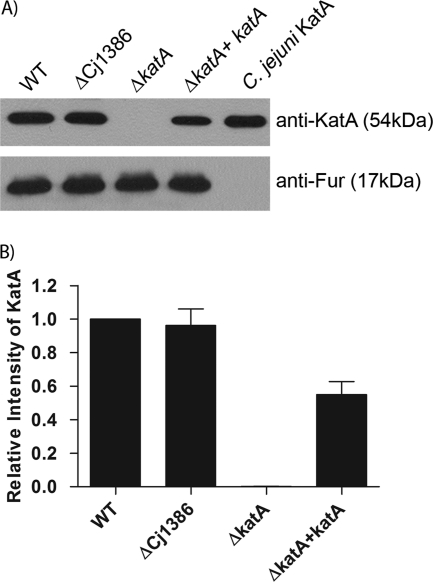
The observed decrease in catalase activity in the ΔCj1386 mutant could be explained either by a direct role for Cj1386 in KatA function or by an effect of Cj1386 on KatA expression. To determine whether the reduction in catalase activity seen in the ΔCj1386 mutant was the result of a decrease in KatA expression, qRT-PCR and Western blot analysis were performed. No significant difference in katA transcript abundance could be detected in the ΔCj1386 mutant strain compared to the wild-type C. jejuni by qRT-PCR analysis (data not shown). Additionally, Western blot analysis using anti-KatA antisera to compare the relative KatA protein levels in whole-cell extracts from wild-type C. jejuni, ΔCj1386 mutant, ΔkatA mutant, and the complemented strains indicated no significant decrease in the amount of KatA in the ΔCj1386 mutant strain compared to the wild-type strain (Fig. 4). The lack of a band in the ΔkatA mutant confirmed the specificity of the anti-KatA antibody (Fig. 4A, lane 3). Equal loading of each protein lysate sample was confirmed using an anti-Fur antibody (Fig. 4A, lower panel). Quantification of the results is seen in Fig. 4B. These results demonstrate that the decrease in catalase activity seen in the ΔCj1386 mutant strain is not due to a decrease in either katA transcript or KatA protein level.
Fig 4.
KatA expression is not affected in the ΔCj1386 mutant, as shown by Western blot analysis of KatA protein content in wild-type C. jejuni; ΔCj1386, ΔkatA, and ΔkatA+katA mutants; and affinity-purified C. jejuni KatA. (A) Five micrograms of protein lysate or 100 ng of purified protein was loaded into each lane followed by immunoblotting using anti-KatA or anti-Fur antiserum. (Top) KatA protein level as detected by anti-KatA antiserum. (Bottom) Loading control of total protein content detected by anti-Fur antiserum. (B) Quantification of KatA protein level in wild-type C. jejuni, ΔCj1386, ΔkatA, and ΔkatA+katA strains. Relative intensity of KatA was determined by quantifying each band from the immunoblot and standardizing it against WT KatA. Error bars represent the standard errors of 3 biological replicates.
Cj1386 is involved in heme trafficking to KatA.
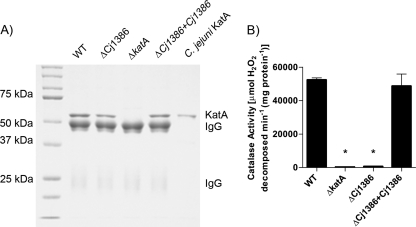
The catalase activity and KatA expression quantification results for the ΔCj1386 strain suggest that Cj1386 is playing a direct role in KatA function. To date, the reaction catalyzed by catalase has been well documented and involves a two-step oxidation and reduction reaction to detoxify hydrogen peroxide. Central to this reaction is a heme prosthetic group, which plays an important role in electron transfer to facilitate the redox reactions that decompose the hydrogen peroxide molecules (15). Given that there was a significant decrease in catalase activity, but no decrease in KatA expression in the ΔCj1386 strain, we hypothesized that the decrease in KatA activity might be due to a decrease in the heme cofactor in the expressed KatA protein. To test this hypothesis, the KatA from the wild-type C. jejuni NCTC1168, ΔkatA, ΔCj1386, and ΔCj1386+Cj1386 strains was immunoprecipitated and assayed for catalase activity, and the heme content of the protein was quantified. Figure 5A shows typical results of the immunoprecipitation experiments with elution in 50 mM glycine, pH 2.8. The specificity of the immunoprecipitated KatA can be seen in the ΔkatA mutant strain in which the band corresponding to 54-kDa KatA is absent. Catalase activity assays were performed on the immunoprecipitated samples that were eluted in soft-elution buffer (the soft-elution buffer did not decrease the activity of wild-type KatA [data not shown]). Interestingly, the catalase activity of the immunoprecipitated strains revealed that the ΔCj1386 mutant KatA had a severe reduction in catalase activity (625 U/mg protein) relative to the wild-type C. jejuni KatA (5.2 × 104 U/mg protein) (Fig. 5B). The catalase activity was restored in the complemented ΔCj1386+Cj1386 KatA protein (4.9 × 104 U/mg protein).
Fig 5.
KatA immunoprecipitated from the ΔCj1386 mutant has decreased catalase activity relative to wild-type C. jejuni. (A) Immunoprecipitated KatA from wild-type C. jejuni, ΔCj1386, ΔkatA, and ΔCj1386+Cj1386 strains eluted in 50 mM glycine, pH 2.8. Five microliters of each immunoprecipitated sample and 0.5 μg of purified C. jejuni KatA were loaded into each lane and separated on a 10% denaturing SDS-PAGE gel. (B) Catalase activity of KatA immunoprecipitated samples eluted in soft-elution buffer from wild-type C. jejuni, ΔCj1386, ΔkatA, and ΔCj1386+Cj1386 strains. KatA protein concentrations were determined by densitometry from the SDS-PAGE gel (not shown), and 250 μg of KatA was assayed for activity. Catalase activity is expressed as μmol of hydrogen peroxide decomposed per minute per mg of protein. Error bars indicate standard errors of the means (n = 4). An asterisk indicates a P value of <0.05 using the Student t test.
Next, we quantified the heme content of the immunoprecipitated KatA samples from each strain. To do this, the heme was extracted from the immunoprecipitated KatA protein samples under acidic conditions by suspending the KatA-conjugated beads in 50 mM glycine solution at pH 2.8. Exposing heme-containing proteins to low-pH conditions allows for extraction of heme from proteins (4, 46) and also elutes the proteins bound to the beads. Following elution from the beads, the immunoprecipitated samples were brought to neutral pH by addition of 1 M Tris, pH 7.4. No detectable catalase activity was observed in the immunoprecipitated samples (data not shown).
Quantification of the heme content from the immunoprecipitated KatA samples (Table 4) revealed a significant reduction in the concentration of heme present in the ΔCj1386 mutant strain (0.19 ± 0.1 nM) relative to the wild-type C. jejuni (6.99 ± 0.19 nM). The concentration of heme was restored to wild-type levels in the complemented ΔCj1386+Cj1386 strain (6.41 ± 0.54 nM). Furthermore, the ratio of heme per KatA subunit (54 kDa) was close to a 1:1 ratio (0.94, Table 4) as would be expected for tetrameric, monofunctional, heme-containing catalases (15). The ΔCj1386 immunoprecipitated KatA-heme ratio was significantly reduced at approximately 0.03 heme group per KatA subunit. This result suggests that 97% of the immunoprecipitated KatA from the ΔCj1386 mutant strain is lacking the heme prosthetic group, providing an explanation for the significant reduction in enzymatic activity of this protein as assayed in Fig. 5B. These results suggest an important role for Cj1386 in heme trafficking to KatA.
Table 4.
Quantification of heme content from KatA protein immunoprecipitated from wild-type C. jejuni NCTC11168, ΔCj1386, and ΔCj1386 + Cj1386 strainsa
| Source of immunoprecipitated KatA | Concn (nM) of: |
Ratio (heme/KatA) | |
|---|---|---|---|
| KatA (subunits) | Heme | ||
| C. jejuni NCTC11168 | 7.4 | 6.99 ± 0.19 | 0.94 |
| ΔCj1386 strain | 7.4 | 0.19 ± 0.10* | 0.03 |
| ΔCj1386 + Cj1386 strain | 7.4 | 6.41 ± 0.54 | 0.87 |
The amount of heme is represented as the mean concentration of heme detected ± the standard error (nM) for equal starting concentrations of KatA protein for each strain tested. Experiments were repeated in biological quadruplicate. The Student t test was used to determine statistical significance with P values of <0.05 (*) considered significant.
Chick colonization model.
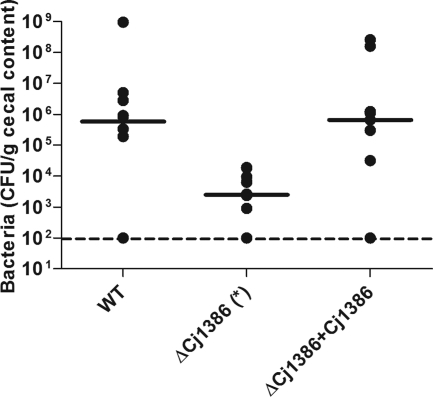
To determine whether Cj1386 is important in the colonization of chick ceca, 1-day-old specific-pathogen-free broiler chicks were inoculated with wild-type C. jejuni NCTC11168, ΔCj1386 mutant, and the ΔCj1386+Cj1386 strains. As shown in Fig. 6, the wild-type C. jejuni colonized the ceca at a level of approximately 106 CFU per gram. The ΔCj1386 mutant was significantly affected in its ability to colonize the ceca relative to the wild-type strain and colonized at a level of approximately 2.5 × 103 CFU per gram (P < 0.05). Complementation in trans of ΔCj1386 with Cj1386 restored the colonization ability of the mutant strain to levels comparable to that of the wild type. These results indicate an important role for Cj1386 in chick colonization.
Fig 6.
The ΔCj1386 mutant is defective for cecal colonization of chicks. Chicks were grouped into two sets of 10 (WT and ΔCj1386 + Cj1386) and one set of 12 (ΔCj1386). Data points correspond to the levels of colonization of the ceca per chick. The dashed line represents the limit of detection for the assay. Solid bars indicate the median colonization level of bacteria for each strain. The asterisk indicates a P value of <0.05 using a nonparametric Mann-Whitney rank sum test.
Neonate piglet infectious model.
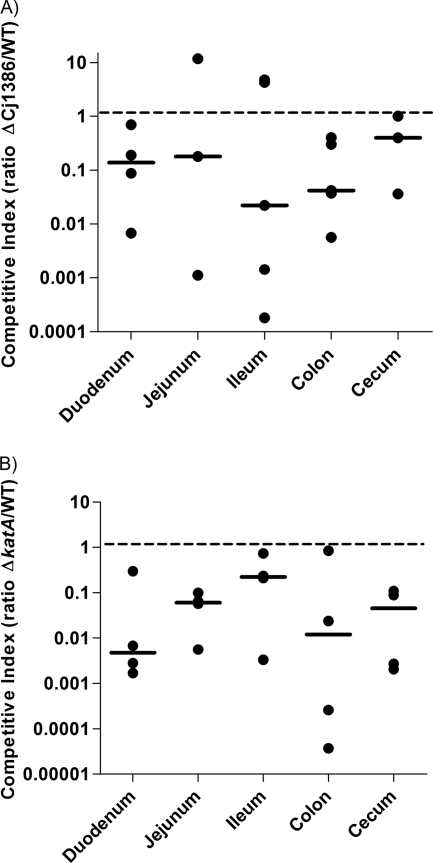
It has been previously shown that upon infection with C. jejuni, piglets become ill and display symptoms which are similar to those observed in humans affected by campylobacteriosis (6). Therefore, neonate piglets were used as an infectious model to assess the importance of Cj1386 and KatA in pathogenesis and colonization of piglet intestines. Colostrum-deprived piglets were inoculated with a suspension containing a 1:1 ratio of wild-type C. jejuni to ΔCj1386 or ΔkatA mutant (approximately 108 viable C. jejuni bacteria). The piglets were euthanized 2 days after infection, and the competitive index was calculated (as described in Materials and Methods) for each intestinal segment. A competitive index of less than 1 indicates that the ΔCj1386 mutant was outcompeted by the wild-type C. jejuni. As shown in Fig. 7A, the median for the competitive index corresponding to the five different intestinal segments from the ΔCj1386–wild-type competition was less than 1 (P < 0.0013), indicating that the ΔCj1386 mutant was significantly outcompeted by the wild-type C. jejuni in the colonization of piglet intestines. Figure 7B shows that the ΔkatA mutant was significantly outcompeted by the wild-type C. jejuni, in colonization of all five intestinal segments (P < 0.0001). Importantly, in vitro competitive growth assays of wild-type C. jejuni and ΔCj1386 mutant strains and wild-type C. jejuni and ΔkatA mutant strains found that the ΔCj1386 and ΔkatA mutants were not outcompeted by the wild-type strain (data not shown). These results indicate that the outcompetition of ΔCj1386 and ΔkatA mutants by the wild-type strain (Fig. 7) in vivo is not due to an in vitro growth difference between the mutants and the wild-type strain. The data from the neonate competitive piglet assay suggest that Cj1386 and KatA play important roles in colonization of the piglet intestine.
Fig 7.
ΔCj1386 and ΔkatA mutants are affected for intestinal colonization of piglets. Competitive index in neonate piglets. Each data point represents the competitive index for C. jejuni NCTC11168 and the ΔCj1386 (A) or ΔkatA (B) mutant in five intestinal segments from one piglet. The dashed line represents the ratio at which wild-type C. jejuni and the ΔCj1386 (A) or ΔkatA (B) mutant are colonizing the intestinal segment at similar levels (one is not outcompeting the other). Solid bars represent the medians for each segment.
DISCUSSION
Hydrogen peroxide is a harmful by-product of metabolic bacterial activities, mainly from the electron transfer chains (28). In the presence of iron, the Fenton reaction process converts H2O2 to hydroxyl radicals, the most powerful oxidizing species in the biosphere (28). Efficient and rapid removal of H2O2 is therefore vital for all living organisms. Hydrogen peroxide is primarily detoxified by catalases, which degrade H2O2 to water and oxygen (39). Alternatively, cytochrome c peroxidases and peroxiredoxins could also catalyze the reduction of H2O2 to water using cytochrome c or reduced pyridine dinucleotides as the electron donor. C. jejuni NCTC11168 contains one catalase (KatA), two cytochrome c peroxidases (Cj0358 and Cj0020c), and three peroxiredoxins (AhpC, Tpx, and Bcp) (34). KatA, Tpx, and Bcp have been shown to contribute, at least to some extent, to H2O2 detoxification in C. jejuni (5, 17), and Cj0358 and Cj0020c have been demonstrated to have peroxidase activity (11). AhpC has also been demonstrated to scavenge H2O2 in E. coli (39) and likely performs a similar function in C. jejuni. In this study, we identified and characterized one additional member of the C. jejuni anti-H2O2 defense, Cj1386, and demonstrated that KatA is localized in both the cytoplasmic and periplasmic fractions. A ΔCj1386 mutant exhibited increased sensitivity toward H2O2. Complementation in trans of the ΔCj1386 mutant with Cj1386 restored the wild-type levels of sensitivity, confirming that the observed phenotype was due to deletion of Cj1386 and not due to polar effects in surrounding genes (in particular, katA). These observations indicate that Cj1386 functions to protect the cell from exposure to H2O2. Cj1386 is an ankyrin repeat-containing protein of 156 amino acids (34). An ankyrin repeat is comprised of a series of 33 amino acids, and these repeats mediate protein-protein interactions (9). Cj1386 is located directly downstream from katA within the C. jejuni NCTC11168 genome (34). Although katA and Cj1386 are not cotranscribed, the ΔkatA, ΔCj1386, and ΔkatA ΔCj1386 mutants all displayed similar sensitivities toward H2O2, indicating that KatA and Cj1386 are involved in the same H2O2 defense pathway within the cell. Another important finding is the presence of KatA-mediated catalase activity in both the periplasmic and cytoplasmic fractions. Periplasmic catalase activity might endow C. jejuni with enhanced resistance toward aggressive H2O2 production during host inflammation by detoxifying H2O2 before it reaches and damages essential cytoplasmic proteins. The ΔCj1386 mutant has reduced catalase activity in both compartments (the reduction in the periplasmic fraction, however, was not statistically significant), suggesting that Cj1386 is required for proper KatA function and not for KatA localization. Furthermore, the KatA enzyme in the ΔCj1386 mutant strain was found to have a significant decrease in heme content. This finding indicates that Cj1386 is involved in heme trafficking within C. jejuni. Although to date there is little known about heme incorporation and folding of catalase enzymes in bacteria, we favor this explanation over a role for Cj1386 in KatA protein folding, as experiments performed with purified, denatured bovine liver catalase have demonstrated complete protein refolding and restoration of activity without the aid of a chaperone or additional factors (35). In addition, preliminary data from experiments carried out on the large (90-kDa) catalase in the fungus Aspergillus terreus suggest that apocatalase enzymes can be partially reconstituted with heme under in vitro conditions to regain catalase activity (46). These observations suggest that catalase enzymes are able to fold without the aid of chaperones and that heme may play an important role in the folding process. Given that Cj1386 contains ankyrin repeats, it is tempting to hypothesize that KatA and Cj1386 may interact and that this interaction is required for heme transfer to the KatA protein. However, the precise function that Cj1386 plays in heme trafficking remains to be investigated.
Cj1386 homologs are found within several species of Campylobacter, including C. coli, C. lari, C. fetus, and C. showae. The genomic orientation of these Cj1386 homologs downstream from katA is also conserved. Additionally, homologs of Cj1386 are also present in many pathogenic bacteria, including Streptomyces avermitilis, Pseudomonas putida, Vibrio cholerae, and Helicobacter hepaticus, and are similarly located downstream of their respective catalase. The conservation of Cj1386 and its genetic organization among diverse microorganisms underscores its importance in bacterial physiology. Studies involving genes that affect catalase activity have been carried out in other bacterial species such as Helicobacter pylori (20) and Pseudomonas aeruginosa (25); however, our findings demonstrate a novel role for Cj1386 compared to these previous studies.
Helicobacter pylori possesses a gene named kapA which is located directly downstream from its katA gene. KapA, like Cj1386, is important in defense against hydrogen peroxide, and its gene genomic orientation is similar to what is observed in C. jejuni (with respect to katA and Cj1386) (21). A kapA-deficient mutant had decreased catalase activity in the periplasmic fraction relative to the wild-type H. pylori strain while no differences were observed in the cytosolic fraction (20). Although there is no significant amino acid identity between KapA and Cj1386, mutations in both of these genes disrupt the normal catalase activity levels within the cell. However, KapA is required only for the periplasmic catalase activity (20) and has a distinctly different function from Cj1386. Interestingly, the N-terminal sequence of KapA contains a twin-arginine translocase (TAT)-like signal sequence and was proposed to function as a transport accessory protein involved in the translocation of KatA via the TAT secretion system (20). This TAT-like motif is absent from Cj1386. Moreover, Cj1386 was not identified as a potential substrate of C. jejuni TAT secretion machinery (23, 36). However, the absence of secretion motifs in both Cj1386 and KatA raises the question of how KatA and possibly Cj1386 are translocated into the periplasmic space.
Cj1386 shares 31% identity at the amino acid level with AnkB from Pseudomonas aeruginosa, which is also critical for optimal H2O2 resistance (25). ankB is located directly downstream from the gene for its catalase enzyme, KatB, and codes for an ankyrin-like protein. An ankB-deficient mutant was shown to exhibit an increased sensitivity toward hydrogen peroxide and a decrease in KatB-mediated catalase activity (determined using whole-cell extracts) (25). Interestingly, Howell et al. (25) reported that AnkB is found primarily within the periplasmic space of P. aeruginosa. AnkB contains a single membrane-spanning domain with a small 3-amino-acid C terminus located within the cytoplasmic space, while the remaining N terminus of the protein is hydrophilic and predicted to be located in the periplasmic space (25). The specific catalase activity of the ΔankB mutant in the cytoplasmic and periplasmic spaces was not reported; however, KatB activity was found in both of these two cellular compartments. It is, therefore, not known whether the catalase activity is affected in both the cytoplasmic and periplasmic compartments of P. aeruginosa (as is observed in the ΔCj1386 mutant) or whether the reduction in catalase activity primarily occurs in the periplasmic space as is reported in H. pylori. Howell et al. suggested that AnkB may be interacting with KatB either to localize KatB to the inner membrane surface (to protect H2O2-sensitive biological proteins or molecules) or to stabilize or enhance KatB activity (25). Use of the same transmembrane-spanning domain prediction program (TopPred 2.0 [47]) did not indicate the presence of any putative transmembrane domains in Cj1386. Further experiments will be required to determine the localization of Cj1386 within the cell as well as any interaction of Cj1386 with KatA.
We have previously shown that the ability of C. jejuni to cope with oxidative stress is fundamental for its colonization of the chick gastrointestinal tract. A katA-deficient mutant was significantly altered in its ability to colonize the chick cecum compared to the wild-type strain (32). It is, therefore, not surprising that our ΔCj1386 mutant exhibited a similar colonization defect. We further confirmed the importance of oxidative stress defense in host colonization by testing our ΔkatA mutant in the neonate piglet infectious model, revealing that the ΔkatA mutant was significantly outcompeted by the wild-type C. jejuni strain. Additionally, the ΔCj1386 mutant was outcompeted by the wild-type strain in the neonate piglet infectious model, further highlighting the importance of Cj1386 function for gut colonization. An analogous result was reported in H. pylori, where KapA was found to be essential to sustain a long-term colonization in the murine model of H. pylori infection (22). It is likely that the lower catalase activity in the ΔCj1386 mutant is the cause of the reduced colonization in both animal models, as the mutant is more susceptible to the damaging effects of exogenously produced hydrogen peroxide (either from the host or from the host intestinal flora). In the absence of Cj1386, H2O2 will be detoxified at lower rates, allowing the oxidant to damage DNA, protein, and lipids.
In summary, we demonstrated that Cj1386 is important in defense against hydrogen peroxide within the cell by promoting optimal levels of catalase activity and by contributing to heme trafficking to KatA. Cj1386 also plays an important role in the colonization of the gastrointestinal tract in the chick colonization model and in the piglet pathogenesis model. Future experiments will determine the heme binding ability of Cj1386 and the proteins involved in this process.
ACKNOWLEDGMENTS
This work was supported by the Canadian Institute of Health Research (to A.S.) and the Ontario Graduate Scholarship of Science and Technology (to A.F.).
We also thank J.-F. Couture (Ottawa Institute of Systems Biology, University of Ottawa) for providing the pGST vector and TEV protease.
Footnotes
Published ahead of print 11 November 2011
REFERENCES
- 1. Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5: 28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Annuk H, et al. 2003. Characterization of intestinal lactobacilli as putative probiotic candidates. J. Appl. Microbiol. 94: 403–412 [DOI] [PubMed] [Google Scholar]
- 3. Antrobus R, Borner GH. 2011. Improved elution conditions for native co-immunoprecipitation. PLoS One 6: e18218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ascoli F, Fanelli MR, Antonini E. 1981. Preparation and properties of apohemoglobin and reconstituted hemoglobins. Methods Enzymol. 76:72–87 [DOI] [PubMed] [Google Scholar]
- 5. Atack JM, Harvey P, Jones MA, Kelly DJ. 2008. The Campylobacter jejuni thiol peroxidases Tpx and Bcp both contribute to aerotolerance and peroxide-mediated stress resistance but have distinct substrate specificities. J. Bacteriol. 190: 5279–5290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babakhani FK, Bradley GA, Joens LA. 1993. Newborn piglet model for campylobacteriosis. Infect. Immun. 61: 3466–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Beers RF, Jr, Sizer IW. 1952. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J. Biol. Chem. 195: 133–140 [PubMed] [Google Scholar]
- 8. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340: 783–795 [DOI] [PubMed] [Google Scholar]
- 9. Bennett V. 1992. Ankyrins. Adaptors between diverse plasma membrane proteins and the cytoplasm. J. Biol. Chem. 267: 8703–8706 [PubMed] [Google Scholar]
- 10. Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. 2002. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc. Natl. Acad. Sci. U. S. A. 99: 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bingham-Ramos LK, Hendrixson DR. 2008. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect. Immun. 76: 1105–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. 1988. Experimental Campylobacter jejuni infection in humans. J. Infect. Dis. 157: 472–479 [DOI] [PubMed] [Google Scholar]
- 13. Blaser MJ. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl 2):S103–S105 [DOI] [PubMed] [Google Scholar]
- 14. Brown SM, Howell ML, Vasil ML, Anderson AJ, Hassett DJ. 1995. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J. Bacteriol. 177: 6536–6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chelikani P, Fita I, Loewen PC. 2004. Diversity of structures and properties among catalases. Cell. Mol. Life Sci. 61: 192–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dasti JI, Tareen AM, Lugert R, Zautner AE, Gross U. 2010. Campylobacter jejuni: a brief overview on pathogenicity-associated factors and disease-mediating mechanisms. Int. J. Med. Microbiol. 300: 205–211 [DOI] [PubMed] [Google Scholar]
- 17. Day WA, Jr, Sajecki JL, Pitts TM, Joens LA. 2000. Role of catalase in Campylobacter jejuni intracellular survival. Infect. Immun. 68: 6337–6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Farr SB, Kogoma T. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55: 561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grant KA, Park SF. 1995. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology 141: 1369–1376 [DOI] [PubMed] [Google Scholar]
- 20. Harris AG, Hazell SL. 2003. Localisation of Helicobacter pylori catalase in both the periplasm and cytoplasm, and its dependence on the twin-arginine target protein, KapA, for activity. FEMS Microbiol. Lett. 229: 283–289 [DOI] [PubMed] [Google Scholar]
- 21. Harris AG, Hinds FE, Beckhouse AG, Kolesnikow T, Hazell SL. 2002. Resistance to hydrogen peroxide in Helicobacter pylori: role of catalase (KatA) and Fur, and functional analysis of a novel gene product designated ‘KatA-associated protein’, KapA (HP0874). Microbiology 148: 3813–3825 [DOI] [PubMed] [Google Scholar]
- 22. Harris AG, et al. 2003. Catalase (KatA) and KatA-associated protein (KapA) are essential to persistent colonization in the Helicobacter pylori SS1 mouse model. Microbiology 149: 665–672 [DOI] [PubMed] [Google Scholar]
- 23. Hitchcock A, et al. 2010. Roles of the twin-arginine translocase and associated chaperones in the biogenesis of the electron transport chains of the human pathogen Campylobacter jejuni. Microbiology 156: 2994–3010 [DOI] [PubMed] [Google Scholar]
- 24. Holmes K, et al. 2005. Campylobacter jejuni gene expression in response to iron limitation and the role of Fur. Microbiology 151: 243–257 [DOI] [PubMed] [Google Scholar]
- 25. Howell ML, et al. 2000. AnkB, a periplasmic ankyrin-like protein in Pseudomonas aeruginosa, is required for optimal catalase B (KatB) activity and resistance to hydrogen peroxide. J. Bacteriol. 182: 4545–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Humphrey T, O'Brien S, Madsen M. 2007. Campylobacters as zoonotic pathogens: a food production perspective. Int. J. Food Microbiol. 117: 237–257 [DOI] [PubMed] [Google Scholar]
- 27. Imlay JA. 2008. Cellular defenses against superoxide and hydrogen peroxide. Annu. Rev. Biochem. 77: 755–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Imlay JA. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57: 395–418 [DOI] [PubMed] [Google Scholar]
- 29. Klotz MG, Kim YC, Katsuwon J, Anderson AJ. 1995. Cloning, characterization and phenotypic expression in Escherichia coli of catF, which encodes the catalytic subunit of catalase isozyme CatF of Pseudomonas syringae. Appl. Microbiol. Biotechnol. 43: 656–666 [DOI] [PubMed] [Google Scholar]
- 30. Nachamkin I. 2002. Chronic effects of Campylobacter infection. Microbes Infect. 4: 399–403 [DOI] [PubMed] [Google Scholar]
- 31. Naikare H, Palyada K, Panciera R, Marlow D, Stintzi A. 2006. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infect. Immun. 74: 5433–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Palyada K, et al. 2009. Characterization of the oxidative stress stimulon and PerR regulon of Campylobacter jejuni. BMC Genomics 10: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Palyada K, Threadgill D, Stintzi A. 2004. Iron acquisition and regulation in Campylobacter jejuni. J. Bacteriol. 186: 4714–4729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parkhill J, et al. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403: 665–668 [DOI] [PubMed] [Google Scholar]
- 35. Prakash K, Prajapati S, Ahmad A, Jain SK, Bhakuni V. 2002. Unique oligomeric intermediates of bovine liver catalase. Protein Sci. 11: 46–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajashekara G, et al. 2009. Functional characterization of the twin-arginine translocation system in Campylobacter jejuni. Foodborne Pathog. Dis. 6: 935–945 [DOI] [PubMed] [Google Scholar]
- 37. Reid AN, Pandey R, Palyada K, Naikare H, Stintzi A. 2008. Identification of Campylobacter jejuni genes involved in the response to acidic pH and stomach transit. Appl. Environ. Microbiol. 74: 1583–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schnell S, Steinman HM. 1995. Function and stationary-phase induction of periplasmic copper-zinc superoxide dismutase and catalase/peroxidase in Caulobacter crescentus. J. Bacteriol. 177: 5924–5929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seaver LC, Imlay JA. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183: 7173–7181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Segal AW. 2005. How neutrophils kill microbes. Annu. Rev. Immunol. 23: 197–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sellars MJ, Hall SJ, Kelly DJ. 2002. Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184: 4187–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheffield P, Garrard S, Derewenda Z. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15: 34–39 [DOI] [PubMed] [Google Scholar]
- 43. Smith MA, Finel M, Korolik V, Mendz GL. 2000. Characteristics of the aerobic respiratory chains of the microaerophiles Campylobacter jejuni and Helicobacter pylori. Arch. Microbiol. 174: 1–10 [DOI] [PubMed] [Google Scholar]
- 44. Tao H, Bausch C, Richmond C, Blattner FR, Conway T. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181: 6425–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Vliet AH, Baillon ML, Penn CW, Ketley JM. 1999. Campylobacter jejuni contains two fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181: 6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vatsyayan P, Goswami P. 2011. Acidic pH conditions induce dissociation of the haem from the protein and destabilise the catalase isolated from Aspergillus terreus. Biotechnol. Lett. 33: 347–351 [DOI] [PubMed] [Google Scholar]
- 47. von Heijne G. 1992. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225: 487–494 [DOI] [PubMed] [Google Scholar]
- 48. Wang Y, Taylor DE. 1990. Natural transformation in Campylobacter species. J. Bacteriol. 172: 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Woodbury W, Spencer AK, Stahman MA. 1971. An improved procedure using ferricyanide for detecting catalase isozymes. Anal. Biochem. 44: 301–305 [DOI] [PubMed] [Google Scholar]
- 50. Yao R, Alm RA, Trust TJ, Guerry P. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130: 127–130 [DOI] [PubMed] [Google Scholar]
- 51. Yumoto I, et al. 2000. Purification and characterization of a catalase from the facultatively psychrophilic bacterium Vibrio rumoiensis S-1(T) exhibiting high catalase activity. J. Bacteriol. 182: 1903–1909 [DOI] [PMC free article] [PubMed] [Google Scholar]