Abstract
We describe the first direct comparison of the reverse and traditional syphilis screening algorithms in a population with a low prevalence of syphilis. Among 1,000 patients tested, the results for 6 patients were falsely reactive by reverse screening, compared to none by traditional testing. However, reverse screening identified 2 patients with possible latent syphilis that were not detected by rapid plasma reagin (RPR).
TEXT
The diagnosis of syphilis is challenging and is typically made by serologic testing. Traditionally, syphilis screening has been performed using a nontreponemal test, such as rapid plasma reagin (RPR), with screen-reactive samples being tested by a treponemal assay (e.g., fluorescent treponemal antibody [FTA]) for confirmation. In recent years, many clinical laboratories have implemented a reverse screening algorithm, in which sera are screened using an automated, treponemal test (e.g., enzyme immunoassay [EIA]). Samples that are reactive by EIA are then tested by RPR to assess disease and treatment status and provide a supplemental marker of infection (1, 2, 6). The implementation of reverse screening has allowed clinical laboratories to meet increasing demands for syphilis testing by providing an automated screening test that yields an objective interpretation of results (4). However, the results of reverse screening often cause confusion and anxiety among health care providers and patients, especially when the results of EIA screening and RPR are discordant (e.g., reactive by EIA and non-reactive by RPR). Although these results typically reflect past, successfully treated syphilis, they may also occur in patients with (i) no syphilis (e.g., a falsely reactive EIA) or (ii) early or late/latent syphilis, when the sensitivity of RPR is low (2, 6).
To assist in the interpretation of discordant EIA and RPR results, the Centers for Disease Control and Prevention (CDC) recently recommended that sera testing reactive by EIA but nonreactive by RPR be analyzed by the Treponema pallidum particle agglutination (TP-PA) assay (2). A nonreactive TP-PA result would suggest that the results of EIA screening were falsely reactive, while a reactive TP-PA result would support an interpretation of either (i) past, successfully treated syphilis or (ii) late/latent syphilis (2).
To assess the potential impact of reverse screening on the diagnosis of syphilis, the CDC analyzed the results of 140,176 sera screened by a treponemal EIA or chemiluminescence immunoassay (CIA) (2). The samples were collected from patients living in areas with either low or high prevalence for syphilis. Among the 140,176 samples tested by EIA/CIA, 4,834 (3.4%) were reactive by the screening test, and of these screen-reactive sera, 2,743 (56.7%) were subsequently nonreactive by RPR. Importantly, these discordant samples (n = 2,743) were tested by TP-PA or FTA, which gave nonreactive results for 833 (31.6%) sera, suggesting a falsely reactive EIA/CIA screening result (2). These findings supported past work (3) suggesting that reverse screening may detect a higher rate of screen-reactive patients compared to traditional testing by RPR. In addition, these data raised concerns that reverse screening may yield an elevated percentage of falsely reactive results.
Although these findings are critical in assessing the performance of the reverse screening algorithm, an inherent limitation of these initial studies was the lack of parallel RPR screening on all samples. Therefore, it is impossible to conclude from these data whether reverse screening yielded a higher false-reactive rate than screening by the traditional screening algorithm. Because of these limitations, we sought to directly compare the reverse and traditional screening algorithms in a patient population with a low prevalence of syphilis.
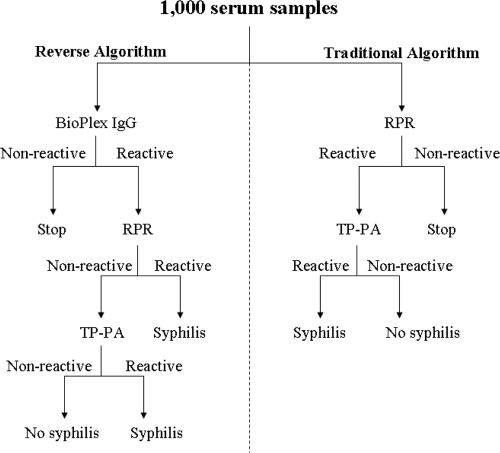
Prospectively collected sera (n = 1,000; one sample per patient) were submitted for routine syphilis testing in our laboratory, which consists of reverse screening using the BioPlex 2200 syphilis IgG multiplex flow immunoassay (MFI) (Bio-Rad Laboratories, Hercules, CA) (4). Samples testing reactive by the BioPlex assay were tested by RPR (Becton Dickinson, Franklin Lakes, NJ); if RPR gave a positive result, the titer of the serum sample was determined to an endpoint. In addition, sera testing reactive by the BioPlex were also analyzed by TP-PA (Fujirebio Diagnostics, Malvern, PA). In addition to the reverse screening algorithm, each sample was also screened by RPR, with the performing technologist unaware of the results of other testing. The titers of sera that were reactive by RPR were determined to an endpoint and subsequently tested by TP-PA (Fig. 1). All testing was performed according to the manufacturers' recommendations.
Fig 1.
Study design. One thousand prospective serum samples (one per patient) were tested by both the reverse and traditional syphilis screening algorithms in a blinded fashion. RPR, rapid plasma reagin; TP-PA, Treponema pallidum particle agglutination.
Among the 1,000 samples tested, 15 (1.5%) were reactive by reverse screening (BioPlex IgG) compared to 4 (0.4%) by the traditional screening test (RPR) (P = 0.01). The four samples that were reactive by RPR were confirmed to be positive by TP-PA; notably, these same four patients were also detected by the reverse screening algorithm (Table 1, patients 1 to 4). Patient 1 had underlying HIV infection and was subsequently diagnosed and treated for neurosyphilis based on a positive venereal disease research laboratory (VDRL) result from a cerebrospinal fluid (CSF) sample. Patients 2 to 4 each had a past diagnosis of syphilis, and sera were submitted to monitor their response to therapy. Each of these patients had a low RPR titer (reciprocal endpoint of 1) and were confirmed to be positive by TP-PA.
Table 1.
Clinical data and results of traditional and reverse syphilis screening algorithms for patients with a positive screening result (n = 15)
| Patient | Traditional algorithma |
Reverse algorithma |
Interpretationb | Reason for testingc | |||
|---|---|---|---|---|---|---|---|
| RPR (titer) | TP-PA | BioPlex | RPR (titer) | TP-PA | |||
| 1 | + (128) | + | + | + (128) | + | Active syphilis (treated) | HIV, neurosyphilis |
| 2 | + (1) | + | + | + (1) | + | Past, treated syphilis | Past syphilis |
| 3 | + (1) | + | + | + (1) | + | Past, treated syphilis | HIV, neurosyphilis |
| 4 | + (1) | + | + | + (1) | + | Past, treated syphilis | HIV, past syphilis |
| 5 | − | N/A | + | − | + | Past, treated syphilis | STD screen |
| 6 | − | N/A | + | − | + | Past, treated syphilis | Pretransplant exam |
| 7 | − | N/A | + | − | + | Past, treated syphilis | Penile discharge |
| 8 | − | N/A | + | − | + | Latent syphilis (treated) | Immigration exam |
| 9 | − | N/A | + | − | + | Latent syphilis (treated) | Pretransplant exam |
| 10 | − | N/A | + | − | − | Falsely reactive IgG | Cognitive disorder |
| 11 | − | N/A | + | − | − | Falsely reactive IgG | Cognitive disorder |
| 12 | − | N/A | + | − | − | Falsely reactive IgG | Cognitive disorder |
| 13 | − | N/A | + | − | − | Falsely reactive IgG | Pretransplant exam |
| 14 | − | N/A | + | − | − | Falsely reactive IgG | Urinary incontinence |
| 15 | − | N/A | + | − | − | Falsely reactive IgG | Vaginal discharge |
The results of traditional and reverse syphilis screening algorithms are shown. The tests are abbreviated as follows: RPR, rapid plasma reagin; TP-PA, Treponema pallidum particle agglutination; BioPlex, BioPlex syphilis IgG immunoassay. The test results are shown as follows: +, reactive; −, nonreactive; N/A, not applicable. If the rapid plasma reagin test gave a reactive result, the reciprocal endpoint titer is provided in parentheses.
Patients that were treated as a result of testing are indicated in parentheses. An interpretation of “Past, treated syphilis” was based on provider documentation or patient communication of past treatment for syphilis.
STD, sexually transmitted disease.
In addition to these 4 patients, the results of reverse screening were reactive in an additional 11 patients that were not detected by RPR. A review of each patient's medical records was performed to determine the reason for testing and the final clinical interpretation of results. Three patients (Table 1, patients 5 to 7) had a history of past, successfully treated syphilis and were not retreated based on these results. All three patients had a reactive TP-PA result, which supported the findings of the BioPlex IgG assay. Two additional patients (patients 8 and 9) were reactive by the BioPlex IgG assay and TP-PA but nonreactive by RPR. These patients were examined as a component of routine immigration or pretransplant evaluation and had no history of syphilis or treatment. Both patients were diagnosed with possible latent syphilis and were treated appropriately. Finally, 6 patients (patients 10 to 15) were reactive by the BioPlex IgG assay but nonreactive by RPR and TP-PA. These findings were interpreted as falsely reactive screening results based on an alternative diagnosis and/or negative TP-PA result, and these patients were not treated for syphilis (Table 1).
This is the first direct comparison of the reverse and traditional syphilis screening algorithms in a population with a low prevalence of syphilis. The results suggest that reverse screening yields a higher false-reactive rate than traditional testing does (0.6% versus 0.0%, respectively; P = 0.03). Interestingly, the overall false-reactive rate by reverse screening in our study (6/1,000 [0.6%]) was the same as the rate previously reported by the CDC (866/140,176 [0.6%]) (2). Despite the increased false-reactive rate, the reverse syphilis screening algorithm detected two patients with possible latent syphilis that went undetected by RPR screening. Whether or not these two patients had (i) latent, untreated syphilis, or (ii) past, resolved syphilis that had been treated unknowingly is impossible to determine. However, our findings support prior data suggesting that reverse screening may enhance the sensitivity for detection of early or late/latent disease (5). Taken together, these data underscore the importance of performing a second treponemal assay (e.g., TP-PA) when the results of reverse screening and RPR are discordant. Future studies should expand on these findings by directly comparing both algorithms in a population with a high prevalence of syphilis.
ACKNOWLEDGEMENT
We have no conflicts of interest.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Association of Public Health Laboratories 2009. Laboratory diagnostic testing for Treponema pallidum - expert consultation meeting summary report, January 13-15, 2009 Atlanta, GA Association of Public Health Laboratories, Silver Spring, MD: http://www.aphl.org/aphlprograms/infectious/std/Documents/LaboratoryGuidelinesTreponemapallidumMeetingReport.pdf [Google Scholar]
- 2. Centers for Disease Control and Prevention 2011. Discordant results from reverse sequence syphilis screening — five laboratories, United States, 2006–2010. MMWR Morb. Mortal. Wkly. Rep. 60:133–137 [PubMed] [Google Scholar]
- 3. Centers for Disease Control and Prevention 2008. Syphilis testing algorithms using treponemal tests for initial screening — four laboratories, New York City, 2005–2006. MMWR Morb. Mortal. Wkly. Rep. 57:872–875 [PubMed] [Google Scholar]
- 4. Gomez E, Jespersen DJ, Harring JA, Binnicker MJ. 2010. Evaluation of the Bio-Rad BioPlex 2200 syphilis multiplex flow immunoassay for the detection of IgM- and IgG-class antitreponemal antibodies. Clin. Vaccine Immunol. 17:966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mishra S, et al. 2011. The laboratory impact of changing syphilis screening from the rapid-plasma reagin to a treponemal enzyme immunoassay: a case-study from the Greater Toronto Area. Sex. Transm. Dis. 38:190–196 [DOI] [PubMed] [Google Scholar]
- 6. Sena AC, White BL, Sparling PF. 2010. Novel Treponema pallidum serologic tests: a paradigm shift in syphilis screening for the 21st century. Clin. Infect. Dis. 51:700–708 [DOI] [PubMed] [Google Scholar]