Abstract
Neisseria meningitidis is a leading pathogen of epidemic bacterial meningitis and fulminant sepsis worldwide. Twelve different N. meningitidis serogroups have been identified to date based on antigenic differences in the capsular polysaccharide. However, more than 90% of human cases of N. meningitidis meningitis are the result of infection with just five serogroups, A, B, C, W135, and Y. Efficient methods of detection and genogrouping of N. meningitidis isolates are needed, therefore, in order to monitor prevalent serogroups as a means of disease control and prevention. The capsular gene complex regions have been sequenced from only seven out of the 12 serogroups. In this study, the capsular gene complexes of the remaining five serogroups were sequenced and analyzed. Primers were designed that were specific for N. meningitidis species and for the 12 individual serogroups, and a multiplex PCR assay using these specific primers was developed for N. meningitidis detection and genogrouping. The assay was tested using 15 reference strains covering all 12 serogroups, 143 clinical isolates, and 21 strains from closely related species or from species that cause meningitis. The assay could detect N. meningitidis serogroups and was shown to be specific, with a detection sensitivity of 1 ng of genomic DNA (equivalent to ∼4 × 105 genomes) or 3 × 105 CFU/ml in noncultured mock cerebrospinal fluid (CSF) specimens. This study, therefore, describes for the first time the development of a molecular protocol for the detection of all N. meningitidis serogroups. This multiplex PCR-based assay may have use for the clinical diagnosis and epidemiological surveillance of N. meningitidis.
INTRODUCTION
Neisseria meningitidis is an encapsulated Gram-negative bacterium and the leading pathogenic cause of epidemic bacterial meningitis and fulminant sepsis worldwide. Infections with N. meningitidis are a significant cause of mortality and morbidity in young children and adolescents (16, 23). The bacterial capsular polysaccharide (CPS) is the most important virulence factor for N. meningitidis (14, 20, 26, 28, 32). Twelve different serogroups, A, B, C, 29E, H, I, K, L, W135, X, Y, and Z, have been identified based on antigenic variation in the CPSs (6, 22). More than 90% of human cases of N. meningitidis meningitis are the result of infection with serogroups A, B, C, W135, and Y (21). Rare meningitis cases caused by serogroup K and Z infections have also been reported over the last few decades (11, 25). Detection and genogrouping of N. meningitidis isolates are important to monitor changes in the population of prevalent circulating serogroups for the purposes of disease control and prevention, vaccination strategy, and contact management (3, 12, 30).
Several genes, which include ctrA, porA, crgA, and 16S rRNA, have been widely used as the gene targets for N. meningitidis detection by PCR-based assays (7, 16, 33). In particular, genes such as sacB and siaD have been used to genogroup the most prevalent A, B, C, W135, and Y serogroups (3, 4, 10). However, to our knowledge, no molecular method that distinguishes all 12 N. meningitidis serogroups has ever been developed. The conventional serogrouping method that uses specific antisera is still in routine practice in many diagnostic laboratories; however, it is labor-intensive, and antisera for all serogroups are very hard to obtain. Therefore, developing a rapid, accurate, easy to use, and inexpensive method for the detection and genogrouping of N. meningitidis is in urgent need.
Variations in genes clustered in the chromosomal capsular gene complex are responsible for CPS differences in the N. meningitidis serogroups. A typical N. meningitidis capsular gene complex consists of regions A, B, C, D, and E. Regions A and C are located between the galE and tex genes on the chromosome and are transcribed independently (31). Region B is upstream of region A or downstream of region C (35). Genes in region A are responsible for the synthesis of the polysaccharide. Genes in region B, which include lipA and lipB, are involved in translocation and surface expression of capsule polymers. Region C contains four genes (ctrA, -B, -C, and -D) that are necessary for the transport of the capsule to the membrane (35). Region D is comprised of a cassette of genes, which include rmlA, -B, and -C and galE, and is not involved in capsule expression but is responsible for meningococcal lipooligosaccharide (LOS) biosynthesis (13). Region E contains only one gene, tex, that has been proposed to regulate CPS synthesis (19). The gene sequences in region A of the complex are different in individual serogroups, and those in regions B, C, D, and E are highly conserved between serogroups (32).
Out of the 12 serogroups, only the seven capsular gene complexes from serogroups A, B, C, 29E, W135, X, and Y have been characterized previously (8, 27). In this study, the capsular gene complexes of the five uncharacterized serogroups (H, I, K, L, Z) were sequenced, which made it possible to develop primers to differentiate genes specific for all 12 N. meningitidis serogroups. A multiplex PCR-based assay for N. meningitidis detection and genogrouping was developed based on “screening out” specific genes and primers for each N. meningitidis serogroup. This assay was tested on 179 bacterial isolates and proved to be specific, accurate, and sensitive.
MATERIALS AND METHODS
Bacterial strains.
The 179 strains used in this study are listed in Table 1. They include 15 reference strains and 143 clinical strains of the 12 targeted N. meningitidis serogroups (A, B, C, 29E, H, I, K, L, W135, X, Y, and Z) and 15 strains of other Neisseria species, 5 strains of Streptococcus pneumoniae, and one strain of Haemophilus influenzae. N. meningitidis strains were grown on chocolate agar plates at 37°C in the presence of 5% CO2 in air for 24 h.
Table 1.
Bacterial strains used in this studya
| Serogroup, sample type, or species | Strain no.b (source of strain) | Total no. of strains |
|---|---|---|
| Neisseria meningitidis reference strain (n = 15) serogroup | ||
| A | 29010 (CMCC), 29019 (CMCC) | 2 |
| B | 29011 (CMCC), 29061 (CMCC) | 2 |
| C | 29012 (CMCC) | 1 |
| 29E | 29034 (CMCC) | 1 |
| H | 29031 (CMCC) | 1 |
| I | 29044 (CMCC) | 1 |
| K | 29047 (CMCC) | 1 |
| L | 43828 (ATCC) | 1 |
| W135 | 29037 (CMCC), 29057 (CMCC) | 2 |
| X | M8210 (ICDC) | 1 |
| Y | 29038 (CMCC) | 1 |
| Z | 35562 (ATCC) | 1 |
| N. meningitidis clinical isolate (n = 143) sample type | ||
| CSF | 23 (ICDC), 2 (SCDC) | 25 |
| Blood | 4 (ICDC) | 4 |
| TS | 61 (ICDC), 6 (SCDC) | 67 |
| CP | 30 (SCDC) | 30 |
| BP | 4 (SCDC) | 4 |
| Unknown | 9 (ICDC), 4 (SCDC) | 13 |
| Strains from other species (n = 21) | ||
| Neisseria lactamica | 3719 (CCM), 29114 (CMCC) | 2 |
| Neisseria flavescens | 2827 (CCM) | 1 |
| Neisseria subflava | 3482 (CCM), 4392 (CCM), 4400 (CCM), 29109 (CMCC), 29110 (CMCC), 29111 (CMCC), 29121 (CMCC) | 7 |
| Neisseria mucosa | 3483 (CCM) | 1 |
| Neisseria sicca | 4404 (CCM), 4405 (CCM) | 2 |
| Neisseria gonorrhoeae | No. 1 (DDTGH), no. 2 (DDTGH) | 2 |
| Streptococcus pneumoniae 6A | G1839 (NU) | 1 |
| Streptococcus pneumoniae 1 | G1863 (NU) | 1 |
| Streptococcus pneumoniae 19A | G1921 (NU) | 1 |
| Streptococcus pneumoniae 23F | G1934 (NU) | 1 |
| Streptococcus pneumoniae 6B | G1940 (NU) | 1 |
| Haemophilus influenzae | 58528 (CMCC) | 1 |
CMCC, China's Medical Culture Center; ATCC, American Type Culture Collection; ICDC, Chinese Center for Disease Control and Prevention, Beijing, China; SCDC, Shanghai Center for Disease Control and Prevention; CCM, Czech Collection of Microorganisms; DDTGH, Dermatological Department of Tianjin General Hospital; NU, Nankai Univeristy, China; CSF, cerebrospinal fluid; CP, chocolate agar plate; BP, blood agar plate; TS, throat swab.
For N. meningitidis clinical isolate sample types, the number of strains is given instead of strain number.
Genomic DNA extraction.
Genomic DNA was extracted with a DNA extraction kit (QIAamp DNA minikits; Qiagen, Hilden, Germany).
Capsular gene complex amplification, sequencing, and analysis.
Primers wl-14395 (5′-CGCCATTTCTTCCGCCAACACCA-3′) and wl-22861 (5′-CCAGCCCGAAAGTAGCCGATGC-3′) were designed based on the sequences of galE and gltS genes, respectively, and were used to amplify the capsular gene complexes of serogroups D, H, and K. The primers wl-14396 (5′-GCGTTTGCGTGCAGCATCGACT-3′) and wl-27419 (5′-GCCGAGAACCTTAACTGGCATATTGTGAA-3′) were designed based on tex and lipB gene sequences, respectively, and were used to amplify the capsular gene complexes of serogroups I, L, and Z. Long, high-fidelity PCR was carried out under the following conditions: denaturation step at 95°C for 15 s, annealing step at 62°C for 30 s, and extension step at 68°C for 10 min for 32 cycles. The PCR products were gel purified on UNIQ-10 columns (Sangon, Shanghai, China), and the sample DNA was sheared at speed code 2 (20 cycles) to the desired fragment length of 2 to 3 kb by HydroShear fragmentation (GeneMachines, CA). The fragments were then cloned into the pUC18 vector after end-blunting reaction with T4 polymerase and Klenow fragments (New England BioLabs, MA) to produce shotgun banks.
The samples were sequenced on an ABI 3730 automated DNA sequencer (Applied Biosystems, CA), and sequencing data were assembled by use of the Staden package software (29). The Artemis program (www.sanger.ac.uk) was used to identify open reading frames (ORFs) and annotations (24). BLAST and PSI-BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) were used to search several databases (1), which included GenBank (www.ncbi.nlm.nih.gov/GenBank), the Clusters of Orthologous Groups (COG; www.ncbi.nlm.nih.gov/COG/), and Pfam (pfam.sanger.ac.uk) protein motif databases (2, 34).
Primer design.
All the primers used in this study are listed in Table 2. The genes porA and ctrA were used as the gene targets for N. meningitidis species-specific primers. Sequences of N. meningitidis porA and ctrA genes were obtained from the GenBank database and aligned using ClustalX software (www.clustal.org). The universal primers were designed based on the most conserved regions of these two genes. Serogroup-specific primers were designed based on the sacD gene sequence for serogroup A, the siaD gene sequence for serogroups B and C, the synG/synF gene sequence for W135/Y, the cap29EH gene sequence for serogroup 29E, the wnmB gene sequence for serogroups I/K, the capZC gene sequence for serogroup Z, the lcbB gene sequence for serogroup L, the xcbA gene sequence for serogroup X, and the wnmA gene sequence for serogroup H. In addition, the primers used to differentiate serogroups I and K were based on the lipA gene sequence (sense primer) and galE gene sequence (antisense primer) of serogroup I. The primers that were used to differentiate serogroups W135 and Y were based on synF and synG gene sequences. At least two pairs of primers were designed for screening each serogroup.
Table 2.
Primers used in this study
| Primer pair | Targeted gene | GenBank accession no. | Amplicon size (bp) | Targeted serogroup | Sequence of primer pairs (5′→ 3′) |
|---|---|---|---|---|---|
| wl-35663/38969 | ctrA | GU391296 | 257 | All | GTCGCGGTGATGTGGTTA/AATCTCTGCCTCACTGCCAT |
| wl-40218/40170 | porA | AY319969 | 158 | All | CTCATAGCCGCCCGTCA/GCGGTTTTGCCGGGAACTAT |
| wl-37416/37417 | sacD | NC_003116 | 470 | A | TTTTATTCTTAGATGTTGACGTTTT/ATGCCAGAAATGTTTAGGAGTT |
| wl-37410/37411 | siaD | NC_003112 | 555 | B | TTTTTAGCATATTCAGGAAAGG/TTCAATGTGGTTGACAACATCT |
| wl-37469/37470 | siaD | NC_008767 | 381 | C | TGTGCTAATCCCGCCTGA/AGAAAGCCGGGAATCGTT |
| wl-39429/39430 | cap29EH | AJ576117 | 694 | 29E | TTGGCGGTTGAAACCTTAC/GCGTATCATGCTCCATTACCA |
| wl-39433/39434 | wnmB | HQ437687 | 893 | I/K | AATGTGTGTGCAGGAGCTTG/ATGCTTCGGTTGCCTGTC |
| wl-37709/37710 | capZC | HQ437689 | 656 | Z | TATTGGCCTGAGCACCG/AGGCGTACCGTCTGTAACTG |
| wl-37406/37407 | lcbB | HQ437688 | 988 | L | TTTGAATGTACCCTCTCCTCTG/TAACAGTCCTGATATCACTCCGTA |
| wl-39431/39432 | xcbA | AY289931 | 777 | X | CTCTCGTCTACCAAATGGCTAT/GTGAGGGTGACATCCGCTA |
| wl-36794/36795 | wnmA | HQ437685 | 1454 | H | TGATCTACCCAAGGCACATAC/CAATCGGCTTATTGAGGGT |
| wl-39435/39436 | synG | EU038215 | 520 | W/Y | AAGGTGAATCTTCCGAGCAG/CCATTGGAAATTTTCGCTT |
| wl-32526/45168 | galE/lipA | HQ437687 | 5030 | I | TATTGGTTCGCATACTGTCATT/GTCATAGACGATGGAATAGGGA |
| wl-37473/37475 | synG | EU038216 | 476 | W135 | ACGGTATCTGATGAAATGCTG/TCATATACAACGATTGGAATATC |
Specificity of primers.
Primers based on porA and ctrA gene sequences were used to amplify the DNA templates of N. meningitidis reference strains from the 12 serogroups, from Neisseria flavescens, Neisseria subflava, Neisseria mucosa, Neisseria lactamica, Neisseria sicca, Neisseria gonorrhoeae strains, and from S. pneumoniae and H. influenzae. The PCRs were carried out in a reaction mixture that contained 1× PCR buffer (50 mM KCl, 10 mM Tris-HCl [pH 8.3]), 2.5 mM MgCl2, 0.4 mM deoxynucleoside triphosphate (dNTP), 1 U rTaq DNA polymerase, 0.1 μM each primer, and 50 to 100 ng of DNA template in a final volume of 20 μl. The PCR program used was as follows: a denaturation step at 94°C for 5 min and then 31 cycles of a denaturation step at 94°C for 30 s, an annealing step of 56°C for 45 s, and an extension step of 72°C for 1 min, with a final extension at 72°C for 5 min.
Development of the multiplex PCR-based assay.
This assay consisted of three multiplex PCRs. PCR 1 was used to detect serogroups A, B, C, 29E, and I/K, and PCR 2 was used to detect serogroups Z, L, X, H, and W135/Y. Reaction 3 was used to differentiate serogroups I and K and W135 and Y, which could not be distinguished by PCRs 1 and 2 (see Fig. S1 in the supplemental material). The PCR mixture for reactions 1, 2, and 3 contained the same reagents except for the primers, and the details are described in the supplemental material. The PCR program for reactions 1 and 2 was as follows: a denaturation step at 94°C for 5 min and then 24 cycles of a denaturation step at 94°C for 30 s, an annealing step at 59°C for 45 s, and an extension step at 72°C for 1 min, with a final extension at 72°C for 5 min. The PCR program for reaction 3 was the same as that for reactions 1 and 2 except that the annealing step was 54°C for 45 s and the extension step was 72°C for 3 min.
Nucleotide sequence accession numbers.
The DNA sequences of the capsular gene complexes of N. meningitidis serogroups H, I, K, L, and Z have been deposited in the GenBank database under the accession numbers HQ437685, HQ437686, HQ437687, HQ437688, and HQ437689, respectively.
RESULTS
Capsular gene complexes of six N. meningitidis serogroups.
The capsular gene complexes of the remaining five serogroups, H, I, K, L, and Z, were sequenced in this study. DNA sequences of 23,488, 24,676, 27,709, 18,453, and 21,871 bp in length were obtained from serogroups H, I, K, L, and Z, respectively. These gene complexes contained 19, 19, 22, 14, and 15 ORFs, respectively (see Fig. S2 in the supplemental material). The average GC contents of the sequences were 45.13, 48.04, 45.03, 45.62, and 46.97% in serogroups H, I, K, L, and Z, respectively. The functions of each ORF in these capsular gene complexes were predicted based on sequence homology by search of the listed databases and are summarized in Tables S1 to S5 in the supplemental material.
Genes specific for N. meningitidis detection and genogrouping.
ctrA or porA genes have been used individually to identify N. meningitidis (15, 16). However, we found that our primers based on the ctrA sequence produced PCR products from both N. meningitidis and N. lactamica, and those based on the porA sequence gave PCR products from both N. meningitidis and N. gonorrhoeae (data not shown). Therefore, it was necessary to use a combination of these two genes as the targets to distinguish N. meningitidis from other species. Serogroup-specific genes were chosen based on the capsular gene sequences of all 12 N. meningitidis serogroups from those genes responsible for different CPS structures. These genes included ones that encode glycosyltransferase (sacD, wnmB, lcbB), polysialyltransferase (siaD), polymerase (synF, synG), phosphotransferase (capZC, wnmA, xcbA), and d-arabinose-5-phosphate isomerase (cap29EH). However, the gene sequences responsible for CPS synthesis in serogroups I and K, and those in serogroups W135 and Y, were too similar to be useful for differentiation. Genes lipA and galE, whose location is different in serogroups I and K, were used to differentiate the two serogroups. For serogroups W135 and Y, a minor difference between synF and synG sequences was used to differentiate these two serogroups.
Primers based on the ctrA and porA gene sequences gave both of the two expected PCR products in N. meningitidis, but only one or no PCR product in other species. Primers based on serogroup-specific genes could amplify the expected PCR products in the corresponding serogroup(s) but not in other serogroups. Primers that differentiated the serogroups I and K, or serogroups W135 and Y, gave PCR products only for serogroups I and W135, respectively, and no PCR product was detected for serogroups K or Y. The positions of the serogroup-specific primers are shown in Fig. S2 and S3 in the supplemental material.
Development of a multiplex PCR-based assay.
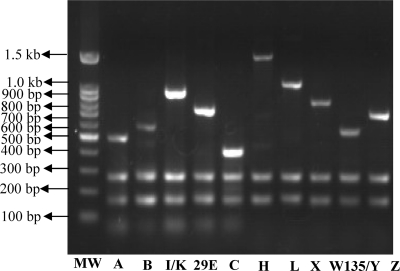
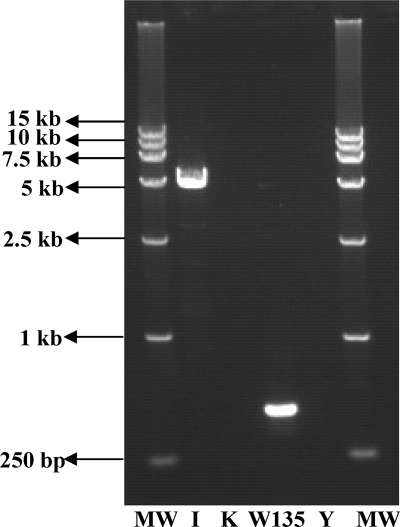
Three groups of primer sets were made based on the screened specific primers. The primer pair efficiency in each group was determined on the basis of achieving amplicons of the expected sizes at a range of different primer concentrations. The primer concentration that resulted in high-signal products was used as described in Materials and Methods. At the optimized primer concentration ratio, the DNA of strains that belonged to serogroups A, B, C, 29E, and I/K produced the expected PCR products of distinct sizes (470 bp for serogroup A, 555 bp for B, 381 bp for C, 694 bp for 29E, 893 bp for I/K, and 257 and 158 bp for ctrA and porA of all the tested serogroups) in the group 1 multiplex PCRs (Fig. 1). The DNA from serogroups Z, L, X, H, and W135/Y produced the expected PCR products (656 bp for Z, 988 bp for L, 777 bp for X, 1,454 bp for H, 520 bp for W135/Y, and 257 and 158 bp for ctrA and porA of all the tested serogroups) in the group 2 multiplex PCRs (Fig. 1). Serogroups I and W135 gave expected PCR products of 5,030 bp and 476 bp, respectively, in the group 3 multiplex PCRs (Fig. 2).
Fig 1.
Agarose gel electrophoresis of the PCR products obtained from multiplex PCRs 1 and 2 in the assay. Lane MW, 100-bp DNA ladder marker; lane A, serogroup A; lane B, serogroup B; lane I/K, serogroup I/K; lane 29E, serogroup 29E; lane C, serogroup C; lane H, serogroup H; lane L, serogroup L; lane X, serogroup X; lane W135/Y, serogroup W135/Y; lane Z, serogroup Z.
Fig 2.
Agarose gel electrophoresis of the PCR products obtained from multiplex PCR 3. Lanes MW, DNA marker DL15,000; lane I, serogroup I; lane K, serogroup K; lane W135, serogroup W135; lane Y, serogroup Y.
Specificity of the multiplex PCR-based assay.
The developed multiplex PCR-based assay was tested with the 15 reference strains that belonged to the 12 N. meningitidis serogroups, with 97 clinical strains that belonged to A, B, C, W135, 29E, X, or Y or nongroupable isolates from different locations and years; with 15 strains of closely related species such as N. flavescens, N. subflava, N. mucosa, N. lactamica, N. sicca, and N. gonorrhoeae; and with six strains from species also reported to cause meningitis that included five S. pneumoniae isolates (belonging to serotypes 1, 23F, 6A, 6B, and 19A, reported to be more virulent) and one H. influenzae isolate (Table 1; see also Table S6 in the supplemental material). All the reference strains were detected with 100% specificity. Among the 97 clinical isolates, 90 of known serotype were detected and confirmed by the specific antiserum serogrouping. Five out of the seven strains that were classified as nongroupable by antisera could be classified as belonging to a specific serogroup by the multiplex PCR assay (see Table S6). For strains that belonged to other Neisseria species, only one or no PCR products that corresponded to segments of ctrA or porA genes could be detected. There was no PCR product obtained for S. pneumoniae and H. influenzae strains. These results indicate that the multiplex PCR-based assay is specific for individual N. meningitidis serogroups.
Double-blinded test to verify the multiplex PCR assay.
A double-blinded test was performed in order to verify the stability and specificity of the assay. Forty-six clinical isolates were used for the test (see Table S6 in the supplemental material). The isolates were also characterized by specific antisera at the Center for Disease Control and Prevention, Shanghai, China. The detection results of the PCR assay were consistent with the results by antiserum serogrouping except for the isolate NM033 (see Table S6). The reason for the different detection results for isolate NM033 using antisera and the multiplex PCR assay are unknown, and the strain needs to be further analyzed.
Detection sensitivity of the multiplex PCR assay.
The detection sensitivities for genomic DNA and isolates in noncultured mock CSF specimens were both determined. Serial dilutions of genomic DNA (10 ng, 1 ng, 100 pg, 10 pg, and 1 pg per ml) of N. meningitidis serogroups A, B, C, 29E, and Z reference strains were used to test the sensitivity of the multiplex assay. As little as 1 ng of DNA (equivalent to ∼4 × 105 genomes) could be detected by the assay. Pure cultures of strain NM055 (serogroup C, isolated from a patient) were mixed with 1 ml CSF specimens from healthy people with the concentrations of 107, 106, 105, 104, to 103 CFU/ml, respectively, and tested with the multiplex assay. The lower limit of detection was determined to be 3 × 105 CFU/ml. All the reference strains of other serogroups could also be detected by this assay at the level of ∼4 × 105 genomes or 3 × 105 CFU/ml. Therefore, ∼4 × 105 genomes or 3 × 105 CFU/ml strains in the mock CSF specimen were the lower limits for the detection using this assay.
DISCUSSION
Molecular protocols for the detection of N. meningitidis have been reported previously. However, serogrouping of N. meningitidis is dependent mainly on methods that use specific antisera. The capsular gene complexes from seven out of the 12 N. meningitidis serogroups have been identified previously. The complexes of the remaining five serogroups were sequenced in this study, and these sequences enabled the design of primers for further genogrouping. As far as we know, this study is the first to describe a molecular protocol for the detection and genogrouping of all 12 N. meningitidis serogroups.
In the multiplex PCR-based assay developed in this study, primers for ctrA and porA genes were combined to be used for N. meningitidis identification. Genes that encode glycosyltransferase, polysialyltransferase, polymerase, phosphotransferase, and d-arabinose-5-phosphate isomerase in the capsular gene complexes were used as the serogroup-specific genes, as these genes were responsible for differences in the CPSs between the serogroups. For serogroups W135 and Y, it was reported that the amino acid 310 in synF and synG was responsible for the different capsular forms of the two serogroups (5). A primer pair based on minor divergence between synF and synG sequences was used to differentiate serogroups W135 and Y. The antisense primer covered three consistently different positions (L324V, A325V, N327D) between the two genes, and the fragment amplified contained six of the nine consistently different positions (including G310P) of the two genes.
A total of 179 strains, which included the N. meningitidis reference strains, clinical isolates from different locations and years, those of closely related species, and those of species that can be detected in blood or CSF causing meningitis, were used to characterize the specificity of the assay. The results corresponded well with the findings by the traditional serology-based method. Some isolates previously classified as nongroupable were also able to be genogrouped by our assay. The total time to perform this assay was only 2 to 4 h, following bacterial isolation, and a PCR block is the only major equipment necessary. The accurate and efficient detection of a pathogenic microorganism is crucial for the prevention and effective treatment of disease, especially in the case of pathogen outbreaks. Therefore, this multiplex PCR-based assay has the advantage for applications in clinical diagnosis and epidemiological surveillance.
Although the detection sensitivity (4 × 105 genomes or 3 × 105 CFU/ml) is close to the level of N. meningitidis in most clinical CSF specimens (105 CFU/ml) (17), a better sensitivity will be helpful for the assay to be applied. It was reported that real-time PCR assays for N. meningitidis had a sensitivity less than 100 genomes (250 fq) (9, 18); therefore, real-time PCR assays for the detection and genogrouping of all N. meningitidis serogroups will be more applicable and can be developed in future studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Chinese Center for Disease Control and Prevention for kindly providing the bacteria of CMCC29031, -29044, and -29047.
This study was supported by grants from the National Natural Science Foundation of China (NSFC) Key Program (no. 31030002) and General Program (no. 30900255, 30788001, 81071392, 30870070, 31111120026), the National 973 program of China (no. 2009CB522603, 2011CB504900, 2012CB721001, and 2012CB721101), Tianjin Research Program of Application Foundation and Advanced Technology (no. 10JCYBJC10100), and the Fundamental Research Funds for the Central Universities (no. 65020121, 65020061).
Footnotes
Published ahead of print 16 November 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3398–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bateman A, et al. 2002. The Pfam Protein Families Database. Nucleic Acids Res. 30:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bennett DE, Cafferkey MT. 2006. Consecutive use of two multiplex PCR-based assays for simultaneous identification and determination of capsular status of nine common Neisseria meningitidis serogroups associated with invasive disease. J. Clin. Microbiol. 44:1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Borrow R, et al. 1998. siaD PCR ELISA for confirmation and identification of serogroup Y and W135 meningococcal infections. FEMS Microbiol. Lett. 159:209–214 [DOI] [PubMed] [Google Scholar]
- 5. Claus H, Stummeyer K, Batzilla J, Muhlenhoff M, Vogel U. 2009. Amino acid 310 determines the donor substrate specificity of serogroup W-135 and Y capsule polymerases of Neisseria meningitidis. Mol. Microbiol. 71:960–971 [DOI] [PubMed] [Google Scholar]
- 6. Claus H, Vogel U, Muhlenhoff M, Gerardy-Schahn R, Frosch M. 1997. Molecular divergence of the sia locus in different serogroups of Neisseria meningitidis expressing polysialic acid capsules. Mol. Gen. Genet. 257:28–34 [DOI] [PubMed] [Google Scholar]
- 7. Corless CE, et al. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dolan-Livengood JM, Miller YK, Martin LE, Urwin R, Stephens DS. 2003. Genetic basis for nongroupable Neisseria meningitidis. J. Infect. Dis. 187:1616–1628 [DOI] [PubMed] [Google Scholar]
- 9. Dolan Thomas J, et al. 2011. sodC-based real-time PCR for detection of Neisseria meningitidis. PLoS One 6:e19361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Drakopoulou Z, et al. 2008. Simultaneous single-tube PCR-based assay for the direct identification of the five most common meningococcal serogroups from clinical samples. FEMS Immunol. Med. Microbiol. 53:178–182 [DOI] [PubMed] [Google Scholar]
- 11. Grahlow WD, Kuzemenska P. 1989. Meningitis in a child due to Neisseria meningitidis group K. Infection 17:309–310 [DOI] [PubMed] [Google Scholar]
- 12. Guarner J, et al. 2004. Pathogenesis and diagnosis of human meningococcal disease using immunohistochemical and PCR assays. Am. J. Clin. Pathol. 122:754–764 [DOI] [PubMed] [Google Scholar]
- 13. Hammerschmidt S, et al. 1994. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol. Microbiol. 11:885–896 [DOI] [PubMed] [Google Scholar]
- 14. Hammerschmidt S, et al. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J. 15:192–198 [PMC free article] [PubMed] [Google Scholar]
- 15. Jordens JZ, Heckels JE. 2005. A novel porA-based real-time PCR for detection of meningococcal carriage. J. Med. Microbiol. 54:463–466 [DOI] [PubMed] [Google Scholar]
- 16. Jordens JZ, Williams JN, Jones GR, Heckels JE. 2002. Detection of meningococcal carriage by culture and PCR of throat swabs and mouth gargles. J. Clin. Microbiol. 40:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. La Scolea LJ, Jr, Dryja D. 1984. Quantitation of bacteria in cerebrospinal fluid and blood of children with meningitis and its diagnostic significance. J. Clin. Microbiol. 19:187–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mothershed EA, et al. 2004. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J. Clin. Microbiol. 42:320–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Petering H, et al. 1996. Genes associated with meningococcal capsule complex are also found in Neisseria gonorrhoeae. J. Bacteriol. 178:3342–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raskin DM, Seshadri R, Pukatzki SU, Mekalanos JJ. 2006. Bacterial genomics and pathogen evolution. Cell 124:703–714 [DOI] [PubMed] [Google Scholar]
- 21. Richardson AR, Yu Z, Popovic T, Stojiljkovic I. 2002. Mutator clones of Neisseria meningitidis in epidemic serogroup A disease. Proc. Natl. Acad. Sci. U. S. A. 99:6103–6107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roberts IS. 1996. The biochemistry and genetics of capsular polysaccharide production in Bacteria. Annu. Rev. Microbiol. 50:285–315 [DOI] [PubMed] [Google Scholar]
- 23. Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. 2001. Meningococcal disease. N. Engl. J. Med. 344:1378–1388 [DOI] [PubMed] [Google Scholar]
- 24. Rutherford K, et al. 2000. Artemis: sequence visualisation and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 25. Ryan NJ, Hogan GR. 1980. Severe meningococcal disease caused by serogroups X and Z. Am. J. Dis. Child. 134:1173. [DOI] [PubMed] [Google Scholar]
- 26. Schoen C, et al. 2008. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A. 105:3473–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Snyder LA, Davies JK, Ryan CS, Saunders NJ. 2005. Comparative overview of the genomic and genetic differences between the pathogenic Neisseria strains and species. Plasmid 54:191–218 [DOI] [PubMed] [Google Scholar]
- 28. Spinosa MR, et al. 2007. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun. 75:94–3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Staden R. 1996. The Staden sequence analysis package. Mol. Biotech. 5:233–241 [DOI] [PubMed] [Google Scholar]
- 30. Stollenwerk N, Maiden MC, Jansen VA. 2004. Diversity in pathogenicity can cause outbreaks of meningococcal disease. Proc. Natl. Acad. Sci. U. S. A. 101:10229–10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Swartley JS, Ahn JH, Liu LJ, Kahler CM, Stephens DS. 1996. Expression of sialic acid and polysialic acid in serogroup B Neisseria meningitidis: divergent transcription of biosynthesis and transport operons through a common promoter region. J. Bacteriol. 178:4052–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swartley JS, et al. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. U. S. A. 94:271–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Taha MK, et al. 2005. Interlaboratory comparison of PCR-based identification and genogrouping of Neisseria meningitidis. J. Clin. Microbiol. 43:144–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tatusov RL, et al. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tzeng YL, et al. 2005. Translocation and surface expression of lipidated serogroup B capsular polysaccharide in Neisseria meningitidis. Infect. Immun. 73:1491–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.