Abstract
Diauxic growth was observed in anaerobic C4-dicarboxylate-adapted cells of “Aromatoleum aromaticum” EbN1 due to preferred benzoate utilization from a substrate mixture of a C4-dicarboxylate (succinate, fumarate, or malate) and benzoate. Differential protein profiles (two-dimensional difference gel electrophoresis [2D DIGE]) revealed dynamic changes in abundance for proteins involved in anaerobic benzoate catabolism and C4-dicarboxylate uptake. In the first active growth phase, benzoate utilization was paralleled by maximal abundance of proteins involved in anaerobic benzoate degradation (e.g., benzoyl-coenzyme A [CoA] reductase) and minimal abundance of DctP (EbA4158), the periplasmic binding protein of a predicted C4-dicarboxylate tripartite ATP-independent periplasmic (TRAP) transporter (DctPQM). The opposite was observed during subsequent succinate utilization in the second active growth phase. The increased dctP (respectively, dctPQM) transcript and DctP protein abundance following benzoate depletion suggests that repression of C4-dicarboxylate uptake seems to be a main determinant for the observed diauxie.
INTRODUCTION
Next to the glycosyl moiety, the benzene ring is the second most abundant terrestrial organic structure, present in lignin (30% of the organic carbon in the biosphere) (4), proteins, plant metabolites, and fossil carbon (e.g., up to 30% of crude oil) (46). Wide usage in industrial chemical synthesis further contributes to the distribution of aromatic compounds in the environment. Anoxic conditions prevail in many habitats (e.g., freshwater and marine sediments or deep soil layers), emphasizing the significance of anaerobic biodegradation. Most monoaromatic compounds are degraded via the central benzoyl-coenzyme A (benzoyl-CoA) pathway, with the CoA-activated form of benzoate representing the central intermediate in the anaerobic degradation of aromatic compounds (5, 19). Mixed substrate conditions are typical for natural habitats. The rhizosphere, for example, contains an ample mixture of small molecules arising from plant roots (e.g., amino acids and dicarboxylates) (12), lignin degradation (3-phenylpropanoids), or fermentation (e.g., acetate). When mixtures of utilizable carbon sources are present, microorganisms display cometabolic or sequential substrate utilization, often depending on the concentrations of individual compounds (24). Sequential substrate utilization in batch cultures often results in diauxic growth comprising two active growth phases separated by an intermediary (diauxic) lag phase (32). Generally, the preferred substrate permits a higher specific growth rate and prevents utilization of the less favored substrate(s) (24).
The selective utilization of carbohydrates mediated by carbon catabolite repression (CCR) is best understood in standard microorganisms such as Escherichia coli and Bacillus subtilis (22). However, little is known about the mechanisms of CCR in aromatic compound degradation by nonstandard microorganisms. In Pseudomonas putida and Acinetobacter baylyi, succinate strongly represses catabolic operons for aerobic aromatic compound degradation by an alternative mechanism (22). Conversely, catechol (central intermediate of aerobic benzoate degradation) represses acoE expression required for acetate utilization in Ralstonia eutropha (1), and naphthalene inhibits glucose uptake in P. putida CSV86 (3). In the denitrifying Azoarcus sp. strain CIB, low-molecular-weight organic acids (e.g., succinate and acetate) repressed anaerobic catabolism of benzoate (30).
The denitrifying betaproteobacterium “Aromatoleum aromaticum” EbN1 degrades 28 different aromatic compounds under anaerobic conditions, including petroleum hydrocarbons (e.g., ethylbenzene) (39), phenolic solvents (e.g., p-cresol) (49), and plant-derived 3-phenylpropanoids (e.g., p-coumarate; K. Trautwein, H. Wilkes, and R. Rabus, unpublished data). Despite the good understanding of the architecture and substrate-specific regulation of the catabolic network (29, 37, 49, 50), the effect of low-molecular-weight organic acids on the anaerobic degradation of aromatic compounds in strain EbN1 is unknown.
The aim of this study was to investigate and compare the effects of different aliphatic organic acids (C4-dicarboxylates and acetate) on the anaerobic degradation of benzoate, phenylacetate, and p-hydroxybenzoate in strain EbN1 and in three close relatives of strain EbN1 (“Aromatoleum” strains HxN1 and ToN1, as well as Thauera aromatica K172).
MATERIALS AND METHODS
Media and cultivation.
The denitrifying bacterium “A. aromaticum” EbN1 was cultivated in a phosphate-buffered mineral medium (modified from reference 47) under aerobic or nitrate-reducing conditions at 28°C. The basal medium had the following composition: KH2PO4, 1.1 g liter−1; K2HPO4, 5.6 g liter−1; NH4Cl, 0.8 g liter−1; Na2SO4, 0.23 g liter−1; and NaCl, 1.0 g liter−1. The medium was composed as described previously (39); additionally, 1 ml of a solution containing MgCl2 (324 g liter−1) and CaCl2 (29.4 g liter−1) was added per liter of medium. Oxic medium was prepared in 1-liter Duran bottles (Schott AG, Mainz, Germany) containing 250 ml medium, sealed with butyl rubber stoppers and screw caps, with the headspace containing enough oxygen for complete substrate oxidation. Aerobic cultivation was performed on magnetic stirrers at 250 rpm. Anoxic media with 10 mM nitrate as the electron acceptor and 10 mM NaHCO3 were prepared as described previously in butyl rubber-sealed 500-ml (or 250-ml) flat glass bottles, containing 400 ml (or 200 ml) of medium under an N2-CO2 (9:1 [vol/vol]) atmosphere (39). Anaerobic cultivation of the related denitrifying “Aromatoleum” strains HxN1 and ToN1, as well as Thauera aromatica K172, was carried out under the same conditions. Adaptation substrates and cosubstrates were added from sterile, aqueous stock solutions. (The respective concentrations in the medium are given in Table 1.) The chemicals used were of analytical grade. Strains EbN1, HxN1, and ToN1 have been maintained in our laboratory since their isolation, and strain K172 was obtained from DSMZ (Braunschweig, Germany).
Table 1.
Growth of “A. aromaticum” EbN1 and related strains with aliphatic and aromatic substrates
| Strain(s) and condition | Substratea |
Result forb: |
Further detailsd | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Single substrate |
Mixture of SAd and SCoc |
|||||||||||
| SAd |
SCo |
Phase 1 |
Phase 2 |
Phase 3 |
||||||||
| SAd | SCo | ODmax | μmax (h−1) | ODmax | μmax (h−1) | ODmax | μmax (h−1) | Duration of diauxic lag phase (h) | ODmax | μmax (h−1) | ||
| “A. aromaticum” EbN1 | ||||||||||||
| Benzoate as cosubstrate | ||||||||||||
| Anaerobic | Succinate (5 mM) | Benzoate (1 mM) | 0.48 | 0.25 | ND | ND | 0.30 | 0.21 | 3.3 ± 0.6 | 0.69 | 0.33 | Fig. 1 |
| Fumarate (5 mM) | Benzoate (1 mM) | 0.57 | 0.20 | 0.27 | 0.18 | 0.30 | 0.19 | 6.4 ± 0.8 | 0.70 | 0.25 | Fig. S1 | |
| d,l-Malate (5 mM) | Benzoate (1 mM) | 0.53 | 0.12 | 0.27 | 0.19 | 0.28 | 0.15 | 2.9 ± 0.0 | 0.69 | 0.12 | Fig. S1 | |
| Oxaloacetate (6 mM) | Benzoate (1 mM) | 0.43 | 0.22 | 0.27 | 0.19 | 0.60 | 0.25 | None | None | None | Fig. S1 | |
| Acetate (8 mM) | Benzoate (1 mM) | 0.41 | 0.24 | ND | ND | 0.62 | 0.24 | None | None | None | Fig. 1 | |
| Aerobic | Succinate (5 mM) | Benzoate (1 mM) | 1.14 | 0.19 | ND | ND | 0.40 | 0.20 | 1.0 ± 0.0 | 1.16 | 0.24 | Fig. S1 |
| Other cosubstrates | Succinate (5 mM) | Acetate (4 mM) | 0.53 | 0.23 | 0.21 | 0.21 | 0.27 | 0.23 | 3.0 ± 0.0 | 0.55 | 0.27 | Fig. S2 |
| Succinate (5 mM) | Phenylacetate (1 mM) | 0.53 | 0.23 | 0.29 | 0.19 | 0.58 | 0.19 | None | None | None | Fig. S2 | |
| Succinate (5 mM) | p-Hydroxybenzoate (1 mM) | 0.53 | 0.23 | 0.19 | 0.19 | 0.66 | 0.20 | None | None | None | Fig. S2 | |
| Related strains (anaerobic) | ||||||||||||
| Benzoate as cosubstrate | ||||||||||||
| “Aromatoleum” strain HxN1 | Succinate (5 mM) | Benzoate (1 mM) | 0.44 | 0.25 | 0.26 | 0.15 | 0.65 | 0.24 | None | None | None | Fig. S3 |
| “Aromatoleum” strain ToN1 | Succinate (5 mM) | Benzoate (1 mM) | 0.42 | 0.25 | 0.23 | 0.21 | 0.52 | 0.15 | None | None | None | Fig. S3 |
| T. aromatica K172 | Succinate (5 mM) | Benzoate (1 mM) | 0.42 | 0.24 | 0.20 | 0.20 | 0.61 | 0.22 | None | None | None | Fig. S3 |
Preferentially utilized substrates are highlighted in boldface. Applied substrate concentrations are indicated in parentheses. SAd, adaptation substrate; SCo, cosubstrate.
Values for maximal optical densities (ODmax) and maximum specific growth rates (μmax) are based on at least two replicate cultures (three for each substrate mixture) yielding standard deviations of <5%. μmax was calculated from the slope of the active growth phase (m): μmax = m × 1/ΔOD. If present, the duration of the diauxic lag phase (phase 2) is indicated with standard deviation. ND, not determined.
In the case of diauxic growth, phase 1 corresponds to the first active growth phase. In the case of monophasic growth, phase 1 corresponds to the only observed active growth phase. If present, phase 3 corresponds to a second active growth phase, with phase 2 representing the diauxic lag between phases 1 and 3 (see Fig. 2).
Fig. S1, S2, and S3 are in the supplemental material.
Growth experiments.
The growth behavior of strain EbN1 and related strains was analyzed in the presence of different substrate mixtures as listed in Table 1. The substrate mixtures were composed such that complete substrate oxidation would theoretically yield about twice as many electrons from the adaptation substrate than from the cosubstrate. Cells were cultivated with the respective adaptation substrate for at least 5 passages before a freshly grown culture was used to inoculate cultures for growth experiments. The inoculum sizes were 2% (vol/vol) in the case of anaerobic cultures and 4% (vol/vol) in the case of aerobic cultures. Three replicate cultures were performed for mixed substrate and two replicate cultures for single-substrate conditions. Growth was monitored by measuring the optical density at 660 nm (OD660) (UV-mini 1202; Shimadzu, Duisburg, Germany) and by high-performance liquid chromatography (HPLC) measurement of substrate concentrations. For sampling, 3 ml was removed from each culture with sterile syringes (N2 flushed in the case of anaerobic cultures): 1 ml was used for monitoring the OD, and 2 ml was immediately filtered (Spartan 13/0.2 RC; Schleicher & Schuell, Dassel, Germany) and stored at −20°C. Negative controls lacking the inoculum were treated in the same way.
Mass cultivation.
To obtain sufficient cell material for proteomic, enzymatic, and RNA analyses, strain EbN1 was grown with a mixture of succinate (5 mM) and benzoate (1 mM) in a 5-liter Duran bottle (Ochs, Bovenden, Germany) filled with 4 liters anoxic medium under an N2-CO2 (9:1 [vol/vol]) atmosphere. The bottle contained three openings for gassing, inoculation or sampling, and harvesting. Samples were treated as described above. Cells were harvested during diauxic growth in phase 1 (benzoate utilization; OD, ∼0.17), phase 2 (diauxic lag phase; OD, ∼0.28), and phase 3 (succinate utilization; OD, ∼0.43). Cells grown anaerobically with 5 mM succinate (400-ml culture volume) were harvested at an OD of 0.20 (corresponding approximately to the half-maximal optical density [½ ODmax]) or shortly after transition into the stationary growth phase due to succinate depletion (OD, ∼0.50; ODmax). Cells grown anaerobically with 4 mM benzoate were harvested at an OD of 0.20. Harvesting of cells was performed as described previously (9). Cell pellets for proteomic analyses were immediately frozen in liquid nitrogen and stored at −80°C until further analysis. For enzyme activity measurements, culture volumes of 4 to 8 liters per growth condition were anoxically harvested, and cell pellets were stored in butyl rubber-sealed anoxic glass vials at −80°C until further analysis. Cell pellets for RNA extraction were immediately processed.
Chemical analysis.
Concentrations of organic acids (excluding benzoate) were determined with an HPLC system (Sykam GmbH, Fürstenfeldbruck, Germany) equipped with an anion-neutral precolumn (4 by 20 mm; Sykam) and an Aminex HPX-87H separation column (300 by 7.8 mm; Bio-Rad, Munich, Germany) that was temperature controlled to 60°C. With 5 mM H2SO4 in HPLC-grade water as the eluent and a flow rate of 0.6 ml min−1, organic acids were detected using a refractive index (RI) or UV detector (210 nm). The retention times (with detection limits in parentheses) of RI-detected compounds were as follows: succinate, 11.2 min (0.2 mM); malate, 9.0 min (0.2 mM); oxaloacetate, 9.4 min (0.5 mM); and acetate, 14.3 min (0.2 mM). The retention times (with detection limits in parentheses) of UV-detected compounds were as follows: fumarate, 12.2 min (0.25 μM); phenylacetate, 37.9 min (50 μM); and p-hydroxybenzoate, 48.6 min (50 μM).
Benzoate was determined with an HPLC system (Sykam) previously described for the determination of nitrate and nitrite (39) at a retention time of 3.2 min and with a detection limit of 20 μM benzoate. The eluent was composed of 20 mM NaCl in 45% (vol/vol) ethanol.
Soluble protein fraction: 2D DIGE and MALDI-TOF-MS/MS.
Preparation of cell-free protein extracts and two-dimensional difference gel electrophoresis (2D DIGE) were carried out as previously reported (20, 38). Cells were disrupted with the PlusOne sample grinding kit (GE Healthcare, Munich, Germany), and the protein concentration was determined as described by Bradford (7). Isoelectric focusing (IEF) was performed using the IPGphor system (GE Healthcare) and commercial 24-cm immobilized pH gradient (IPG) strips with a nonlinear pH gradient of 3 to 11 (GE Healthcare). The EttanDalt II system (GE Healthcare) was used for separation according to the molecular mass in 12.5% acrylamide gels. Pre-electrophoretic labeling with different fluorescent dyes allows coseparation of three samples in a single gel, representing the reference state (Cy5 labeled), the test state (Cy3 labeled), and the (pooled) internal standard (Cy2 labeled). Protein extracts from cells adapted to anaerobic growth with succinate served as the reference state. Individual test states were prepared from protein extracts of cells during diauxic growth: phase 1, benzoate utilization; phase 2, diauxic lag phase; and phase 3, succinate utilization. The internal standard was composed of equal amounts of protein extracts of the reference and all test states.
2D DIGE gels were scanned immediately after electrophoresis with a Typhoon 9400 scanner (GE Healthcare). Cropped images were analyzed with the DeCyder software (version 5.0, GE Healthcare). The parameters for codetection of spots were described previously (49). Spot matching was manually controlled for differentially regulated spots, which had to fulfill the following criteria: average ratio (n-fold change in protein abundance) of ≤−2.5 or ≥2.5; analysis of variance (ANOVA) P value of <0.05, and t test value of <10−4, and matching in at least 75% of the analyzed gels. To achieve statistical confidence, at least three parallel gels were included in the analysis for each tested state. Overall, the DeCyder analysis included 11 gels (representing 33 gel images). The master gel contained 1,050 detected spots. Differentially regulated protein spots were manually excised from preparative two-dimensional gel electrophoresis (2DE) gels (300-μg protein load) stained with colloidal Coomassie brilliant blue (cCBB) (13). The high reproducibility of spot patterns allowed mass spectrometry (MS)-based identification of differentially regulated proteins as determined by 2D DIGE/DeCyder.
Tryptic digest of excised 2DE-separated proteins was performed as previously described (28). Peptide masses were determined by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF-MS) (TopLab GmbH, Martinsried, Germany). Protein identification and genome analysis were based on the published list of annotated genes from the genome sequence of strain EbN1 (37). Peptide mass fingerprints were mapped to the in silico digests of the predicted proteins by using the MS-Digest program (10).
Membrane protein fraction: SDS-PAGE and LC-ESI-MS/MS.
Following mechanic breakage of cells (∼120 mg wet weight), membrane preparations were obtained by ultracentrifugation and treatment with ice-cold carbonate, essentially as described elsewhere (35). Proteins were solubilized from the prepared membranes by treatment with hot (95°C) SDS (1% [wt/vol]). Protein concentrations were determined with the detergent-compatible RC DC protein assay (Bio-Rad). Separation of proteins (30 μg per lane) was achieved by conventional SDS-PAGE (12.5% acrylamide) and gel staining with cCBB.
Gel bands were excised, cut into smaller pieces, and washed twice with 50 mM NH4HCO3 and 50 mM NH4HCO3–acetonitrile (ACN) (1:1 [vol/vol]) prior to rehydration and tryptic digest with 5 ng μl−1 trypsin solution (Promega, Mannheim, Germany) in 25 mM NH4HCO3. Following incubation overnight at 37°C, peptides were extracted once with 50% ACN and 1% trifluoroacetic acid (TFA) and once with 90% ACN and 1% TFA. The supernatants were pooled prior to analysis.
Separation of peptides was performed with an UltiMate3000 nano-liquid chromatography (nano-LC) system (Dionex GmbH, Idstein, Germany). Peptides were loaded on a trap column at a flow rate of 6 μl min−1 (PepMap nanoTrap: C18, 100 Å, 3-μm bead size, 75-μm internal diameter, 2-cm length; Dionex [solvent, 0.05% TFA]) prior to separation with an analytical column (PepMapRSLC: C18, 100 Å, 2-μm bead size, 75-μm internal diameter, 15-cm length; Dionex) and a 180-min gradient at a flow rate of 0.3 μl min−1 (solvents with 0.05% TFA): 0 to 5% ACN for 5 min, 5 to 22% ACN for 107 min, 22 to 26% ACN for 24 min, 26 to 32% ACN for 20 min, 32 to 55% ACN for 24 min, and 95% ACN for 15 min.
Mass spectrometric analysis of the nano-LC effluent was performed with an online-coupled ion trap mass spectrometer (amaZon ETD; Bruker Daltonik GmbH, Bremen, Germany). For electrospray ionization, a distal coated fused SilicaTip (NewObjective, Woburn, MA) with an inner diameter of 10 μm was used at a voltage of 1.6 kV. The MS method consisted of a full MS scan (mass range, 300 to 1,500 m/z) with subsequent MS/MS of the three most intense MS peaks (successive exclusion after 2 spectra for 0.3 min).
Protein identification was performed with the software ProteinScape (Bruker Daltonik) on a Mascot server searching against the genomic database of strain EbN1, translated into amino acid sequences. The following settings were applied: taxonomy, all; enzyme, trypsin; may missed cleavage, 1; fixed modification, carbamidomethyl; variable modification, oxidation; peptide mass tolerance, 0.4 Da; fragment mass tolerance, 0.4 Da; mass values, monoisotopic; instrument type, ESI-trap; and inclusion of peptide decoy.
Shotgun proteomics: LC-ESI-MS/MS.
For shotgun proteomics (TopLab), the cell pellet (110 to 160 mg wet weight) was resuspended in 200 μl lysis buffer (30 mM Tris-HCl [pH 8.5], 2 M thiourea, 7 M urea) containing a protease inhibitor mix (Complete protease inhibitor cocktail tablets; Roche Diagnostics, Basel, Switzerland). Cells were lysed by ultrasonification (6 times 10 s) on ice and subsequently shaken (1,000 rpm) for 20 min at 20°C. Then, cell debris was removed by centrifugation (30 min, 20,000 × g, 20°C). The protein concentration in the supernatant was determined as described by Bradford (7). Fifty micrograms of protein was diluted in urea-containing buffer (8 M urea, 0.4 M NH4HCO3) to obtain a total volume of 50 μl. Disulfide bonds were reduced at 55°C for 30 min using 45 mM dithiothreitol. After cooling, free cysteines were alkylated for 15 min with 100 mM iodoacetamide. For digestion, the protein solution was diluted with water (HPLC grade) in order to obtain a concentration of 2 M urea prior to addition of 1 μg modified porcine trypsin (Serva Electrophoresis GmbH, Heidelberg, Germany) at a ratio of 50:1. The digestion was performed overnight at 37°C.
Separation of tryptic peptides (1 to 2 μg total protein) was performed on a NanoLC-Ultra 2D chromatography system (Eksigent Technologies, Dublin, CA). Peptides were loaded on a reversed-phase chromatography (RPC) trap column (ReproSil C18, 0.3 by 10 mm) (Dr. Maisch HPLC GmbH, Ammerbuch-Entringen, Germany) and subsequently separated with an analytical column (PepMap100: C18, 100 Å, 3-μm bead size, 75-μm internal diameter, 15-cm length; Dionex), using a 120-min linear gradient (A, 0.1% formic acid in water; B, 0.1% formic acid in 84% ACN) at a flow rate of 280 nl min−1. The gradient used was 1 to 30% B in 80 min, 30 to 60% B in 20 min, and 100% B for 10 min.
Mass spectrometry was performed on a linear ion trap mass spectrometer (Thermo LTQ Orbitrap, Thermo Fisher Scientific, Dreieich, Germany) online coupled to the nano-LC system. For electrospray ionization, a distal-coated SilicaTip (FS-360-50-15-D-20) and a needle voltage of 1.4 kV were used. The LTQ-Orbitrap was operated in parallel mode, performing precursor mass scanning in the Orbitrap (60,000 full-width half-maximum [FWHM] resolution at m/z 400) and isochronous acquisition of three data-dependent collision-induced dissociation (CID) MS/MS scans in the LTQ ion trap (35-eV collision energy).
The database queries were performed using the Mascot program searching against the genomic database of strain EbN1. The following parameters were used: taxonomy, all; enzyme, trypsin; maximum missed cleavage, 1; fixed modification, carbamidomethyl; variable modification, oxidation; peptide mass tolerance, 2 Da; fragment mass tolerance, 0.8 Da; mass values, monoisotopic; instrument type, ESI-trap.
In vitro benzoyl-CoA reductase assay.
For the preparation of crude extracts, 1 g of frozen cells was suspended in 2 ml of 20 mM triethanolamine hydrochloride (TEA)–KOH buffer (pH 7.8) containing 4 mM MgCl2, 0.5 mM sodium dithionite, and 0.1 mg of DNase I. Cell lysates were acquired by French press treatment. For preparation of soluble protein fractions, extracts were centrifuged at 100,000 × g (1 h, 4°C). All enzyme assays were performed at 30°C under an N2-H2 (95:5 [vol/vol]) atmosphere (strictly anoxic conditions). The discontinuous assay monitored benzoyl-CoA consumption and product formation by HPLC analysis, as described previously (45). The assay mixture contained 190 μl buffer (100 mM MOPS [morpholinepropanesulfonic acid]-KOH, 15 mM MgCl2 [pH 7.3]), 5 mM ATP, 5 mM Ti(III) citrate as an electron donor, 0.2 mM benzoyl-CoA, and 0.2 to 0.4 mg of cell extract. Benzoyl-CoA was synthesized as described previously (42).
Preparation of RNA.
RNA was isolated immediately from harvested cell pellets (see mass cultivation) using the RNeasy minikit and Midi kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Aliquots were run on agarose gels and stained to confirm RNA integrity.
Northern hybridization analysis.
Isolated total RNA was separated on 1.4% agarose gels containing 3% formaldehyde and transferred onto nylon membranes by capillary blotting (41). Specific probes derived from the dctP (ebA4158) and dctM (ebA4162) genes via PCR were 33P radiolabeled using the DecaLable DNA labeling kit (Fermentas GmbH, St. Leon-Rot, Germany). Hybridization signals were recorded with a Typhoon 9400 scanner with preinstalled settings (GE Healthcare); their relative quantification was performed with the software ImageQuant (GE Healthcare).
RESULTS
Anaerobic growth with mixtures of aliphatic and aromatic substrates.
The growth behavior and substrate utilization preference of “Aromatoleum” strains EbN1, HxN1, and ToN1, as well as T. aromatica K172, were investigated in the presence of binary substrate mixtures containing an aliphatic acid (C4-dicarboxylate or acetate) and an aromatic acid (benzoate, phenylacetate, or p-hydroxybenzoate).
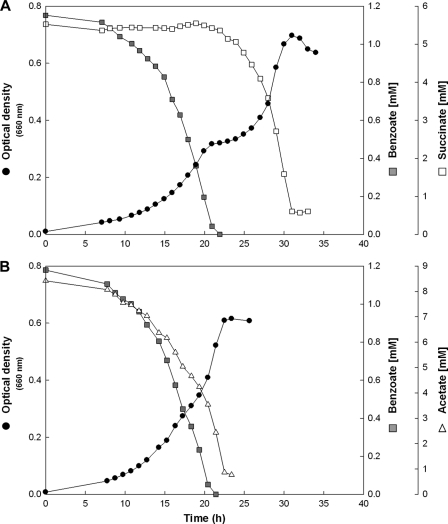
Anaerobic succinate-adapted cells of strain EbN1 growing with a mixture of succinate and benzoate displayed diauxic growth due to sequential substrate utilization (Fig. 1A; Table 1). During the first active growth phase (phase 1), the cells exclusively and completely consumed benzoate (1 mM), yielding an optical density of 0.30. Following a 3.3-h-long diauxic lag phase (phase 2), utilization of succinate (5 mM) coincided with the second active growth phase (phase 3). Preference of benzoate and diauxic growth were also observed in aerobic succinate-adapted cells; however, the diauxic lag phase was considerably shorter under oxic than under anoxic conditions (Table 1; see Fig. S1D in the supplemental material). In contrast, anaerobic acetate-adapted cells simultaneously utilized acetate and benzoate, resulting in monophasic growth (Fig. 1B; Table 1).
Fig 1.
Anaerobic growth of “A. aromaticum” EbN1 with mixtures of succinate or acetate plus benzoate. (A) Succinate-adapted cells displayed diauxic growth with preferred utilization of benzoate. (B) Acetate-adapted cells immediately coutilized acetate and benzoate.
In addition to succinate, anaerobic fumarate-, malate-, or oxaloacetate-adapted cells of strain EbN1 were shifted to a mixture of the respective C4-dicarboxylate and benzoate (Table 1; see Fig. S1A to C in the supplemental material). Diauxic growth with preferential utilization of benzoate was observed in fumarate- and malate-adapted cells. In contrast, oxaloacetate and benzoate were concomitantly utilized, even though the former was more rapidly consumed than the latter, suggesting a certain preference for oxaloacetate.
In addition to benzoate, further cosubstrates were tested for possible repressive effects on anaerobic succinate utilization in strain EbN1 (Table 1; see Fig. S2 in the supplemental material). Among the tested cosubstrates, only acetate fully repressed succinate utilization, resulting in diauxic growth. The aromatic cosubstrate phenylacetate inhibited succinate utilization initially; however, below a concentration of 0.5 mM phenylacetate, both substrates were coutilized. In contrast, succinate and p-hydroxybenzoate were immediately coutilized.
To test whether the inhibition of succinate utilization by benzoate is restricted to strain EbN1, succinate-adapted cells of “Aromatoleum” strains HxN1 and ToN1, as well as those of T. aromatica K172, were shifted to a mixture of succinate and benzoate. None of the tested strains displayed true diauxic growth; nevertheless, distinct substrate utilization patterns were observed for each strain (Table 1; see Fig. S3 in the supplemental material). Strain HxN1 initially preferred benzoate, while coutilization of succinate was delayed until the concentration of benzoate decreased below 0.7 mM. The opposite was observed in strain ToN1, which did not utilize benzoate until the concentration of succinate decreased below 2 mM. In the more distantly related strain K172, immediate coutilization of succinate and benzoate was observed.
Proteome dynamics in the course of diauxic growth of strain EbN1 with a mixture of succinate and benzoate.
The differential protein signatures of cells from three different phases of the diauxic growth curve (phases 1 to 3) (Fig. 2) were determined by comparison to succinate-adapted cells (reference state for 2D DIGE analysis). Across phases 1 to 3, a total of 62 proteins displayed significantly changed abundances, 52 of which were also identified in a complementary whole-cell shotgun proteomic analysis (see Table S1 in the supplemental material).
Fig 2.
Proteomic response of “A. aromaticum” EbN1 during anaerobic diauxic growth with succinate plus benzoate. (A) Samples (green circles; test states) for differential proteomic analysis (2D DIGE) were retrieved from a culture growing diauxically during phase 1 (benzoate utilization), phase 2 (diauxic lag phase), and phase 3 (succinate utilization) and from cultures growing anaerobically with succinate as a single substrate (red circle; reference state). (B) 2D DIGE gel of benzoate-utilizing cells (phase 1) in comparison to the reference state. Green-colored spots are more abundant during phase 1, whereas red-colored spots are more abundant in the reference state. Selected differentially regulated proteins (or their subunits) are annotated. (C) Fold changes in protein abundance during phases 1 to 3 in comparison to the succinate-grown reference state. Protein names are as follows: BclA, benzoate-CoA ligase; BcrCBDA, benzoyl-CoA reductase; Oah, 6-oxo-cyclohex-1-ene-carbonyl-CoA hydrolase; Dch, dienoyl-CoA hydratase; Had, 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase; DctP, periplasmic solute-binding protein of C4-dicarboxylate TRAP transporter.
(i) Proteins of benzoate catabolism.
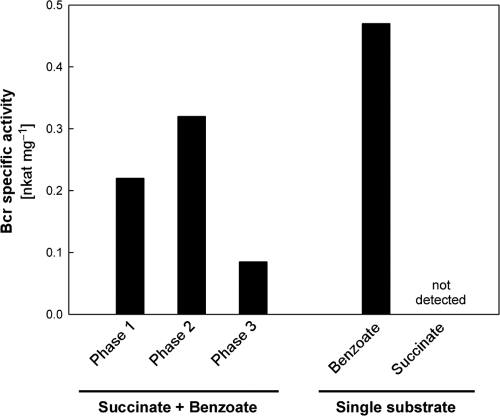
The proteomic analysis revealed dynamic regulation of all enzymes required for conversion of benzoate to 3-hydroxypimelyl-CoA via the anaerobic benzoyl-CoA pathway (BclA, BcrCBDA, Dch, Had, and Oah). The largest increases in abundance (up to 10.9-fold) were observed during benzoate utilization (phase 1), declining markedly during succinate utilization in phase 3 (Fig. 2 and Table 2). Likewise, the key enzyme benzoyl-CoA reductase (BcrCBDA) displayed much higher specific activities in phases 1 and 2 than in phase 3 (Fig. 3). Similar abundance variation was detected for further proteins related to anaerobic benzoate degradation (Orf3, BzdV, BzdZ, and GcdH) as well as for proteins related to other degradation pathways (benzoate, aerobic; m-hydroxybenzoate, aerobic and anaerobic; see Table S1 in the supplemental material). Notably, these proteins (except for BzdV and GcdH) were also detected by whole-cell shotgun proteomic analysis in succinate-adapted cells (see Table S1). Furthermore, the predicted benzoate permease BenK (EbA5311) was identified from the membrane fraction of anaerobic succinate-grown cells (see Fig. S4 in the supplemental material).
Table 2.
Changes in abundance of selected proteins during diauxic growth of anaerobic succinate-adapted “A. aromaticum” EbN1 cells with succinate plus benzoatea
| Proteinb | Predicted pathway and functionb | Fold change in phasec: |
||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| Anaerobic benzoate degradation | ||||
| BclA (EbA5301) | Benzoate-CoA ligase | 4.9 | 6.2 | 3.1 |
| BcrC (EbA5282) | Benzoyl-CoA reductase, C subunit | 8.3 | 8.0 | 5.0 |
| BcrB (EbA5284) | Benzoyl-CoA reductase, B subunit | 10.9 | 9.1 | 4.5 |
| BcrD (EbA5286) | Benzoyl-CoA reductase, D subunit | 5.2 | 5.0 | 3.2 |
| BcrA (EbA5287) | Benzoyl-CoA reductase, A subunit | 9.0 | 8.7 | 5.3 |
| Dch (EbA5296) | Dienoyl-CoA hydratase | 5.1 | 4.9 | 3.3 |
| Had (EbA5297) | 6-Hydroxy-cyclohex-1-ene-1-carboxyl-CoA dehydrogenase | 5.1 | 5.1 | 3.8 |
| Oah (EbA5298) | 6-Oxo-cyclohex-1-ene-carbonyl-CoA hydrolase | 10.8 | 10.0 | 6.4 |
| Other proteins related to anaerobic benzoate degradation | ||||
| Orf3 (EbA5292) | Putative regulatory protein (conserved in anaerobic benzoate degradation gene clusters) | 17.5 | 15.6 | 10.0 |
| BzdV (EbA5294) | Subunit of oxidoreductase (conserved in Azoarcus-type benzoate degradation gene clusters) | 2.7 | 1.8 | −1.6 |
| BzdZ (EbA5300) | Putative dehydrogenase | 6.9 | 6.3 | 4.3 |
| GcdH (EbA2993) | Glutaryl-CoA dehydrogenase | 6.8 | 8.0 | 4.5 |
| Transport-related proteins | ||||
| EbA1033 | DctP-type periplasmic solute-binding protein | −3.7 | 1.9 | 2.0 |
| DctP (EbA4158) | Periplasmic binding protein of C4-dicarboxylate TRAP transporter | −5.7 | −2.3 | 1.1 |
| EbA4994 | DctP-type periplasmic solute-binding protein | 2.2 | 2.2 | 3.0 |
| EbA1861 | Periplasmic protein similar to uncharacterized protein probably involved in high-affinity Fe2+ transport | 42.8 | 34.6 | 23.6 |
| EbA3939 | Putative TonB-dependent siderophore receptor | 15.9 | 9.6 | 3.5 |
| EbA4918 | Putative periplasmic binding protein of ferric iron uptake ABC transporter | 3.9 | 3.4 | 3.1 |
| EbA5333 | Putative TonB-dependent siderophore receptor | 10.6 | 6.5 | 2.7 |
| EbA2115 | Putative periplasmic binding protein of taurine uptake ABC transporter | 7.1 | 6.4 | 5.0 |
For more detail, see Fig. 2. For changes in abundance of further proteins, see Table S1 in the supplemental material.
Protein designations and predicted functions are as described previously (37).
If proteins were identified from more than one protein spot, the fold change of the major protein spot (i.e., largest spot volume) is displayed. Changes marked in boldface are above the set threshold of significant protein regulation (>|2.5|), as determined by 2D DIGE.
Fig 3.
Benzoyl-CoA reductase (Bcr) activity in anaerobically grown cells of “A. aromaticum” EbN1 with a mixture of succinate and benzoate (phases 1 to 3 of diauxic growth; Fig. 2A), or with benzoate and succinate as single substrates.
Several iron uptake-related proteins displayed abundance profiles similar to those of the detected enzymes of benzoate catabolism: i.e., their largest abundance increases (up to 42.8-fold) occurred during phase 1 (Table 2; see Table S1 in the supplemental material). Among them are a predicted periplasmic high-affinity ferrous iron-binding protein (EbA1861) and two putative TonB-dependent siderophore receptors (EbA3939 and EbA5333). This might indicate an increased demand for iron during benzoate catabolism, which may be due to the synthesis of iron-sulfur cluster-containing benzoyl-CoA reductase (6) or its electron donor, ferredoxin.
During benzoate utilization (phase 1), enzymes of the tricarboxylic acid (TCA) cycle (e.g., α-subunit of 2-oxoglutarate:NADP+ oxidoreductase, KorA) and the glyoxylate shunt (isocitrate lyase [AceA] and malate synthase G [GlcB]) displayed increased abundance, as expected from previous reports (15, 30) (see Table S1, footnote f, in the supplemental material).
(ii) Effect of benzoate on C4-dicarboxylate TRAP transporter.
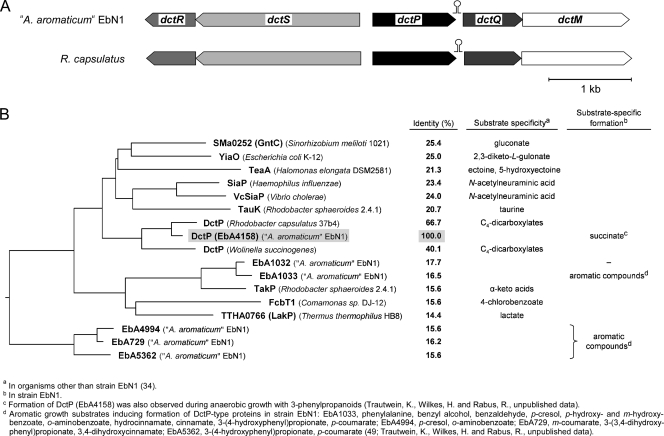
The periplasmic solute-binding protein DctP (EbA4158) and two membrane-integral proteins DctQ (EbA4159) and DctM (EbA4162) constitute a tripartite ATP-independent periplasmic (TRAP) transporter in strain EbN1. The coding genes form an operon-like structure (dctPQM) on the chromosome and colocalize in trans with dctR (ebA4154) and dctS (ebA4156), predicted to encode a two-component sensory regulatory system (37). The same gene order (Fig. 4A), together with high protein sequence identities (66.7, 52.0, and 80.0% for DctP, DctQ, and DctM, respectively) to the characterized C4-dicarboxylate TRAP transporter from Rhodobacter capsulatus (18, 44, 48), suggests that the DctPQM system from strain EbN1 shares the same substrate specificity. This is further supported by the markedly lower sequence identities (<25.4%) of DctP (EbA4158) to described DctP-type proteins recognizing, e.g., monocarboxylates or ectoine (Fig. 4B) (34) and by the absence of alternative known C4-dicarboxylate transporters (e.g., DctA) (27) in the genome of strain EbN1.
Fig 4.
The DctPQM C4-dicarboxylate TRAP transporter in “A. aromaticum” EbN1. (A) Gene organization of the dctPQM orthologs in strain EbN1 and R. capsulatus 37b4. The C4-dicarboxylate-responsive sensor kinase (dctS) and response regulator (dctR) are encoded in trans. In both organisms, a potential RNA stem-loop structure in the dctP-dctQ intercistronic region (see Fig. S5 in the supplemental material) could possibly regulate the abundance of the soluble (DctP) and the membrane-integral transporter components (DctQM). (B) Tree derived from an alignment of the DctP polypeptide sequences using ClustalV of the DNASTAR program (Lasergene). The illustrated relationships are tentative. Database accession no. (from top to bottom) are as follows: NP_435380, AP_004213, YP_003899343, ZP_05849124, NP_231414, YP_345189, CAA45385, YP_159380, NP_907973, YP_157566, YP_157567, ABA79272, AAF16407, YP_144032, YP_159864, YP_157394, and YP_160070.
DctP (EbA4158) displayed decreased abundance during benzoate utilization (phase 1; −5.7-fold) and accounted for more than 5% of the total 2DE-resolved soluble proteome in succinate-utilizing cells of strain EbN1. In addition to dctP (ebA4158), the genome of strain EbN1 encodes five other DctP-type proteins (Fig. 4B). Two of them displayed differing abundance changes during diauxic growth (Table 2): −3.7-fold for EbA1033 during benzoate utilization (phase 1) and 3.0-fold for EbA4994 during succinate utilization (phase 3). However, both proteins share only low sequence identity (16.5 and 15.6%) with DctP (EbA4158). Since EbA1033 and EbA4994 were also detected during anaerobic growth of strain EbN1 with various aromatic compounds (49; K. Trautwein, H. Wilkes, and R. Rabus, unpublished data), their substrate specificity remains unknown.
Subsequent attempts to identify DctQ and DctM in the SDS-PAGE-resolved membrane protein fractions succeeded only for DctQ, identified by nano-LC-ESI-MS/MS in succinate-utilizing cells (single substrate or phase 3 of diauxic growth; see Fig. S4 in the supplemental material). No identification was obtained for DctQ from benzoate-adapted cells or in cells from phases 1 and 2 during diauxic growth. Since DctQ was present only in a faint gel band at a size of ∼20 kDa and ranked only 8th (3 peptides of 16 theoretical peptides matched) among proteins identified from such bands, one may assume that in contrast to DctP, this protein is far less abundant, which probably applies also to DctM.
Transcript profile of dctP in the course of diauxic growth of strain EbN1 with a mixture of succinate and benzoate.
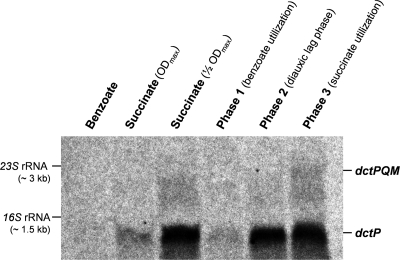
The transcript profile of dctP (Fig. 5) was analyzed by Northern blot hybridization using 33P-labeled DNA probes generated for the dctP (ebA4158) gene, the product of which displayed decreased abundance during benzoate utilization (described above). Total RNA was isolated from cells harvested during phases 1 to 3 of anaerobic diauxic growth (Fig. 2). RNA from cells adapted to anaerobic growth with benzoate or succinate served as a control. The dctP transcript was detected at a size of 1 kb (matching the expected size of 1,005 bp); the signal intensity was standardized using the 16S rRNA signal. Maximal dctP transcript abundance was observed during succinate utilization (phase 3 and succinate-grown control at ½ ODmax), whereas only 2 to 8% of the maximal dctP transcript level was detected in benzoate-utilizing cells (phase 1 and benzoate-grown control). At the beginning of the diauxic lag phase (phase 2), dctP transcript levels immediately started to increase (data not shown), reaching the maximal abundance at the end. The expression of dctP apparently also depended on the succinate concentration, indicated by the 80% reduced level of dctP transcript due to succinate depletion in succinate-adapted cells at the maximal optical density (ODmax).
Fig 5.
Variation of dctP (ebA4158) transcript abundance during anaerobic diauxic growth of “A. aromaticum” EbN1 with succinate plus benzoate (phases 1 to 3), compared to that in cells grown anaerobically with benzoate or succinate (½ ODmax and ODmax) as single substrates. Two transcripts were detected in succinate-utilizing cells (succinate, ½ ODmax; phase 3) with dctP-specific probes: a major dctP transcript (1 kb) and a minor dctPQM transcript (3 kb).
Remarkably, using dctP-specific (Fig. 5) or dctM-specific (data not shown) DNA probes, an additional RNA band with a size of 3 kb was detected. The transcript size matched the expected full-length dctPQM transcript (3,006 bp), suggesting that the three genes form a transcription unit. However, compared to the dctP transcript, the abundance of the dctPQM transcript is low (6% of dctP transcript). The difference in dctP and dctPQM abundances could be explained by the potential of the dctP-dctQ intercistronic region to form an extensive stem-loop structure in the RNA (see Fig. S5 in the supplemental material). A similar RNA structure was previously detected in the dctP-dctQ intercistronic region of R. capsulatus, which is suggested to be involved in rho-independent transcription termination, or in increasing the stability of the dctP transcript (44). Either of the two mechanisms is likely to be responsible for a higher abundance of the dctP than of the dctPQM transcript, thus possibly regulating the abundance of the soluble and membrane components in R. capsulatus and in strain EbN1.
DISCUSSION
Transcriptional regulation of the anaerobic benzoyl-CoA pathway in the absence of benzoate.
Rhodopseudomonas palustris (25) and T. aromatica K172 (43) induce expression of genes involved in anaerobic benzoate degradation, when shifted from an aliphatic substrate to benzoate, with benzoyl-CoA regarded as the inducer. Anaerobic acetate- or glutarate-grown cells of T. aromatica K172 displayed benzoate-CoA ligase activity (26), contrasting with succinate-grown cells of Azoarcus sp. strain CIB lacking benzoate-CoA ligase (BzdA) activity and repressing bzdA expression (30). In strain CIB, benzoyl-CoA was shown to relieve BzdR-mediated transcriptional repression of the catabolic (PN) promoter, thus inducing the expression of the bzd genes encoding the enzymes for anaerobic benzoate degradation (2). The presence of multiple operator sites at the PN promoter and the cooperative nature of BzdR binding are assumed to be responsible for the strong repression of bzd gene expression in strain CIB (14). The bzdR gene is localized directly upstream of the benzoate catabolic gene cluster in strains CIB and EbN1. The respective gene products exhibit high sequence identity (76%), suggesting that BzdR serves similar functions in both strains. Unexpectedly, succinate-adapted cells of strain EbN1 already contained protein components of the benzoyl-CoA pathway, which were however markedly increased in abundance when benzoate became available (see Table S1 in the supplemental material). These protein profiles (Fig. 2) as well as the absence of benzoyl-CoA reductase activity in succinate-adapted cells (Fig. 3) agree with earlier observations from T. aromatica K172 studies (26). Thus, the BzdR homologue in strain EbN1 does not exert a similarly strong repression to that reported for strain CIB.
Anaerobic growth with mixtures of aliphatic and aromatic substrates.
Utilization of succinate is often preferred over that of aromatic compounds (e.g., benzoate), as observed in aerobic degraders like P. putida (33) and A. baylyi (17) or in the anaerobic degrader Azoarcus sp. strain CIB (30). In contrast, anaerobic succinate-adapted cells of strain EbN1 immediately utilized benzoate, phenylacetate, or p-hydroxybenzoate (all degraded via the anaerobic benzoyl-CoA pathway) from the substrate mixture. Remarkably, strain EbN1 displayed different substrate utilization patterns and growth physiologies for these aromatic acids: (i) complete depletion of benzoate prior to onset of succinate utilization (biphasic growth; diauxie); (ii) semidepletion of phenylacetate prior to onset of succinate utilization (monophasic growth); and (iii) immediate coutilization of p-hydroxybenzoate and succinate (monophasic growth). The unexpected diauxie of C4-dicarboxylate-adapted cells of strain EbN1 growing with a mixture of benzoate plus succinate, fumarate, or malate is uncoupled from growth rates: i.e., benzoate (maximum specific growth rate [μmax], 0.19 h−1) is preferred over substrates supporting higher (succinate; μmax, 0.25 h−1), similar (fumarate; μmax, 0.20 h−1), or lower (malate; μmax, 0.12 h−1) maximum specific growth rates (Table 1). This contrasts with the general view that preferred substrates support the highest growth rates (22, 24). Thus, growth rate alone is apparently not the only factor controlling substrate preference in strain EbN1.
The observed diauxie of strain EbN1 does not represent a general growth behavior among members of the “Aromatoleum”/Azoarcus/Thauera cluster. The tested relatives of strain EbN1 displayed monophasic growth and strain-specific substrate utilization patterns (see Fig. S3 in the supplemental material), ranging from immediate (strain K172) (see also reference 26) and delayed (strain HxN1) coutilization to a complete reversal of substrate preference (strain ToN1). Notably, strain ToN1 showed the same substrate preference as its closest relative, Azoarcus sp. strain CIB (99.2% 16S rRNA gene similarity) (30). The results may suggest the presence of different regulatory stringencies and mechanisms within individual subgroups of the “Aromatoleum”/Azoarcus/Thauera cluster. Further research is required to investigate the ecophysiological meaning of such a diversification in the regulation of substrate utilization among closely related strains having similar substrate ranges and inhabiting common ecological niches.
Benzoate-mediated repression of C4-dicarboxylate uptake.
Succinate uptake in strain EbN1 is predicted to proceed via the C4-dicarboxylate TRAP transporter DctPQM. The decrease of dctP transcript and DctP protein abundances during diauxic phase 1 (benzoate utilization) agrees with the concurrent nonutilization of succinate. Conversely, the increase in transcript (dctP and dctPQM) and protein (DctP) abundance during the diauxic lag phase apparently prepares the cells for succinate uptake and utilization in phase 3 (Fig. 2 and 5). Thus, the assumed succinate-responsive DctSR-mediated transcriptional activation of dctPQM expression (23) appears to be negatively controlled in the presence of benzoate, suggesting that repression of C4-dicarboxylate uptake during phase 1 seems to be the main determinant for diauxie in strain EbN1. In analogy, naphthalene and benzyl alcohol mediate repression of an ABC-type periplasmic glucose-binding protein involved in glucose uptake in P. putida CSV86 (3). Further evidence for the causal link between the observed diauxie and DctPQM is the monophasic growth of strain EbN1 with a mixture of benzoate and oxaloacetate, since related DctPQM from R. capsulatus cannot transport C4-dicarboxylates other than succinate, fumarate, or d,l-malate (e.g., d-tartrate) (44, 48).
While the molecular mechanisms of the assumed benzoate-mediated repression are presently unknown, several possibilities could be envisioned that may also have some interplay. (i) A benzoate-responsive global transcriptional regulator could inhibit activation of dctPQM transcription. Homologs of global regulators (e.g., Crp and Crc) discussed to serve such functions in E. coli (11) and Pseudomonas sp. (8; for overview, see reference 40) are encoded on the chromosome of strain EbN1. (ii) Benzoate may directly or indirectly interfere with the C4-dicarboxylate-induced signal transduction of DctSR, e.g., by modulating kinase activity of the DctS sensor via the cytoplasmic PAS domain (16, 21, 31). (iii) The presence of benzoate may induce Hfq-mediated decay of the dctP (respectively, dctPQM) transcript in analogy to ABC-type periplasmic solute-binding proteins (for a review, see reference 36). Notably, identification of Hfq (EbA1255) in protein extracts of strain EbN1 (see Table S1 in the supplemental material) suggests the possibility of such a kind of posttranscriptional control in strain EbN1.
At present, we can only speculate about the metabolic signal triggering repression of C4-dicarboxylate utilization. Aerobic diauxic growth with succinate and benzoate could suggest that benzoate or benzoyl-CoA serves such a function, since both compounds are shared by the otherwise differing aerobic and anaerobic degradation pathways for benzoate in strain EbN1 (49). The observation that acetate in addition to benzoate confers full repression of succinate utilization in strain EbN1, however, questions direct involvement of benzoate or its degradation intermediates. Whether benzoate and acetate mediate repression of succinate uptake by the same or a different molecular mechanism is unknown.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to D. Lange and S. Lahme (both from Bremen) and J. Holert and N. Som (both from Oldenburg) for technical assistance. We are indebted to F. Widdel (Bremen) for general support of proteomic studies in our group.
This study was supported by the University of Oldenburg and the Max-Planck-Gesellschaft.
Footnotes
Published ahead of print 11 November 2011
This article is dedicated to Milton H. Saier, Jr.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1. Ampe F, Lindley N. 1995. Acetate utilization is inhibited by benzoate in Alcaligenes eutrophus: evidence for transcriptional control of the expression of acoE coding for acetyl coenzyme A synthetase. J. Bacteriol. 177:5826–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barragan MJL, et al. 2005. BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 280:10683–10694 [DOI] [PubMed] [Google Scholar]
- 3. Basu A, Shrivastava R, Basu B, Apte SK, Phale PS. 2007. Modulation of glucose transport causes preferential utilization of aromatic compounds in Pseudomonas putida CSV86. J. Bacteriol. 189:7556–7562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annu. Rev. Plant Biol. 54:519–546 [DOI] [PubMed] [Google Scholar]
- 5. Boll M. 2005. Dearomatizing benzene ring reductases. J. Mol. Microbiol. Biotechnol. 10:132–142 [DOI] [PubMed] [Google Scholar]
- 6. Boll M, Fuchs G. 1995. Benzoyl-coenzyme A reductase (dearomatizing), a key enzyme of anaerobic aromatic metabolism. Eur. J. Biochem. 234:921–933 [DOI] [PubMed] [Google Scholar]
- 7. Bradford MM. 1976. Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 8. Busch A, Lacal J, Silva-Jimenez H, Krell T, Ramos JL. 2010. Catabolite repression of the TodS/TodT two-component system and effector-dependent transphosphorylation of TodT as the basis for toluene dioxygenase catabolic pathway control. J. Bacteriol. 192:4246–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Champion KM, Zengler K, Rabus R. 1999. Anaerobic degradation of ethylbenzene and toluene in denitrifying strain EbN1 proceeds via independent substrate-induced pathways. J. Mol. Microbiol. Biotechnol. 1:157–164 [PubMed] [Google Scholar]
- 10. Clauser KR, Baker P, Burlingame AL. 1999. Role of accurate mass measurement (±10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71:2871–2882 [DOI] [PubMed] [Google Scholar]
- 11. Davies SJ, et al. 1999. Inactivation and regulation of the aerobic C4-dicarboxylate transport (dctA) gene of Escherichia coli. J. Bacteriol. 181:5624–5635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dennis PG, Miller AJ, Hirsch PR. 2010. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 72:313–327 [DOI] [PubMed] [Google Scholar]
- 13. Doherty NS, et al. 1998. Analysis of changes in acute-phase plasma proteins in an acute inflammatory response and in rheumatoid arthritis using two-dimensional gel electrophoresis. Electrophoresis 19:355–363 [DOI] [PubMed] [Google Scholar]
- 14. Durante-Rodriguez G, et al. 2010. Biochemical characterization of the transcriptional regulator BzdR from Azoarcus sp. CIB. J. Biol. Chem. 285:35694–35705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ebenau-Jehle C, Boll M, Fuchs G. 2003. 2-Oxoglutarate:NADP+ oxidoreductase in Azoarcus evansii: properties and function in electron transfer reactions in aromatic ring reduction. J. Bacteriol. 185:6119–6129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Etzkorn M, et al. 2008. Plasticity of the PAS domain and a potential role for signal transduction in the histidine kinase DcuS. Nat. Struct. Mol. Biol. 15:1031–1039 [DOI] [PubMed] [Google Scholar]
- 17. Fischer R, Bleichrodt FS, Gerischer UC. 2008. Aromatic degradative pathways in Acinetobacter baylyi underlie carbon catabolite repression. Microbiology 154:3095–3103 [DOI] [PubMed] [Google Scholar]
- 18. Forward JA, Behrendt MC, Wyborn NR, Cross R, Kelly DJ. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179:5482–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuchs G. 2008. Anaerobic metabolism of aromatic compounds. Ann. N. Y. Acad. Sci. 1125:82–99 [DOI] [PubMed] [Google Scholar]
- 20. Gade D, Thiermann J, Markowsky D, Rabus R. 2003. Evaluation of two-dimensional difference gel electrophoresis for protein profiling. J. Mol. Microbiol. Biotechnol. 5:240–251 [DOI] [PubMed] [Google Scholar]
- 21. Garcia PP, Bringhurst RM, Arango Pinedo C, Gage DJ. 2010. Characterization of a two-component regulatory system that regulates succinate-mediated catabolite repression in Sinorhizobium meliloti. J. Bacteriol. 192:5725–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Görke B, Stülke J. 2008. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 6:613–624 [DOI] [PubMed] [Google Scholar]
- 23. Hamblin MJ, Shaw JG, Kelly DJ. 1993. Sequence analysis and interposon mutagenesis of a sensor-kinase (DctS) and response-regulator (DctR) controlling synthesis of the high-affinity C4-dicarboxylate transport system in Rhodobacter capsulatus. Mol. Gen. Genet. 237:215–224 [DOI] [PubMed] [Google Scholar]
- 24. Harder W, Dijkhuizen L. 1982. Strategies of mixed substrate utilization in microorganisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 297:459–480 [DOI] [PubMed] [Google Scholar]
- 25. Harwood CS, Burchhardt G, Herrmann H, Fuchs G. 1998. Anaerobic metabolism of aromatic compounds via the benzoyl-CoA pathway. FEMS Microbiol. Rev. 22:439–458 [Google Scholar]
- 26. Heider J, et al. 1998. Differential induction of enzymes involved in anaerobic metabolism of aromatic compounds in the denitrifying bacterium Thauera aromatica. Arch. Microbiol. 170:120–131 [DOI] [PubMed] [Google Scholar]
- 27. Janausch IG, Zientz E, Tran QH, Kröger A, Unden G. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39–56 [DOI] [PubMed] [Google Scholar]
- 28. Jenö P, Mini T, Moes S, Hintermann E, Horst M. 1995. Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem. 224:75–82 [DOI] [PubMed] [Google Scholar]
- 29. Kühner S, et al. 2005. Substrate-dependent regulation of anaerobic degradation pathways for toluene and ethylbenzene in a denitrifying bacterium, strain EbN1. J. Bacteriol. 187:1493–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lopez Barragan MJ, et al. 2004. The bzd gene cluster, coding for anaerobic benzoate catabolism, in Azoarcus sp. strain CIB. J. Bacteriol. 186:5762–5774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Möglich A, Ayers RA, Moffat K. 2009. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 17:1282–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Monod J. 1942. Recherches sur la croissance des cultures bactériennes. Hermann & Co., Paris, France. [Google Scholar]
- 33. Morales G, et al. 2004. The Pseudomonas putida Crc global regulator controls the expression of genes from several chromosomal catabolic pathways for aromatic compounds. J. Bacteriol. 186:1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mulligan C, Fischer M, Thomas GH. 2011. Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol. Rev. 35:68–86 [DOI] [PubMed] [Google Scholar]
- 35. Nouwens AS, et al. 2000. Complementing genomics with proteomics: the membrane subproteome of Pseudomonas aeruginosa PAO1. Electrophoresis 21:3797–3809 [DOI] [PubMed] [Google Scholar]
- 36. Papenfort K, Vogel J. 2009. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 160:278–287 [DOI] [PubMed] [Google Scholar]
- 37. Rabus R, et al. 2005. The genome sequence of an anaerobic aromatic-degrading denitrifying bacterium, strain EbN1. Arch. Microbiol. 183:27–36 [DOI] [PubMed] [Google Scholar]
- 38. Rabus R, Trautwein K. 2010. Proteogenomics to study the anaerobic degradation of aromatic compounds and hydrocarbons, p 4385–4405 In Timmis KN. (ed), Handbook of hydrocarbon and lipid microbiology. Springer, Berlin, Germany: [Google Scholar]
- 39. Rabus R, Widdel F. 1995. Anaerobic degradation of ethylbenzene and other aromatic hydrocarbons by new denitrifying bacteria. Arch. Microbiol. 163:96–103 [DOI] [PubMed] [Google Scholar]
- 40. Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 34:658–684 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 42. Schachter D, Taggart JV. 1953. Benzoyl coenzyme A and hippurate synthesis. J. Biol. Chem. 203:925–934 [PubMed] [Google Scholar]
- 43. Schühle K, et al. 2003. Benzoate-coenzyme A ligase from Thauera aromatica: an enzyme acting in anaerobic and aerobic pathways. J. Bacteriol. 185:4920–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shaw JG, Hamblin MJ, Kelly DJ. 1991. Purification, characterization and nucleotide sequence of the periplasmic C4-dicarboxylate-binding protein (DctP) from Rhodobacter capsulatus. Mol. Microbiol. 5:3055–3062 [DOI] [PubMed] [Google Scholar]
- 45. Thiele B, et al. 2008. Aromatizing cyclohexa-1,5-diene-1-carbonyl-coenzyme A oxidase: characterization and its role in anaerobic aromatic metabolism. J. Biol. Chem. 283:20713–20721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tissot BP, Welte DH. 1984. Petroleum formation and occurrence, 2nd ed. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 47. Tschech A, Fuchs G. 1987. Anaerobic degradation of phenol by pure cultures of newly isolated denitrifying pseudomonads. Arch. Microbiol. 148:213–217 [DOI] [PubMed] [Google Scholar]
- 48. Walmsley AR, Shaw JG, Kelly DJ. 1992. The mechanism of ligand binding to the periplasmic C4-dicarboxylate binding protein (DctP) from Rhodobacter capsulatus. J. Biol. Chem. 267:8064–8072 [PubMed] [Google Scholar]
- 49. Wöhlbrand L, et al. 2007. Functional proteomic view of metabolic regulation in “Aromatoleum aromaticum” strain EbN1. Proteomics 7:2222–2239 [DOI] [PubMed] [Google Scholar]
- 50. Wöhlbrand L, Wilkes H, Halder T, Rabus R. 2008. Anaerobic degradation of p-ethylphenol by “Aromatoleum aromaticum” strain EbN1: pathway, regulation, and involved proteins. J. Bacteriol. 190:5699–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.