Abstract
Continuing outbreaks of H5N1 highly pathogenic (HP) avian influenza virus (AIV) infections of wild birds and poultry worldwide emphasize the need for global surveillance of wild birds. To support the future surveillance activities, we developed a SYBR green-based, real-time reverse transcriptase PCR (rRT-PCR) for detecting nucleoprotein (NP) genes and subtyping 16 hemagglutinin (HA) and 9 neuraminidase (NA) genes simultaneously. Primers were improved by focusing on Eurasian or North American lineage genes; the number of mixed-base positions per primer was set to five or fewer, and the concentration of each primer set was optimized empirically. Also, 30 cycles of amplification of 1:10 dilutions of cDNAs from cultured viruses effectively reduced minor cross- or nonspecific reactions. Under these conditions, 346 HA and 345 NA genes of 349 AIVs were detected, with average sensitivities of NP, HA, and NA genes of 101.5, 102.3, and 103.1 50% egg infective doses, respectively. Utility of rRT-PCR for subtyping AIVs was compared with that of current standard serological tests by using 104 recent migratory duck virus isolates. As a result, all HA genes and 99% of the NA genes were genetically subtyped, while only 45% of HA genes and 74% of NA genes were serologically subtyped. Additionally, direct subtyping of AIVs in fecal samples was possible by 40 cycles of amplification: approximately 70% of HA and NA genes of NP gene-positive samples were successfully subtyped. This validation study indicates that rRT-PCR with optimized primers and reaction conditions is a powerful tool for subtyping varied AIVs in clinical and cultured samples.
INTRODUCTION
The avian influenza virus (AIV) is a negative-sense, segmented RNA virus and belongs to the family Orthomyxoviridae. AIV has two spike proteins on the virus particle, hemagglutinin (HA) and neuraminidase (NA), which are antigenically divided into 16 (H1 to H16) and 9 (N1 to N9) subtypes, respectively. Each subtype is highly diverse genetically and has unique lineages that are partitioned geographically between global hemispheres (13, 24). All subtypes are harbored in natural reservoir hosts, predominantly within the avian orders of Anseriformes (ducks, geese, and swans) and Charadriiformes (shorebirds, gulls, terns, and auks) (24). Additionally, AIVs are divided into two pathotypes, low pathogenicity (LP) and high pathogenicity (HP), in relation to the severity of disease responses in chickens (18, 24). Because certain subtypes, such as H5 and H7, are more often HP in chickens, rapid and accurate subtyping and pathotyping of AIVs are essential for the diagnosis and surveillance of AIVs.
The hemagglutination inhibition (HI) and neuraminidase inhibition (NI) tests are the current standard protocol for HA and NA subtyping, respectively (24, 29). Although these tests are used worldwide, supply of the reference antisera has limitation because antisera for both North American and Eurasian lineage viruses are required for all HA or NA subtypes, and extensive laboratory support for the broad coverage is needed. Molecular subtyping tests are alternative tests to antiserum-based protocols for subtyping AIVs. Although the microarray technology (5, 11) was first developed for simultaneously subtyping HA and NA genes, the technology requires expensive equipment and specific training and is time-consuming. In contrast, conventional reverse transcriptase PCR (RT-PCR) (3, 10, 19, 25, 26) and real-time RT-PCR (rRT-PCR) (14, 22, 27) are rapid and sensitive and easily applicable to general diagnostic laboratories. Primers containing mixed bases are indispensable for broad detection of genetically diverse HA and NA genes of AIVs and are also useful for sequencing and molecular characterization. H5 and H7 subtyping products contain a molecular marker for pathogenicity in chickens (6, 25, 27), while the NA subtyping products have a molecular marker for adaptation to terrestrial poultry (26). Thus, these molecular subtyping tests have high potential to be an alternative to the standard serological tests. Understanding any limitations of these molecular subtyping tests is required before wide-scale application.
Our extensive study on primer design for H5 and H7 subtypes clarifies some critical points for broad coverage of diverse HA genes (27). Numbers of mixed bases should be less than five positions in each primer, and two primer sets for Eurasian and North American lineages are required for some subtypes. Considering the huge diversity of HA and NA genes in natural ecosystems, it may be required to reevaluate all primer sets reported previously (25, 26) based on these recent findings. Also, to promote the practical use of the molecular subtyping test, comparison of the molecular test with serological subtyping tests is necessary.
Objectives of this study are to determine the specificity, sensitivity, and limitations of a molecular subtyping test and to compare the usefulness of this method with current standard HI and NI tests. To achieve these objectives, we developed a SYBR green-based rRT-PCR test for simultaneous identification of all 16 HA and 9 NA AIV subtypes and the primers and reaction protocol reported previously (25, 26) were comprehensively optimized based on the guideline developed in this study. This validation study, using over 500 AIVs, demonstrated that (i) preferential amplification of subtype-specific HA and NA genes and simultaneous suppression of nonspecific or cross-amplification can be achieved by 30 cycles of amplification of a 1:10 dilution of cDNAs, (ii) genetically diverse HA and NA genes could be detected by using regional primer sets for Eurasian or North American lineage genes, (iii) the program of 40 cycles of amplification allows us to directly subtype AIVs in fecal samples of wild birds, and (iv) the rRT-PCR protocol for subtyping AIVs is superior to the standard HI and NI tests in specificity, sensitivity, and rapidity. These guidelines for primer design and optimized reaction conditions will be useful for the development of rRT-PCR for subtyping AIVs circulating regionally in wild birds.
MATERIALS AND METHODS
Viruses.
A total of 508 avian influenza virus strains were used in this study and were classified into two experimental groups: gene detection and subtyping tests; 349 viruses were used in the gene detection test to determine whether each subtyping primer set was useful for the detection of homologous subtype HA or NA genes, and 159 viruses were utilized in the subtyping test to determine whether full primer sets of H1 to H16 and N1 to N9 genes were useful for subtyping HA and NA genes. Numbers of viruses and HA and NA genes of Eurasian and North American lineages are shown in Table 1. Lineages of HA and NA genes were determined by sequencing and phylogenic analysis of amplified genes. Reference H1 to H16 HA genes and N1 to N9 NA genes of Eurasian and North American lineages, which were chosen from recent duck viruses, and their GenBank accession numbers are shown in Table S4 in the supplemental material.
Table 1.
Numbers and lineages of HA and NA genes of avian influenza viruses used for a gene detection test and a subtyping test
| Test | No. of viruses | No. of: |
|||
|---|---|---|---|---|---|
| HA genes |
NA genes |
||||
| Eurasia | North America | Eurasia | North America | ||
| Gene detection | 349 | 269 | 80 | 236 | 113 |
| Subtyping | 159 | ||||
| Molecular + serological | 104 | 86 | 18 | 98 | 6 |
| Molecular only | 55 | 52 | 3 | 54 | 1 |
| Total | 508 | 407 | 101 | 388 | 120 |
The 349 viruses in the gene detection test included reference and wild duck strains and were obtained from Japan (187 strains), South Korea (70 strains), other Asian countries (11 strains), Australia (2 strains), Europe (10 strains), Africa (1 strain), North America (67 strains), and South America (1 strain). The 349 HA genes contained 269 Eurasian and 80 North American lineage genes (22 H1, 17 H2, 56 H3, 31 H4, 51 H5, 36 H6, 31 H7, 5 H8, 20 H9, 22 H10, 40 H11, 14 H12, 1 H13, 1 H14, 1 H15, and 1 H16). The 349 NA genes included 236 Eurasian and 113 North American lineage genes (36 N1, 81 N2, 55 N3, 11 N4, 20 N5, 44 N6, 14 N7, 54 N8, and 34 N9). For the South Korean strains, cDNAs were prepared in NVRQS in South Korea and rRT-PCR was performed in NIAH in Japan, while cDNAs of Alaskan strains were prepared and tested in Alaska by conventional RT-PCR with the same program as for rRT-PCR described below.
The 159 wild duck-derived AIVs in the subtyping test were composed of 104 viruses isolated in Japan in the 2007/2008 and 2008/2009 wintering seasons (October to March) and 55 viruses in the 2009/2010 wintering season. The 104 viruses were also subtyped serologically by standard tests to compare the usefulness of the molecular subtyping test with the serological ones.
AIVs were propagated in 9- to 11-day-old embryonated chicken eggs for 2 to 4 days, and the allantoic fluids were harvested and stored at −80°C before use. The 50% egg infectious doses (EID50) of AIVs were determined by inoculation of serial 10-fold dilutions of viruses into embryonated chicken eggs (23) and estimated by the method of Reed and Muench (21).
Guidelines for primer design and reaction conditions.
All of the primers for the 16 HA subtypes, 9 NA subtypes, and the NP reported previously (25–27) were reevaluated or newly designed using sequences obtained from publically available databases (Influenza Virus Resource, http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) and those sequenced in our laboratory. The primers were updated using the following guidelines. (i) The primary target genes of the rRT-PCR included both Eurasian and North American hemispheric lineages of each HA and NA subtype gene; however, we focused on Eurasian or North American lineage genes for some subtypes if primers were expected not to cover both lineages. (ii) The number of mixed base positions in each primer was not allowed to exceed five positions as a general rule. (iii) G+C numbers in each primer were less than 8 bases. (iv) The concentration of each primer was optimized experimentally by using several genes in each subtype. (v) A 10-fold dilution of cDNA from cultivated viruses was used. (vi) Thirty cycles of amplification were used for the detection of subtype-specific genes. In addition, a protocol for direct subtyping of AIVs in clinical samples is shown below.
RNA purification and cDNA synthesis.
Viral RNA was extracted from allantoic fluids using a Qiagen viral RNA minikit according to the manufacturer's instructions. Viral RNA was transcribed to cDNA with a PrimeScript reverse transcriptase reagent kit and 6-mer random primers (RR037; Takara, Japan); 15 min of incubation at 37°C was followed by 85°C for 5 s.
rRT-PCR.
The real-time PCR was performed with SYBR Premix Ex Taq (RR041; Takara) and a real-time PCR system (TP800; Takara). The reaction volume was 20 μl, which contained 1 μl of 10-fold dilution of cDNA, 10 μl of SYBR Premix Ex Taq, 2 μl each of forward and reverse primers (5 to 20 pmol each/μl), 0.2 μl of Dye II (×50), and 4.8 μl of water. NP, HA, and NA genes were amplified with the following protocol: 95°C for 30 s and then 30 cycles of 95°C for 30 s, 50°C for 20 s, and 72°C for 32 s, followed by a dissociation cycle (95°C for 15 s, 60°C for 60 s, and 95°C for 15 s). For subtyping of cultured AIVs, 1:10 dilutions of cDNAs were used. Sharp dissociation curves with fluorescence (F) counts of 0 to 199, 200 to 499, and over 500 were considered negative, weak-positive, and positive results, respectively.
Comparison of sensitivities.
To determine the infectivity of AIVs (allantoic fluids) (EID50/0.1 ml), serial 10-fold dilutions of AIVs (18 strains) belonging to H1 to H16 or N1 to N9 subtypes were inoculated into embryonated chicken eggs. Four days after cultivation, the allantoic fluids were tested for the presence of HA activity and the virus infectivity titers were estimated as EID50/0.1 ml by the method of Reed and Muench (21). On the egg inoculation day, the same dilutions were used to prepare viral RNA, followed by cDNA synthesis as described above. The cDNA of each virus dilution was tested for the presence of NP, HA, and NA genes by rRT-PCR in duplicate wells, and the 50% rRT-PCR titers of NP, HA, and NA genes of each virus were estimated as above (21).
Subtyping test of cultured AIVs.
Applicability of the rRT-PCR for subtyping HA and NA genes of AIVs was evaluated by using 104 AIVs (allantoic fluids) isolated from fecal materials of migratory ducks sampled in Ibaraki, Chiba, and Shiga Prefectures in two wintering seasons of October to March in 2007/2008 and 2008/2009. In addition, 55 viruses isolated in 2009/2010 were also used for subtyping genetically. Viral RNA purification, cDNA synthesis, and genetic subtyping of AIVs by rRT-PCR were performed as described above. Some of the amplified HA and NA gene products were sequenced with the same primers as those for rRT-PCR, and criteria for determining negative, weak-positive, and positive samples were the same as described above. Subtype specificities of amplified genes were determined by a BLAST search.
Direct subtyping test of AIVs in fecal samples.
Availability of rRT-PCR for direct subtyping of AIVs in clinical samples was evaluated using 120 fresh fecal samples collected from migratory ducks in October to November 2009 in Ibaraki, Chiba, and Shiga Prefectures. Ten percent fecal sample supernatants were prepared with phosphate-buffered saline (PBS), filtrated with a 0.45-μm membrane, and used for RNA purification followed by cDNA synthesis. The cDNAs were screened for the presence of the NP gene by rRT-PCR, and the NP gene-positive cDNAs were used for subtyping AIVs genetically. A protocol of 40 cycles of amplification of undiluted cDNA was used for direct detection of NP genes and direct subtyping of AIVs in clinical samples. To confirm the consistency of the results of the direct subtyping test, filtrated fecal samples were inoculated into chicken embryonated eggs on the sampling day and the isolated AIVs were subtyped genetically. Sensitivity and specificity of the NP assay were compared with those for the standard virus isolation test as follows: specificity is the number of true negatives/(number of true negatives + number of false positives), and sensitivity is the number of true positives/(number of true positives + number of false negatives).
Serological subtyping test.
The 104 AIVs were subtyped serologically for HA by the HI test as described previously (17) with serial 2-fold dilutions (1:20 to 1:2,560) of 29 reference antisera (see Table S1 in the supplemental material). Three volumes of reference antiserum were mixed with 1 volume of receptor-destroying enzyme (RDE) (Denkaseiken, Niigata, Japan), and the mixture was incubated for 18 h at 37°C. The RDE-treated antisera were diluted to 1:20 with PBS and absorbed with packed chicken red blood cells (CRBC) for 1 h at room temperature, and the absorbed sera were used. The HI antibody titers of the 29 reference antisera to the original AIVs were between 1:640 and 1:2,560. HI antibody titers between 1:20 and 1:80 or more than 1:160 were considered weakly positive or positive, respectively. A virus exhibiting a positive reaction to one HA subtype was classified as “subtyped,“ and a virus indicating positive reactions to two or more HA subtypes was classified as”ambiguous.” A virus exhibiting no reaction to the genetically determined HA subtype was considered “not subtyped.”
The 104 AIVs were also subtyped serologically for NA by the USDA-validated micro-NI test (16). The 1:20 dilutions of nine reference antisera (see Table S1 in the supplemental material) were used after pretreatment with RDE and absorption with CRBC. The antisera had NI antibody titers from 1:800 to 1:6,400 to the original virus strains. A virus sample that showed light or no color in response to antiserum was considered positive. Criteria for subtyped, ambiguous, or not subtyped by the micro-NI test are the same as those for the HI test.
RESULTS
Primer design and establishment of rRT-PCR.
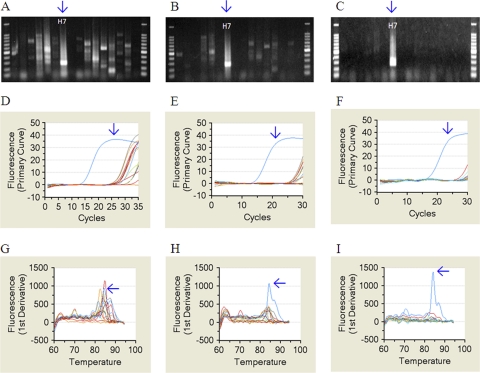
A SYBR green-based rRT-PCR helped us to precisely assess the specificity of primers, based on the shape of the dissociation curve, the temperature, and the height of the dissociation peak, as well as cycle number of amplification plot. After this evaluation of all primers reported previously (25, 26) or designed in this study, new primers (25 HA and 3 NA), including those for the H16 subtype, were updated or developed (Table 2). The updated or newly developed primers contained mixed bases at no more than 5 positions in principle, although 5 primers still contained 6 or 7 mixed-base positions. Although all primers containing mixed bases at more than 6 positions were redesigned and those with fewer than 5 mixed-base positions were tested for broad coverage, these new primers exhibited less sensitivity than primers containing mixed bases at 6 or 7 positions listed in Table 2. Also, both Eurasian and North American primer sets for divergent genes were needed for H1, H6, H7, H9, H10, H12, N8, and N9 subtypes (Table 2). Although previous universal primers were useful for broad detection of these genes, the amplified products were of lower yield than those detected with regional primer sets. Use of regional primer sets was required for preferential amplification of target genes within 30 cycles of amplification. In addition, previous research showed that primer concentration is critical to maintain the high sensitivity of the rRT-PCR (27). Therefore, the concentration of each primer set was determined experimentally using several genetically different genes for each subtype. As a result, optimal primer concentrations were determined to be between 0.5 μM and 2.0 μM (Table 2). Third, use of 30 cycles of amplification of 1:10 dilutions of cDNAs from cultured AIVs in allantoic fluids markedly increased the specificity. For example, many nonspecific or cross-amplification products were observed in A/turkey/Italy/4580/1999 (H7N1) when undiluted cDNA was amplified by 35 cycles (Fig. 1A); the amplification curve indicates that these unexpected products appeared slowly after 30-cycle amplification (Fig. 1D), and after 35-cycle amplification, these products were indistinguishable from the specific product in the dissociation curve (Fig. 1G). On the other hand, these nonspecific products (Fig. 1A, D, and G) could be eliminated dramatically by 30 cycles of amplification (Fig. 1B, E, and H), especially when a 10-fold dilution of the cDNA was used (Fig. 1C, F, and I). Thus, using these guidelines, sensitivity and specificity of the rRT-PCR were substantially improved.
Table 2.
Primers used for detecting NP genes and subtyping HA and NA genes of AIVs by rRT-PCR
| Subtype | Lineagea | Primer identification | Position on genec | Accession no. | Size (bp) | Sequenced | No. of mixed base positions | Final primer concn (pmol/μl) | Dissociation temp (°C) |
|---|---|---|---|---|---|---|---|---|---|
| H1 | Eu | 845b | H1-887Feu | AF091309 | 305 | GATGCTCCRGTYCAYAATT | 3 | 2 | 83–84 |
| 847b | H1-1191Reu | TTTGTGTGCTYTTYTGAT | 2 | ||||||
| Am | 849b | H1-887Fam | AF091309 | 305 | GATRCWCCAGTYCAYAAYT | 5 | 2 | 83–84 | |
| 850b | H1-1191Ram | TCTGYGTGCTYTTYTGRT | 4 | ||||||
| H2 | Eu + Am | 479 | H2-892F | L11127 | 343 | AAMMCTTGAAAAYTGYGA | 4 | 2 | 83–84 |
| 682b | H2-1234R | TTCAATYACRGASTTYACYTTG | 5 | ||||||
| H3 | Eu + Am | 486 | H3-919F | CY006031 | 376 | GYATYACTCCWAATGGAAGC | 3 | 0.5 | 83–84 |
| 683b | H3-1294R | ATYCTYCCTTCYACTTCDGM | 5 | ||||||
| H4 | Eu + Am | 883b | H4-896F | M25283 | 284 | AGYAARTGYCAYACHGACA | 5 | 1 | 84–85 |
| 738b | H4-1179R | GCTGCCTGRGTVGAYTTKAG | 4 | ||||||
| H5 | Eu + Am | 491 | H5-918F | U69277 | 261 | CCARTRGGKGCKATAAAYTC | 5 | 2 | 83–85 |
| 820b | H5-1178R | GTCTGCAGCRTAYCCACTYC | 3 | ||||||
| H6 | Eu | 511 | H6-870F | CY005691 | 387 | TCARAYYTDCCAATHGAGAA | 5 | 2 | 83 |
| 855b | H6-1256Reu | TCGTGRYCKACAGCYTCRA | 5 | ||||||
| Am | 511 | H6-870F | CY005691 | 387 | TCARAYYTDCCAATHGAGAA | 5 | 3 | 83 | |
| 902b | H6-1256Ram | TCRTGSTCKACWGCYTCRA | 6 | ||||||
| H7 | Eu | 815b | H7-937Feu | M31689 | 241 | ATMAATAGCAGRGCARTRGG | 4 | 2 | 83 |
| 496 | H7-1177R | GATCWATTGCHGAYTGRGTG | 4 | ||||||
| Am | 816b | H7-937Fam | M31689 | 241 | ATCAACYCYAGRACWGTKGG | 5 | 2 | 83 | |
| 496 | H7-1177R | GATCWATTGCHGAYTGRGTG | 4 | ||||||
| H8 | Eu + Am | 159 | H8-854F | CY005970 | 376 | GGCAGAATAATYCAAAATGA | 1 | 1 | 83–84 |
| 684b | H8-1229R | TCATYTTRTCAACTATATTR | 3 | ||||||
| H9 | Eu | 781b | H9-952Feu | CY005639 | 312 | AGTAA RTAYGCRTTYGGRA | 5 | 1 | 83–85 |
| 782b | H9-1263Reu | TCRCTGAAYTCATGATCRA | 3 | ||||||
| Am | 779b | H9-919Fam | CY005639 | 312 | AGCAAATAYGCATTTGGGA | 1 | 2 | 83–85 | |
| 780b | H9-1229Ram | TCAYTGAATTCATCATYAA | 2 | ||||||
| H10 | Eu | 856b | H10-935Feu | CY014619 | 311 | AAYYTRTCACCVAGRACAGT | 5 | 2 | 83–85 |
| 501 | H10-1245Reu | TCAGAYTCTATKGAYYCRAAC | 5 | ||||||
| Am | 825b | H10-935Fam | CY014619 | 311 | AATCTTTCMCCRAGAACTGT | 2 | 2 | 83–85 | |
| 826b | H10-1245Ram | TCAGATTCTATGGATTCGAAC | 0 | ||||||
| H11 | Eu + Am | 165 | H11-858F | AY684895 | 410 | GGAAATGGGAAAYTGTTCAG | 1 | 1 | 83–84 |
| 514 | H11-1267R | AATTCRTGTTGHAYAGACTC | 3 | ||||||
| H12 | Eu | 543 | H12-776F | CY005925 | 368 | CARACWGTVAARATACARAC | 5 | 1 | 84–86 |
| 857b | H12-1143Reu | CCYGTTCCCTCRGARTTYTG | 4 | ||||||
| Am | 543 | H12-776F | CY005925 | 368 | CARACWGTVAARATACARAC | 5 | 1 | 84–86 | |
| 903b | H12-1143Ram | CCTGTCCCYTCYGCRTTTTG | 3 | ||||||
| H13 | Eu + Am | 169 | H13-856F | AY684887 | 337 | ATTGARGARTATGGAAAAGG | 2 | 0.5 | 84 |
| 421 | H13-1192R | GTYGAYTCTTTRTCTGCAGC | 3 | ||||||
| H14 | Eu + Am | 171 | H14-863F | CY014604 | 282 | GAGCACAGTGCTTAAAAGTG | 0 | 0.5 | 84 |
| 422 | H14-1144R | GCATTTTGRTGCCTRAATCCATACC | 2 | ||||||
| H15 | Eu + Am | 515 | H15-828F | L43916 | 273 | CCGCTCTAATGCCCCWTCRG | 2 | 0.5 | 84 |
| 423 | H15-1100R | TCGATGAATCCTGCAATTGC | 0 | ||||||
| H16 | Eu + Am | 895b | H16-834F | AY684888 | 371 | GMTAYATHATTGAGAARTACG | 4 | 1 | 85 |
| 897b | H16-1204R | GTTGTTATYTCATYTATKGC | 3 | ||||||
| N1 | Eu + Am | 685 | N1-54F | CY003931 | 245 | TCARTCTGYATGRYAAYTGG | 5 | 0.5 | 81–82 |
| 469 | N1-298R | GGRCARAGAGAKGAATTGCC | 3 | ||||||
| N2 | Eu + Am | 470 | N2-59F | CY005306 | 278 | TYTCTMTAACYATTGCRWCARTA | 6 | 2 | 81–82 |
| 695 | N2-336R | GARTTGTCYTTRGARAAVGG | 5 | ||||||
| N3 | Eu + Am | 687 | N3-79F | AY650272 | 287 | GCCCTTCTYATYGGRRTKGGRAA | 6 | 2 | 82–83 |
| 688 | N3-365R | ACTATDRCRTCYTTGTTYTC | 5 | ||||||
| N4 | Eu + Am | 483 | N4-55F | CY003986 | 236 | AGTGYKAGYATTRTAYTRAC | 6 | 2 | 81–82 |
| 484 | N4-290R | ARGTCTYTYCCACTRGARTA | 5 | ||||||
| N5 | Eu + Am | 509 | N5-115F | CY005693 | 178 | GARTAATATCAGYRACVAAAG | 4 | 1 | 80–82 |
| 87 | N5-279R | GATACATYRCAGAGAGGTTC | 2 | ||||||
| N6 | Eu + Am | 88 | N6-57F | CY005464 | 264 | AGGAATGACACTATCSGTAGTAAG | 1 | 1 | 80–82 |
| 89 | N6-307R | GAYAGRATRTGCCATGAGTTYAC | 4 | ||||||
| N7 | Eu + Am | 472 | N7-53F | CY004347 | 261 | TCWGGAGTGGCMATAGCACT | 2 | 0.5 | 80–82 |
| 473 | N7-313R | CACKACCCAYCCTTCAACWTTG | 3 | ||||||
| N8 | Eu | 107 | N8-93Feu | CY004056 | 137 | CATRTVGTBAGYATYAYARTAAC | 7 | 2 | 79–80 |
| 690 | N8-209R | ACAYTRGYATTGTRCCATTG | 4 | ||||||
| Am | 788b | N8-93Fam | CY004056 | 137 | CAYATAGYTAGYATYACARTAAC | 5 | 2 | 79–80 | |
| 690 | N8-209R | ACAYTRGYATTGTRCCATTG | 4 | ||||||
| N9 | Eu | 691 | N9-64Feu | CY004701 | 227 | GTAATAGGCACRATYGCAGT | 2 | 1 | 81–82 |
| 692 | N9-290Reu | CCTTTRGTYARRTTATTGAA | 4 | ||||||
| Am | 790b | N9-64Fam | CY004701 | 227 | GTRATAGGCAYRATTGCAGT | 3 | 1 | 81–82 | |
| 792b | N9-290Ram | CCTYTRGTCARRTTRTTGAA | 5 | ||||||
| NP | Eu + Am | 551 | NP-1200F | DQ064436 | 330 | AGRTAYTGGGCYATAAGRAC | 4 | 1 | 85 |
| 806 | NP-1529R | GCATTGTCTCCGAAGAAATAAG | 0 |
Eu, Eurasian lineage genes; Am, American lineage genes.
Primer updated in this study.
eu, Eurasion lineage; am, American lineage; neither, common lineage.
Codes for mixed bases (boldface): B, C/G/T; D, A/G/T; H, A/T/C; K, G/T; M, A/C; R, A/G; V, A/C/G; W, A/T; Y, C/T.
Fig 1.
Elimination of nonspecific or cross-reactions of real-time RT-PCR by 30 cycles of amplification of diluted cDNA. The H7 gene of A/turkey/Italy/4580/1999 (H7N1) was amplified by three different protocols of the real-time RT-PCR with H1 to H16 primer sets. The protocols are as follows: protocol I, 35 cycles of amplification of undiluted cDNA (A, D, and G); protocol II, 30 cycles of amplification of undiluted cDNA (B, E, and H); protocol III, 30 cycles of amplification of 1:10 dilution of cDNA (C, F, and I). The HA gene was amplified with each of the H1 to H16 primer sets, the products were electrophoresed in 2.0% agarose gel (A to C), and the H7-specific product was observed in the H7 lane indicated. The amplification (D to F) and dissociation (G to I) curves of each well are shown, and the first lines or high blue peak indicate the H7-specific product.
Gene detection test.
Each of the HA subtyping primer sets was evaluated for the detection of homologous subtype HA genes by rRT-PCR. A total of 349 HA genes, composed of 269 Eurasian and 80 North American lineage genes, were used in this test (22 H1, 17 H2, 56 H3, 31 H4, 51 H5, 36 H6, 31 H7, 5 H8, 20 H9, 22 H10, 40 H11, 13 H12, 1 H13, 1 H14, 1 H15, and 1 H16). After 30 cycles of amplification of 1:10 dilutions of cDNAs, 344 HA genes were positive, and 2 HA genes were weak positive (A/common teal/Korea/SH3-8/2010 [H3N6] and A/wild duck/Korea/CSM27-12/2009 [H7N6] genes). Also, three genes were negative: A/wild duck/Korea/PSC33-16/2009 (H4N6), A/common teal/Korea/CSM4-12/2010 (H6N2), and A/wild duck/Korea/PSC2-10/2009 (H6N6) genes. The newly designed H16 primer set was not cross-reactive to the A/gull/Maryland/704/77 (H13N6) gene despite the close phylogenetic relationship between these genes.
Similarly, each of the NA primer sets including updated or redesigned primers was tested for the detection of NA homologous subtype genes with 349 NA genes. The NA genes included 236 Eurasian and 113 North American lineage genes (36 N1, 81 N2, 55 N3, 11 N4, 20 N5, 44 N6, 14 N7, 54 N8, and 34 N9). As a result, 343 NA genes were positive and 4 NA genes were weak positive (A/duck/Tsukuba/168/2005 [H5N2], A/wild duck/Korea/CSM31-19/2009 [H5N2], A/wild duck/Korea/CSM4-23/2009 [H5N2], and A/duck/Shiga/26/2004 [H4N6] genes). Also, two NA genes derived from A/chicken/Brescia/1902 (H7N1) and A/wild bird feces/Korea/PSC19-38/2008 (H5N2) were negative.
Collectively, these results indicate that the rRT-PCR method using updated or redesigned primer sets can broadly identify HA and NA genes of AIVs in a subtype-specific manner within 30 cycles of amplification.
Comparison of sensitivity with virus isolation test.
The sensitivity for the detection of NP, HA, and NA genes by rRT-PCR was compared with that of the infectivity test using 18 AIVs. The HA and NA rRT-PCR assays (30 cycles of amplification) were almost as sensitive as conventional RT-PCR (35 cycles of amplification) (25, 26). The average detection limits ± standard deviations of the HA and NA assays were 102.3 ± 1.1 and 103.1 ± 0.6 EID50, respectively, while that of the NP assay was 101.5 ± 0.5 EID50 (Table 3).Variations in sensitivity of the rRT-PCR among HA or NA subtypes were not large: 101.5 to 104.0 EID50 for HA genes and 102.0 to 104.0 EID50 for NA genes (Table 3).
Table 3.
Comparison of sensitivities on the basis of infectivity titers with embryonated chicken eggs and rRT-PCR titers
| Virus strain | HA or NA subtype | 50% infectivity titer (log10/0.1 ml)a | 50% rRT-PCR titer (log10/0.14 ml)b |
Infectivity titer vs rRT-PCR titer (log10)c |
||
|---|---|---|---|---|---|---|
| HA or NA | NP | HA or NA | NP | |||
| A/duck/Chiba/9/2009 (H1N2) | H1 | 8.0 | 5.5 | 6.5 | 2.5 | 1.5 |
| A/duck/Shiga/10462-P13/06 (H2N3) | H2 | 7.0 | 5.5 | 6.5 | 1.5 | 0.5 |
| A/duck/Tsukuba/38/2006 (H3N8) | H3 | 7.5 | 4.5 | 6.5 | 3.0 | 1.0 |
| A/duck/Tsukuba/79/2005 (H4N6) | H4 | 7.5 | 3.5 | 5.5 | 4.0 | 2.0 |
| A/duck/Tsukuba/63/2005 (H5N2) | H5 | 7.0 | 5.0 | 5.5 | 2.0 | 1.5 |
| A/duck/Shiga/10418-16/2006 (H6N5) | H6 | 7.5 | 5.5 | 6.5 | 2.0 | 1.0 |
| A/duck/Tsukuba/30/2007 (H7N7) | H7 | 7.5 | 5.5 | 5.5 | 2.0 | 2.0 |
| A/duck/Tsukuba/33/2007 (H8N4) | H8 | 7.5 | 5.5 | 6.5 | 2.0 | 1.0 |
| A/duck/Shimane/1/2005 (H9N2) | H9 | 6.5 | 4.5 | 6.0 | 2.0 | 0.5 |
| A/duck/Tsukuba/578/2006 (H10N3) | H10 | 8.0 | 4.5 | 5.5 | 3.5 | 2.5 |
| A/duck/Tsukuba/2/2005 (H11N2) | H11 | 7.5 | 5.5 | 6.5 | 2.0 | 1.0 |
| A/duck/Tsukuba/212/2006 (H12N5) | H12 | 8.5 | 4.5 | 6.5 | 4.0 | 2.0 |
| A/gull/Maryland/704/1977 (H13N6) | H13 | 8.0 | 6.5 | 6.5 | 1.5 | 1.5 |
| A/mallard/Gurjev/263/1982 (H14N5) | H14 | 8.0 | 6.5 | 6.5 | 1.5 | 1.5 |
| A/duck/Australia/341/1983 (H15N6) | H15 | 8.0 | 6.5 | 6.5 | 1.5 | 1.5 |
| A/b-headed gull/Sweden/5/1999 (H16N3) | H16 | 7.5 | 5.5 | 5.5 | 2.0 | 2.0 |
| Avg | 2.3 ± 1.1 | |||||
| A/duck/Tsukuba/41/2005 (H3N1) | N1 | 8.0 | 4.5 | 6.5 | 3.5 | 1.5 |
| A/duck/Tsukuba/2/2005 (H11N2) | N2 | 7.5 | 4.5 | 6.5 | 3.0 | 1.0 |
| A/duck/Tsukuba/578/2006 (H10N3) | N3 | 8.0 | 4.5 | 5.5 | 3.5 | 2.5 |
| A/duck/Tsukuba/33/2007 (H8N4) | N4 | 7.5 | 3.5 | 6.5 | 4.0 | 1.0 |
| A/duck/Tsukuba/212/2006 (H12N5) | N5 | 8.5 | 5.5 | 6.5 | 3.0 | 2.0 |
| A/duck/Tsukuba/79/2005 (H4N6) | N6 | 7.5 | 4.5 | 5.5 | 3.0 | 2.0 |
| A/duck/Tsukuba/30/2007 (H7N7) | N7 | 7.5 | 5.5 | 5.5 | 2.0 | 20. |
| A/duck/Tsukuba/38/2006 (H3N8) | N8 | 7.5 | 4.5 | 6.5 | 3.0 | 1.0 |
| A/duck/Tsukuba/16420/2005 (H11N9) | N9 | 6.5 | 3.5 | 5.5 | 3.0 | 1.0 |
| Avg | 3.1 ± 0.6 | 1.5 ± 0.5 | ||||
Subtyping test of cultured AIVs with full primer sets.
The allantoic fluids of 104 AIVs derived from migratory ducks in two wintering seasons in Japan (October to March in 2007/2008 and 2008/2009) were used to evaluate the usefulness of rRT-PCR for genetic subtyping of AIVs with all 25 primer sets. Tenfold dilutions of cDNAs synthesized from cultured AIVs were amplified for 30 cycles. As shown in Table 4, 103 of the 104 AIVs, including two weak-positive genes, one N2 (A/duck/Chiba/8/2007 [H4N2] gene) and one N9 (A/duck/Chiba/11/2007 [H11N9] gene), could be subtyped simultaneously for HA and NA genes. However, one N2 gene (A/duck/Chiba/10/2007 [H9N2] gene) was not. Subtype specificities of these NA products were confirmed by sequencing followed by BLAST search analysis. Cross-amplified or nonspecific products were rarely observed.
Table 4.
Simultaneous subtyping of HA and NA genes of avian influenza viruses isolated from migratory ducks wintering in Japan (2007/2008, 2008/2009, 2009/2010) by rRT-PCR
| Subtype | No. of HA genes of type: |
Total no. of NA genes | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 | H13 | H14 | H15 | H16 | ||
| N1 | 11 | 11 | |||||||||||||||
| N2 | 13a | 2 | 4 | 5 | 11 | 12a | 1 | 2 | 50 | ||||||||
| N3 | 6 | 3 | 1 | 10 | |||||||||||||
| N4 | 6 | 6 | |||||||||||||||
| N5 | 1 | 10 | 11 | ||||||||||||||
| N6 | 1 | 2 | 39 | 3 | 45 | ||||||||||||
| N7 | 8 | 8 | |||||||||||||||
| N8 | 11 | 1 | 3 | 15 | |||||||||||||
| N9 | 3 | 3 | |||||||||||||||
| Total no. of HA genes | 20 | 3 | 15 | 43 | 6 | 27 | 8 | 6 | 12 | 0 | 4 | 15 | 0 | 0 | 0 | 0 | 159a |
Two N2 genes, from strains A/duck/Chiba/10/2007 (H9N2) and A/duck/Tsukuba/395/2010 (H1N2), were not detected.
Additionally, 55 viruses, including 45 viruses isolated in a direct subtyping test described below and 10 other viruses isolated in the same 2009/2010 wintering season were used to assess the utility. Consequently, all 55 HA and 54 NA genes, including one H6 weak-positive gene (A/duck/Shiga/S4/2009[H6N6] gene), were genetically subtyped, although one N2 gene (A/duck/Tsukuba/395/2010 [H1N2] gene) was not. Thus, 157 of 159 viruses isolated from migratory birds during three wintering seasons in Japan were subtyped by the rRT-PCR method (Table 4).
Direct subtyping test of AIVs in fecal samples with all primer sets.
We further determined the usefulness of rRT-PCR for direct subtyping of AIVs in clinical samples. A total of 120 fecal samples collected from wintering wild ducks in Japan were used for a virus isolation test, and 45 virus-positive fecal samples were obtained. The HA and NA subtypes of the 45 cultured viruses, except for 1 N2 gene (A/duck/Chiba/10/2007[H9N2] gene), were successfully determined by rRT-PCR, and the N2 subtype was identified serologically by the NI test.
For direct detection of NP genes in clinical samples, a protocol for 40 cycles of amplification of undiluted cDNAs was used because these samples usually contained small amounts of viruses. After 40 cycles of amplification, 40 of 120 fecal samples became NP positive while only 17 samples were NP positive by 30 cycles of amplification. The 40 NP-positive samples consisted of 38 virus-positive samples and two virus-negative samples (Table 5). The 2 NP genes from virus-negative samples had 92 to 94% or 94 to 99% nucleotide sequence identity to those of AIVs isolated in the season. Sensitivity and specificity of the NP assay for clinical samples were 0.84 and 0.97, respectively, compared with the virus isolation test (Table 5). Then, direct subtyping of HA and NA genes of the 40 NP-positive cDNAs was performed by using full primer sets. As a result, 31 HA (78%) and 29 NA (73%) genes of 40 NP-positive cDNAs and 29 HA (64%) and 27 NA (58%) genes of 45 virus-positive fecal samples were successfully subtyped (Table 6). Cross- or nonspecific amplification of these genes was rarely observed after 40 cycles of amplification of clinical samples. The subtypes of AIVs in fecal samples were identical to those of cultured viruses. On the other hand, these virus-positive or NP-positive fecal samples were not available for the current serological subtyping tests.
Table 5.
Comparison of sensitivity and specificity of rRT-PCR for detection of NP genes of avian influenza viruses in wild duck fecal samples with those for the conventional virus isolation test
| rRT-PCR result for NP gene (F > 200)a | No. of samples with a virus isolation test (with chicken embryonated eggs) result of: |
||
|---|---|---|---|
| + | − | Total | |
| + | 38 | 2 | 40 |
| − | 7 | 73 | 80 |
| Total | 45 | 75 | 120 |
Undiluted cDNA was prepared from avian influenza virus from fecal samples, and the NP gene was amplified for 40 cycles by rRT-PCR. Samples showing a clear dissociation curve with fluorescence (F) counts of over 200 are considered positive.
Table 6.
Direct detection of NP genes and direct subtyping of HA and NA genes of AIVs in wild duck fecal samples by rRT-PCR
| Method | No. of positive samples | No. (%) of samples positive by rRT-PCRa for: |
||
|---|---|---|---|---|
| NP gene | HA gene | NA gene | ||
| Virus isolationb | 45 | 38 (84) | 29 (64) | 27 (58) |
| rRT-PCR for NP gene (F > 200)c | 40 | 40 (100) | 31 (78) | 29 (73) |
Viral RNA was purified from wild duck fecal samples and transcribed into cDNA. The undiluted cDNA was used for detecting NP genes and direct subtyping of AIVs by rRT-PCR after 40 cycles of amplification.
AIVs were isolated with embryonated chicken eggs after 4 days of cultivation, and their HA and NA subtypes were determined by rRT-PCR using a 1:10 dilution of cDNA after 30 cycles of amplification.
Samples showing clear dissociation curve with fluorescence counts over 200 were considered positive.
Serological subtyping tests of AIVs.
Serological classification of HA subtypes was conducted by performing the HI test on 104 wild duck isolates (allantoic fluids) using serial 2-fold dilutions of 29 reference antisera (see Table S1 in the supplemental material). The data for the HI test are shown in Table S2 in the supplemental material. Briefly, 47 viruses (45%) were classified as subtyped and 55 strains (53%) were classified as ambiguous. The viruses in the ambiguous group had nonspecific or cross-reactivity with antisera of heterogeneous HA subtypes (HI titer: 1:160 or more). Two H6N2 viruses (2%) belonged to the not-subtyped group by the HI test because two homologous subtype antisera were negative or weak positive (see Table S2 in the supplemental material). We frequently observed cross-reactions by anti-NA antibodies as well as nonspecific reactions in the HI test.
Similarly, the NI test was conducted for serological subtyping of NA of the 104 wild duck viruses with a panel of 9 reference antisera (see Table S1 in the supplemental material). The precise results of the NI test are shown in Table S3 in the supplemental material. Briefly, 70 seven viruses (74%) were classified as subtyped, and 18 of them reacted weakly probably because of antigenic difference. Twenty-seven strains (26%) which reacted positively with more than two reference antisera belonged to the ambiguous group. The 27 viruses in the ambiguous group had the following subtypes: 9 N1, 1 N2, 3 N3, 5 N4, 3 N5, 5 N6, 1 N8, and 1 N9; they could be clearly subtyped by the second NI test using serial 2-fold dilutions of reference antisera (1:20 to 1:2,560).
DISCUSSION
Although the HI and NI tests have long been used as the Office International des Epizooties (OIE) standard tests for subtyping AIVs, these tests often have low levels of specificity and sensitivity, and standardization of the tests and supply of reference antisera are difficult. Because of this, the serological subtyping tests can be done only at the reference laboratories, although test results are needed by many people working for the AI control program worldwide. A specific, sensitive, and simple AIV subtyping test applicable to general laboratories is needed. The genetic subtyping test, rRT-PCR, is substantially superior to the current serological subtyping tests in specificity, sensitivity, and applicability to general laboratories. The guidelines for primer design and optimized reaction conditions established in this study markedly improved specificity and sensitivity of the genetic subtyping tests (25–27). Nonspecific reactions or cross-reactions in the genetic AIV subtyping test were few. Also, the sensitivities of rRT-PCR (30 cycles of amplification) were almost similar to those of conventional RT-PCR (previously 35 cycles of amplification) (25, 26), probably because of better performance of the real-time PCR kit than of the conventional PCR kit. Actual sensitivity of rRT-PCR for subtyping cultured viruses is 1 log lower than those shown in this study because 1:10 dilutions of cDNAs are used. However, rRT-PCR provides enough sensitivity and a broad spectrum: 99% of HA and NA genes of over 500 AIVs were detected or subtyped. This is the first paper to demonstrate that direct subtyping of AIVs in fecal samples can be done by rRT-PCR at a high frequency. Surprisingly, approximately 70% of HA and NA genes in NP gene-positive fecal samples could be subtyped directly, although direct subtyping of AIVs in clinical samples was impossible by serological subtyping tests. It has been shown already that the primers are useful for sequencing amplified products for molecular epidemiology and molecular pathotyping of AIVs (25, 26). Considering the high reliability and applicability to clinical samples as well as the additional genetic information, subtyping of AIVs should be initially determined by genetic tests rather than serological tests. This full-scale and practical rRT-PCR is eligible to become a new standard test for subtyping AIVs.
The guidelines for primer design were established carefully subtype by subtype using over 500 viruses. All of the HA and NA primers reported previously (25, 26) were reevaluated, and consequently 25 HA and 3 NA primers were revised. These primers were designed based on HA and NA genes available in influenza virus resources (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html) and our sequence data of many Asian and some North American isolates. Some of our primer sets were suitable for both Asian and North American viruses, while other primer sets focused on only Eurasian or North American lineage genes (Table 2). Considering that genetic diversity of AIVs preserved in natural ecosystem may be higher than we currently understand it to be, it may be necessary to develop lineage-specific or regional primer sets for all subtypes. Some South Korean HA and NA genes were weak positive or negative (5 in 70 HA genes, 3 in 70 NA genes). Also, we found that 3 HA genes (1 H4, 2 H6) and 4 NA genes (1 N1, 3 N2) were negative and that 3 HA genes (1 H3, 1 H6, 1 H7) and 6 NA genes (4 N2, 1 N6, 1 N9) were weak positive. The N2 primer set may not be optimized to wild duck strains, probably because GenBank data contain many N2 genes derived from chicken H9N2 viruses which are prevalent widely in Asian countries. Therefore, the N2 primer set should be divided into two sets, one for chickens and the other for wild birds. The guidelines developed in this study are useful to establish these lineage-specific or regional primer sets.
Optimal reaction conditions are crucial for high performance of rRT-PCR (Fig. 1). Thirty cycles of amplification of 1:10 dilutions of cDNAs are important for cultured viruses to reduce nonspecific or cross-reactive products, although 35 cycles of amplification may be required for some samples containing small amounts of cDNA. Also, 40 cycles of amplification of undiluted cDNAs were essential for direct subtyping of AIVs in fecal samples because clinical samples contain small amounts of viruses. Interestingly, the HA and NA genes of two virus NP-positive samples could be subtyped by 50 cycles of amplification (data not shown). It is possible that subtyping efficiency of AIVs in clinical samples may be improved by increasing amplification cycle numbers. This high performance of the rRT-PCR partly depends on the high amplification efficiency of the rRT-PCR kit (Takara).
The H5N1 highly pathogenic (HP) AIV has been spreading in Asia, the Middle East, Europe, and parts of Africa, and the role of migratory aquatic birds in carrying the virus has been widely discussed (1, 2, 12, 30, 31). Therefore, surveillance of AIVs in wild aquatic birds has been implemented worldwide to monitor the evolution and potential spread of H5N1 HP AIVs in natural ecosystems. The H5N1 HP AIV was considered not to be maintained in natural reservoirs until 2009 because the viruses have been isolated from wild birds at a very low frequency. However, from 2010 to 2011 (7–9, 28), H5N1 HP AIV of clade 2.3.2 has been isolated frequently from wild birds in China, Mongolia, South Korea, and Japan. From October 2010 to March 2011 in Japan, the clade 2.3.2 virus was frequently isolated from wild birds (30 places in 15 prefectures) and from chickens (24 farms in 9 prefectures) (Ministry of Agriculture, Forestry and Fisheries). These circumstances may increase a risk for HP AIV outbreaks at poultry farms and perpetuation of the virus in wild aquatic birds. It is possible that some waterfowl that winter in Japan and Asian countries may carry the H5N1 virus to Alaska or North America through migration (4). Although the H5N1 HP AIV has never been detected in North America, LP AIVs with Asian lineage genes have been observed in migratory birds in Alaska (15, 20). Thus, the wild bird surveillance is essential for a better understanding of ecology and evolution of LP AIVs in wild birds and of H5N1 virus perpetuation in natural reservoirs. The rRT-PCR method can aid general laboratories in the diagnosis and epidemiological study of LP AIVs and H5N1 HP AIVs in wild birds.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Grant-in-aid Project of the Control of Avian Influenza and other Zoonotic Diseases by the Ministry of Agriculture, Forestry, and Fisheries of Japan.
North American samples were obtained as part of a multiagency surveillance effort including the U.S. Geological Survey, U.S. Fish and Wildlife Service, and the Alaska Department of Fish and Game, and viruses from these samples were isolated by the Diagnostic Virology Laboratory at the USGS National Wildlife Health Center.
Any use of trade names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Published ahead of print 26 October 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Auewarakul P, et al. 2007. Surveillance for reassortant virus by multiplex reverse transcription-PCR specific for eight genomic segments of avian influenza A H5N1 viruses. J. Clin. Microbiol. 45:1637–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen H, et al. 2005. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature 436:191–192 [DOI] [PubMed] [Google Scholar]
- 3. Fereidouni SR, et al. 2009. Rapid molecular subtyping by reverse transcription polymerase chain reaction of the neuraminidase gene of avian influenza A viruses. Vet. Microbiol. 135:253–260 [DOI] [PubMed] [Google Scholar]
- 4. Flint P, et al. 2009. Breeding-season sympatry facilitates genetic exchange among allopatric wintering populations of northern pintails in Japan and California. Condor 111:591–598 [Google Scholar]
- 5. Gyarmati P, et al. 2008. Simultaneous genotyping of all hemagglutinin and neuraminidase subtypes of avian influenza viruses by use of padlock probes. J. Clin. Microbiol. 46:1747–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hoffmann B, et al. 2007. Rapid and highly sensitive pathotyping of avian influenza A H5N1 virus by using real-time reverse transcription-PCR. J. Clin. Microbiol. 45:600–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hu X, et al. 2011. Clade 2.3.2 avian influenza virus (H5N1), Qinghai lake region, China, 2009–2010. Emerg. Infect. Dis. 17:560–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kang HM, et al. 2011. Genetic analyses of H5N1 avian influenza virus in Mongolia, 2009 and its relationship with those of eastern Asia. Vet. Microbiol. 147:170–175 [DOI] [PubMed] [Google Scholar]
- 9. Kim HR, et al. 2011. H5N1 subtype highly pathogenic avian influenza virus isolated form healthy mallard captured in South Korea. Vet. Microbiol. 151:386–389 [DOI] [PubMed] [Google Scholar]
- 10. Lee MS, Chang PC, Shien JH, Cheng MC, Shieh HK. 2001. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J. Virol. Methods 97:13–22 [DOI] [PubMed] [Google Scholar]
- 11. Li J, Chen S, Evans DH. 2001. Typing and subtyping influenza virus using DNA microarrays and multiplex reverse transcriptase PCR. J. Clin. Microbiol. 39:696–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu J, et al. 2005. Highly pathogenic H5N1 influenza virus infection in migratory birds. Science 309:1206. [DOI] [PubMed] [Google Scholar]
- 13. Liu S, et al. 2009. Panorama phylogenetic diversity and distribution of type A influenza virus. PLoS One 4:e5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Monne I, et al. 2008. Development and validation of a one-step real-time PCR assay for simultaneous detection of subtype H5, H7, and H9 avian influenza viruses. J. Clin. Microbiol. 46:1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pearce JM, et al. 2011. Interspecific exchanges of avian influenza virus genes in Alaska: the influence of trans-hemispheric migratory tendency and breeding ground sympatry. Mol. Ecol. 20:1015–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pedersen J. 2008. Neuraminidase-inhibition assay for the identification of influenza A virus neuraminidase subtype or neuraminidase antibody specificity, p. 67–76 In Spackman E. (ed.), Avian influenza viurs. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 17. Pedersen JC. 2008. Hemagglutination-inhibition test for avian influenza virus subtype identification and the detection and quantitation of serum antibodies to the avian influenza virus, p. 53–66 In Spackman E. (ed.), Avian influenza virus. Humana Press, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 18. Peter P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647–1690 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 19. Qiu BF, et al. 2009. A reverse transcription-PCR for subtyping of the neuraminidase of avian influenza viruses. J. Virol. Methods 155:193–198 [DOI] [PubMed] [Google Scholar]
- 20. Ramey AM, et al. 2010. Transmission and reassortment of avian influenza viruses at the Asian-North American interface. Virology 406:352–359 [DOI] [PubMed] [Google Scholar]
- 21. Reed L, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 22. Spackman E, et al. 2002. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 40:3256–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Suzuki K, et al. 2009. Association of increased pathogenicity of Asian H5N1 highly pathogenic avian influenza viruses in chickens with highly efficient viral replication accompanied by early destruction of innate immune responses. J. Virol. 83:7475–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Swayne DE, Halvorso DA. 2003. Influenza, p. 135–160 In Saif YM, et al. (ed.), Diseases of poultry, 11th ed. Iowa State Press, Ames, IA [Google Scholar]
- 25. Tsukamoto K, et al. 2008. Subtyping of avian influenza viruses H1 to H15 on the basis of hemagglutinin genes by PCR assay and molecular determination of pathogenic potential. J. Clin. Microbiol. 46:3048–3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsukamoto K, et al. 2009. Use of reverse transcriptase PCR to subtype N1 to N9 neuraminidase genes of avian influenza viruses. J. Clin. Microbiol. 47:2301–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tsukamoto K, et al. 2010. Broad detection of diverse H5 and H7 hemagglutinin genes of avian influenza viruses by real-time reverse transcription-PCR using primer and probe sets containing mixed bases. J. Clin. Microbiol. 48:4275–4278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Uchida Y, et al. 2008. Highly pathogenic avian influenza virus (H5N1) isolated from whooper swans, Japan. Emerg. Infect. Dis. 14:1427–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Deusen RA, Hinshaw VS, Senne DA, Pellacani D. 1983. Micro neuraminidase-inhibition assay for classification of influenza A virus neuraminidases. Avian Dis. 27:745–750 [PubMed] [Google Scholar]
- 30. Wallensten A, et al. 2007. Surveillance of influenza A virus in migratory waterfowl in northern Europe. Emerg. Infect. Dis. 13:404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Webster RG, Peiris M, Chen H, Guan Y. 2006. H5N1 outbreaks and enzootic influenza. Emerg. Infect. Dis. 12:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.