Abstract
We assessed the performance of a recently validated real-time PCR assay and a commercially available microimmunofluorescence serologic test for the detection of Chlamydophila pneumoniae infection during an outbreak. Evaluation of specimens from 137 individuals suggests that real-time PCR holds greater utility as a diagnostic tool for early C. pneumoniae detection.
TEXT
Chlamydophila pneumoniae is one of the leading causes of community-acquired respiratory tract infections, accounting for approximately 10% of community-acquired pneumonia (CAP) cases and 5% of bronchitis and sinusitis cases in adults and children (18). Most respiratory infections caused by C. pneumoniae are asymptomatic or mild, although severe pneumonia can develop in elderly patients and those with coexisting cardiopulmonary diseases (2). Seroepidemiological studies have shown an antibody prevalence of 50 to 70%, suggesting a high frequency of previous infections, although these data may be misleading for reasons discussed below (12). Nearly all humans can expect to be infected with C. pneumoniae at least once during their lifetimes. Reinfections are common, and persistence of the agent in the host after primary infection is a potential risk for chronic infection (8, 9).
Reliable diagnosis of C. pneumoniae infection remains difficult due to the lack of standardized and commercially available diagnostic tests that are both sensitive and specific. Laboratory methods currently used for the diagnosis of acute C. pneumoniae infection include culture, immunohistochemical assays, serology, and PCR; the latter two are the most often applied (13). Although infections with C. pneumoniae can be identified by direct isolation of the agent, this procedure is laborious and time-consuming and often yields inconsistent results (15). The microimmunofluorescence (MIF) test is currently considered the “gold standard” for the serodiagnosis of C. pneumoniae infection, even though results are often subjective and require specialized training for interpretation (20). In addition, technical complexity, subjective endpoints, and the lack of standardized reagents result in significant intra- and interlaboratory variations in test performance (11). Moreover, the requirement of paired serum samples and the extended persistence of IgG antibody in some adult populations make this test retrospective in nature and unsuitable for timely diagnosis (6). MIF testing is further hampered by poor specificity due to cross-reaction with other chlamydial species and is unable to discriminate between past and persistent infections (3, 7, 17). Molecular analysis-based assays, such as real-time PCR, have recently been developed for the rapid and sensitive detection of C. pneumoniae (14, 22, 23). The overall diagnostic utility of PCR-based assays is currently unknown due to a lack of specimen type, nucleic acid extraction method, and amplification protocol standardization and the unavailability of reliable commercial assays (2).
Outbreaks of C. pneumoniae occur in individuals living in surroundings where they are close to others, such as schools and military barracks (4, 16, 21). C. pneumoniae can also play a significant role in coinfections, as seen in a recent CAP outbreak due to Streptococcus pneumoniae on a military base (5). Recently, a C. pneumoniae CAP outbreak occurred in a prison with male inmates within the southern United States. A validated multiplex real-time PCR assay was used to identify C. pneumoniae as the causative agent of the CAP outbreak (22). After initial real-time PCR testing, a multipathogen PCR-based molecular detection assay was used to rule out coinfection with other pathogens (data not shown) (10). The aim of the present study was to evaluate the performance of a recently validated multiplex real-time PCR assay and a commercially available MIF serologic detection kit for their reliability in detecting C. pneumoniae during this recent outbreak.
Case subjects were defined as those having a fever (temperature of ≥38°C) and a cough that persisted for >3 days or clinical and/or X-ray-confirmed pneumonia. Asymptomatic individuals from shared prison facilities were randomly selected and assigned to a noncase group. Oropharyngeal (OP) and/or nasopharyngeal (NP) swab specimens and serum samples were obtained from symptomatic individuals (n = 38) and from individuals with no reported illness or symptoms (n = 99). Swabs were placed in 2 ml of Universal Transport Medium (BD) and transported frozen or cold (4 to 10°C) for molecular testing. Single serum samples were collected from each individual and transported frozen. Only individuals who provided both a swab and a serum sample were included in this study.
Total nucleic acid extraction was performed on all NP/OP swab specimens following manufacturer's instructions for the Total NA Serum_Plasma_Blood protocol with a 200-μl sample volume and a 100-μl elution volume using MagNa Pure Compact nucleic acid isolation kit I (Roche Applied Science). A validated multiplex real-time PCR assay that tests for C. pneumoniae (CP-arg) along with Mycoplasma pneumoniae (MP181) and Legionella spp. (ssrA) was performed in duplicate on all extracted specimens as previously described (22). Serum samples were tested for reactivity to C. pneumoniae using the commercially available Chlamydia MIF IgG and IgM kits (FOCUS Diagnostics) following the manufacturer's instructions. As indicated by the manufacturer, a probable acute infection was defined by a single serum sample with an IgM titer of ≥1:10 and/or an IgG titer of ≥1:512. The diagnostic sensitivity, specificity, and positive predictive value were calculated for each of these laboratory tests using clinically defined cases as the gold standard (19).
A total of 137 individuals (38 case and 99 noncase subjects) were tested for the presence of C. pneumoniae infection by real-time PCR and by MIF. Table 1 summarizes the sensitivity, specificity, and positive and negative predictive values of each diagnostic assay. Twenty-seven case patients (71%) had positive real-time PCR results, and 37 (97%) had positive results with the IgM + IgG MIF assay. The sensitivity and specificity of the IgM MIF assay were 60% and 77%, respectively, while the IgG MIF had a sensitivity of 82% and a specificity of 40%. While the IgM + IgG MIF assay appeared to be the most sensitive method studied, it had low specificity (30%). The sensitivity (71%) of the real-time PCR assay was lower than that of the IgM + IgG MIF assay (97%), but its specificity (97%) was much higher. Real-time PCR also had a much higher positive predictive value (90%) than the IgM + IgG MIF assay (35%). Table 2 summarizes the isotype distribution of PCR-positive case and noncase individuals. Twenty-seven case individuals were positive by PCR. Of these individuals, 2 (8%), 6 (22%), and 19 (70%) were positive for IgM only, IgG only, and IgM and IgG, respectively. Three individuals from the noncase group had positive PCR results. All three PCR-positive individuals also yielded positive MIF results (2 IgG only and 1 containing both IgM and IgG), suggesting an active but asymptomatic infection for these three individuals.
Table 1.
Diagnostic sensitivities and specificities of real-time PCR and MIF assays for 38 case patients and 99 noncase individualsa
| No. of case subjects | Sensitivity (%) |
No. of noncase subjects | Specificity (%) |
||||||
|---|---|---|---|---|---|---|---|---|---|
| PCR | IgM MIF | IgG MIF | IgM + IgG MIF | PCR | IgM MIF | IgG MIF | IgM + IgG MIF | ||
| 38 | 71 (55–83) | 60 (43–75) | 82 (65–92) | 97 (87–100) | 99 | 97 (91–99) | 77 (67–84) | 40 (30–51) | 30 (22–40) |
Data are percentages (95% confidence intervals). The positive and negative predictive values of the assays are as follows: PCR, 90 and 90%; IgM + IgG MIF, 35 and 97%; IgM MIF, 50 and 82%; IgG MIF, 34 and 85%.
Table 2.
Isotype distribution of real-time PCR-positive case and noncase individuals
| Group | Total no. PCR positive | No. (%) PCR positive for: |
||
|---|---|---|---|---|
| IgM only | IgG only | IgM + IgG | ||
| Case | 27 | 2 (8) | 6 (22) | 19 (70) |
| Noncase | 3 | 0 | 2 (67) | 1 (33) |
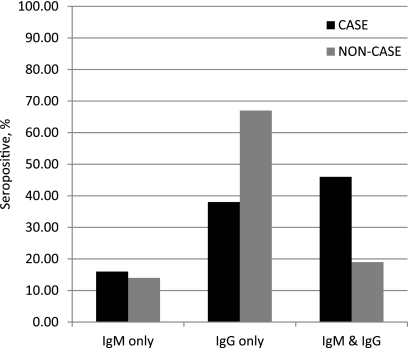
Figure 1 illustrates the isotype distribution among seropositive individuals from both the case and noncase groups that met the required titer indicative of a probable acute infection, as indicated by the manufacturer. Sixty-two percent of seropositive case individuals had IgM only (16%) or both IgG and IgM (46%) isotypes present, suggesting a primary infection, while 38% had IgG only, suggesting the presence of a recurring infection. In contrast, 67% of seropositive noncase individuals had only IgG present. This observation is consistent with the reported high prevalence of persistent IgG titers in some adults and highlights a significant limitation of the MIF assay. The remaining 33% of the seropositive noncase individuals comprised 14% positive for IgM only and 19% positive for both IgG and IgM, suggesting either background seropositivity or cross-reactivity, although a possible asymptomatic infection could be present as well.
Fig 1.
Isotype distribution of 37/38 case patients and 69/99 noncase individuals with positive serologic results as determined by the MIF assay. Seropositivity is defined as an IgM titer ≥1:10 and/or an IgG titer ≥1:512.
In this particular outbreak, serology testing alone would likely have been misleading since the rate of seropositivity among noncase individuals was extremely high. Most studies recommend testing of paired serum samples. However, in an outbreak setting, only a single serum sample would be available for timely diagnostic testing and was therefore used for comparison in the present study. This limitation was minimized by testing serum specimens collected from the noncase group in order to establish the background seropositivity. Although real-time PCR is subject to some limitations as well, it has demonstrated greater utility for rapid and accurate detection of etiologies, especially in outbreak settings (1). Significant challenges continue to confound the accurate and dependable diagnosis of C. pneumoniae infection. Collectively, our analysis of these two commonly used tests for C. pneumoniae detection suggests that real-time PCR is a more useful diagnostic tool to confirm the etiology of C. pneumoniae in an outbreak setting.
ACKNOWLEDGMENTS
The opinions expressed in this report are ours and do not necessarily represent the position of the Federal Bureau of Prisons or the U.S. Department of Justice.
Footnotes
Published ahead of print 26 October 2011
REFERENCES
- 1. Al-Mohri HA, Tadros MA, Louie L, Vearncombe M, Simor AE. 2008. Utility of direct, real-time PCR in the management of a nosocomial outbreak of vancomycin-resistant Enterococcus faecium (vanB genotype). Eur. J. Clin. Microbiol. Infect. Dis. 27:321–322 [DOI] [PubMed] [Google Scholar]
- 2. Blasi F, Tarsia P, Aliberti S. 2009. Chlamydophila pneumoniae. Clin. Microbiol. Infect. 15:29–35 [DOI] [PubMed] [Google Scholar]
- 3. Bourke SJ, Carrington D, Frew CE, Stevenson RD, Banham SW. 1989. Serological cross-reactivity among chlamydial strains in a family outbreak of psittacosis. J. Infect. 19:41–45 [DOI] [PubMed] [Google Scholar]
- 4. Csángó PA, Haraldstad S, Pedersen JE, Jagars G, Foreland I. 1997. Respiratory tract infection due to Chlamydia pneumoniae in military personnel. Scand. J. Infect. Dis. Suppl. 104:26–29 [PubMed] [Google Scholar]
- 5. Dawood FS, et al. 2011. Outbreak of pneumonia in the setting of fatal pneumococcal meningitis among US Army trainees: potential role of Chlamydia pneumoniae infection. BMC Infect. Dis. 11:157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dowell SF, et al. 2001. Standardizing Chlamydia pneumoniae assays: recommendations from the Centers for Disease Control and Prevention (USA) and the Laboratory Centre for Disease Control (Canada). Clin. Infect. Dis. 33:492–503 [DOI] [PubMed] [Google Scholar]
- 7. Gnarpe J, Naas J, Lundback A. 2000. Comparison of a new commercial EIA kit and the microimmunofluorescence technique for the determination of IgG and IgA antibodies to Chlamydia pneumoniae. APMIS 108:819–824 [DOI] [PubMed] [Google Scholar]
- 8. Grayston JT. 2000. Background and current knowledge of Chlamydia pneumoniae and atherosclerosis. J. Infect. Dis. 181(Suppl 3):S402–S410 [DOI] [PubMed] [Google Scholar]
- 9. Hammerschlag MR. 2002. The intracellular life of chlamydiae. Semin. Pediatr. Infect. Dis. 13:239–248 [DOI] [PubMed] [Google Scholar]
- 10. Kodani M, et al. 2011. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J. Clin. Microbiol. 49:2175–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar S, Hammerschlag MR. 2007. Acute respiratory infection due to Chlamydia pneumoniae: current status of diagnostic methods. Clin. Infect. Dis. 44:568–576 [DOI] [PubMed] [Google Scholar]
- 12. Kuo CC, Jackson LA, Campbell LA, Grayston JT. 1995. Chlamydia pneumoniae (TWAR). Clin. Microbiol. Rev. 8:451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marston EL, et al. 2002. Newly characterized species-specific immunogenic Chlamydophila pneumoniae peptide reactive with murine monoclonal and human serum antibodies. Clin. Diagn. Lab. Immunol. 9:446–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mitchell SL, Budhiraja S, Thurman KA, Lanier TW, Winchell JM. 2009. Evaluation of two real-time PCR chemistries for the detection of Chlamydophila pneumoniae in clinical specimens. Mol. Cell. Probes 23:309–311 [DOI] [PubMed] [Google Scholar]
- 15. Mygind T, Birkelund S, Falk E, Christiansen G. 2001. Evaluation of real-time quantitative PCR for identification and quantification of Chlamydia pneumoniae by comparison with immunohistochemistry. J. Microbiol. Methods 46:241–251 [DOI] [PubMed] [Google Scholar]
- 16. Oktem IM, et al. 2007. PCR and serology were effective for identifying Chlamydophila pneumoniae in a lower respiratory infection outbreak among military recruits. Jpn. J. Infect. Dis. 60:97–101 [PubMed] [Google Scholar]
- 17. Ozanne G, Lefebvre J. 1992. Specificity of the microimmunofluorescence assay for the serodiagnosis of Chlamydia pneumoniae infections. Can. J. Microbiol. 38:1185–1189 [DOI] [PubMed] [Google Scholar]
- 18. Park SH, et al. 2009. Identification of immunogenic antigen candidate for Chlamydophila pneumoniae diagnosis. J. Proteome Res. 8:2933–2943 [DOI] [PubMed] [Google Scholar]
- 19. Pepe MS. 2003. The statistical evaluation of medical tests for classification and prediction, p. 33–65 Oxford University Press, Inc., New York, NY [Google Scholar]
- 20. Persson K, Boman J. 2000. Comparison of five serologic tests for diagnosis of acute infections by Chlamydia pneumoniae. Clin. Diagn. Lab. Immunol. 7:739–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt SM, Muller CE, Mahner B, Wiersbitzky SK. 2002. Prevalence, rate of persistence and respiratory tract symptoms of Chlamydia pneumoniae infection in 1211 kindergarten and school age children. Pediatr. Infect. Dis. J. 21:758–762 [DOI] [PubMed] [Google Scholar]
- 22. Thurman KA, Warner AK, Cowart KC, Benitez AJ, Winchell JM. 2011. Detection of Mycoplasma pneumoniae, Chlamydia pneumoniae, and Legionella spp. in clinical specimens using a single-tube multiplex real-time PCR assay. Diagn. Microbiol. Infect. Dis. 70:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Welti M, et al. 2003. Development of a multiplex real-time quantitative PCR assay to detect Chlamydia pneumoniae, Legionella pneumophila and Mycoplasma pneumoniae in respiratory tract secretions. Diagn. Microbiol. Infect. Dis. 45:85–95 [DOI] [PubMed] [Google Scholar]