Abstract
Porphyromonas gingivalis is associated with the development of periodontitis. Here we describe the development of a highly specific protease-based diagnostic method for the detection of P. gingivalis in gingival crevicular fluid. Screening of a proteolytic peptide substrate library, including fluorogenic dipeptides that contain d-amino acids, led to the discovery of five P. gingivalis-specific substrates. Due to the presence of lysine and arginine residues in these substrates, it was hypothesized that the cleavage was mediated by the gingipains, a group of P. gingivalis-specific proteases. This hypothesis was confirmed by the observation that P. gingivalis gingipain knockout strains demonstrated clearly impaired substrate cleavage efficacy. Further, proteolytic activity on the substrates was increased by the addition of the gingipain stimulators dithiothreitol and l-cysteine and decreased by the inhibitors leupeptin and N-ethylmaleimide. Screening of saliva and gingival crevicular fluid of periodontitis patients and healthy controls showed the potential of the substrates to diagnose the presence of P. gingivalis proteases. By using paper points, a sensitivity of approximately 105 CFU/ml was achieved. P. gingivalis-reactive substrates fully composed of l-amino acids and Bz-l-Arg-NHPhNO2 showed a relatively low specificity (44 to 85%). However, the five P. gingivalis-specific substrates that each contained a single d-amino acid showed high specificity (96 to 100%). This observation underlines the importance of the presence of d-amino acids in substrates used for the detection of bacterial proteases. We envisage that these substrates may improve the specificity of the current enzyme-based diagnosis of periodontitis associated with P. gingivalis.
INTRODUCTION
Periodontitis is an inflammation of the periodontium, the tissues that surround and support the teeth. Severe periodontitis affects at least 10% of the general population and involves progressive loss of the alveolar bone around the teeth (17, 18, 29). If left untreated, periodontitis can lead to loosening and subsequent loss of teeth (45). Periodontitis is induced by microorganisms that adhere to and grow at the gingival margin, along with an overly aggressive immune response against these microorganisms (9). The microorganisms implicated in periodontitis include Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, Prevotella intermedia, Tannerella forsythia, and Treponema denticola (11, 13, 26, 31). Periodontal pathogens, in particular, T. denticola, T. forsythia, and P. gingivalis, secrete protease virulence factors that allow these bacteria to invade the host's tissues (13, 16, 21, 25, 37). By liberating amino acids from host proteins, secreted proteases are actively involved in the (anaerobic) metabolism of these bacteria. In addition to being a major cause of tooth loss, periodontal disease has also been associated with several systemic diseases. Animal- and population-based studies have demonstrated associations between periodontal disease and diabetes, cardiovascular disease, rheumatoid arthritis, stroke, and adverse pregnancy outcomes (4, 8, 38). Although the mechanisms that cause these associations are far from being completely understood, more reliable detection of early stage periodontal disease could have widespread health benefits, even beyond the prevention of tooth loss.
Despite today's widespread occurrence of periodontal disease, currently available diagnostic tests are limited in their sensitivity and specificity (46). The best available diagnostic aid is measurement of the depth of the tooth pocket, but this provides a retrospective analysis only and mostly when tooth attachment is already lost. Moreover, this is a mere effect measurement and this does not help to identify causative agents. Culture-based, nucleic acid-based, and antibody-based diagnostic methods are available to help identify the periodontal pathogens involved, which is important to determine suitable therapeutic procedures invoking the highest chance of therapeutic success (6, 14, 19). Still, cultivation or using nucleic acid-based and immunochemical tests to identify periodontal pathogens can be very laborious and time-consuming. So far, direct detection and identification of periodontal pathogens in situ have proven difficult.
Therefore, the goal of this study was to develop a rapid and simple diagnostic technology that would enable the dental practitioner to perform a “chair-side” test to identify the presence of periodontal pathogens with a focus on P. gingivalis. The application of such a test based on the enzymatic diagnosis of periodontal pathogens has already been described by Loesche and coworkers (27). Using the benzoyl-dl-arginine-naphthylamide (BANA) substrate, it is possible to detect T. denticola, T. forsythia, and P. gingivalis by their proteolytic activity in plaque samples (28). To start suitable treatment of periodontitis, it is important to know which pathogen is involved. Nevertheless using the BANA substrate, it is not possible to distinguish among these three bacterial species and exclusively detect P. gingivalis.
We showed in a previous study that the introduction of a d-amino acid residue in a peptide substrate enhanced the specificity of a test that was geared toward the detection of proteolytic activity specifically associated with a certain bacterial species (20). The all-l-amino-acid peptide fluorescein isothiocyanate (FITC)-Leu-Leu-Dabcyl (KDbc) was cleaved by a wide range of bacterial proteases. However, when one of the leucines within the substrate was substituted by its d-enantiomer, the peptide was exclusively cleaved by Bacillus ssp. and not by any of the other species tested (20). Further, in contrast to the all-l-amino-acid parental substrate, the variant was not degraded by enzymes present in serum and saliva. Essentially, the introduction of d-amino acids in a peptide aids to design substrates that are bacterial species specific due to their presence as a component of the bacterial cell wall (43). In contrast, d-amino acids are, with only a few exceptions, not metabolized in eukaryotic cells (33) In our view, this approach has opened a novel, attractive avenue to develop more bacterial species-specific substrates that can be applied in the diagnosis of infectious agents in complex matrices. Based on the study on d-amino-acid-containing peptides for the detection of Bacillus spp., we developed a substrate library containing fluorogenic dipeptides that contain no, one, or two d-amino acids. We used this approach to shotgun screen for substrates that react specifically with P. gingivalis proteases.
MATERIALS AND METHODS
Bacteria.
The bacterial strains used in this study are listed in Table 1. Bacteria were grown in 15 ml brain heart infusion (BHI) medium (BioTrading, Mijdrecht, The Netherlands) under anaerobic conditions at 37°C. All P. gingivalis cultures were supplemented with 1 μg/ml hemin and 0.5 μg/ml menadione (Sigma, Zwijndrecht, The Netherlands). After 72 h of culturing, the bacteria were pelleted by centrifugation for 10 min at 10,000 × g. The supernatant, containing secreted enzymes, was sterilized by filtration through a 0.22-μm filter (Millipore, Amsterdam, The Netherlands). Crude samples were used immediately or stored at −20°C for later use.
Table 1.
Bacterial strains used in this study
| Strain | Characteristic | Reference(s) |
|---|---|---|
| Porphyromonas gingivalis W50 | ||
| Porphyromonas gingivalis HG91 | Nonencapsulated K− | 23, 24 |
| Porphyromonas gingivalis W83 | Capsular serotype K1 | 23, 24 |
| Porphyromonas gingivalis HG184 | Capsular serotype K2 | 23, 24 |
| Porphyromonas gingivalis A7A1-28 | Capsular serotype K3 | 23, 24 |
| Porphyromonas gingivalis ATCC 49417 | Capsular serotype K4 | 23, 24 |
| Porphyromonas gingivalis HG1690 | Capsular serotype K5 | 23, 24 |
| Porphyromonas gingivalis HG1691 | Capsular serotype K6 | 23, 24 |
| Porphyromonas gingivalis 34-4 | Capsular serotype K7 | 3 |
| Porphyromonas gingivalis K1A | ΔKgp | 1 |
| Porphyromonas gingivalis KDP133 | ΔRgpA ΔRgpB | 41 |
| Porphyromonas gingivalis KDP136 | ΔKgp ΔRgpA ΔRgpB | 41 |
| Actinomyces naeslundii ATCC 12104 | ||
| Actinomyces odontolyticus HG472 | ||
| Aggregatibacter actinomycetemcomitans NCTC 9710 | ||
| Fusobacterium nucleatum subsp. periodontium ATCC 33693 | ||
| Fusobacterium nucleatum subsp. nucleatum ATCC 25586 | ||
| Fusobacterium nucleatum subsp. polymorphum ATCC 10953 | ||
| Peptostreptococcus micros ATCC 33270 | ||
| Prevotella intermedia ATCC 25611 | ||
| Prevotella nigrescens NCTC 9336 | ||
| Streptococcus mitis I SK 95 | ||
| Streptococcus oralis SK 1477 | ||
| Tannerella forsythia ATCC 43037 | ||
| Treponema denticola ATCC 35405 |
Fluorescence resonance energy transfer (FRET) substrates.
The 115 novel fluorogenic substrates used were purchased at PepScan Presto B.V. (Lelystad, The Netherlands) and were >90% pure (20, 36). The identity of the substrates was confirmed by mass spectrometry. The P. gingivalis-specific substrates were designated “BikKams” (Table 2). Assays were performed in Blackwell clear-bottom 96-well plates (Corning, Lowell, MA). Proteolytic activity was determined by the addition of 1 μl of substrate (800 μM) to 50 μl of filtered culture supernatant or whole culture for the sensitivity assay. Culture broth was used as a negative control. Plates were read for 60 min at 37°C with 2-min intervals on a fluorescence microplate reader (Fluostar Galaxy, BMG Laboratories, Offenburg, Germany) with an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Relative fluorescence (RF) values were obtained after correction against the culture broth control. The protease activity was defined in RF per minute (RF/min). An RF/min value higher than 5 was considered positive.
Table 2.
Sequences of FRET library peptides cleaved by P. gingivalis culture supernatant
| Peptide | Sequence |
|---|---|
| BikKam9 | FITC–Arg–d-Asp–KDbc |
| BikKam10 | FITC–Arg–d-Glu–KDbc |
| BikKam11 | FITC–Arg–d-His–KDbc |
| BikKam12 | FITC–Arg–d-Lys–KDbc |
| BikKam13 | FITC–Arg–d-Arg–KDbc |
| BikKam14 | FITC-Lys-Lys-KDbc |
| BikKam15 | FITC-Phe-Arg-KDbc |
| BikKam16 | FITC-Arg-Arg-KDbc |
Cleavage characteristics of P. gingivalis substrates.
P. gingivalis strain W50 culture supernatant was prepared as described above. Culture supernatant was incubated with 16 μM of each substrate in the presence of various concentrations of five different chemicals which are known to influence the cleavage activity of gingipains (5, 7). Proteolytic activity was measured as described previously. The compounds used were leupeptin, dithiothreitol (DTT), N-ethylmaleimide (NEM), l-cysteine and glycyl-glycine. All chemicals were obtained from Sigma (Zwijndrecht, The Netherlands).
Sensitivity testing of P. gingivalis substrates.
P. gingivalis W50 was cultured in BHI medium supplemented with 1 μg/ml hemin and 0.5 μg/ml menadione for 72 h under anaerobic conditions at 37°C. The number of bacteria was determined by plating 10-fold serial dilutions on Trypticase soy agar plates (BioTrading, Mijdrecht, The Netherlands). Plates were incubated at 37°C under anaerobic conditions, and bacteria were enumerated after 3 days of incubation. The culture was serially diluted in culture broth (109, 108, 107, 106, and 105 CFU/ml), and 50 μl of each dilution was used to test the sensitivities of the substrates. Cleavage of the substrates was catalyzed by the addition of 5 μl of l-cysteine (50 mM) to each enzyme reaction mixture.
To mimic clinical samples, paper points were spiked with various concentrations of P. gingivalis W50. For this purpose, a P. gingivalis culture was serially diluted as described above. Paper points were incubated in the diluted culture until saturated and placed into reduced transport fluid (RTF)-containing vials (44). Four paper points per dilution were used. After thorough vortexing, 50 μl of suspension was incubated with 1 μl of substrate (800 μM) and 5 μl of l-cysteine (50 mM).
Proteolytic activity was measured as described previously. RF values were obtained after correction against the culture broth or RTF control. The protease activity was defined in RF/min. An RF/min value higher than 5 was considered positive.
Analysis of periodontitis patient paper points.
To study the clinical applicability of the substrates to diagnose P. gingivalis infections in situ, gingival crevicular fluid from 72 patients suffering from periodontitis, sampled using paper points, was utilized. The study was approved by the Institutional Ethical Board of the Academic Hospital Vrije Universiteit at Amsterdam, and informed consent was obtained from all donors. Subgingival samples were taken from four different periodontal pockets per patient using paper points. After sampling, the paper points were transferred into RTF-containing vials. After thorough vortexing, 1 μl of substrate (800 μM) and 5 μl of l-cysteine (50 mM) were added to 50 μl of suspension. Plates were read for 90 min at 37°C with 2-min intervals on a fluorescence microplate reader as described previously. RF values were obtained after correction against the RTF control. The protease activity was defined in RF/min.
Additionally, the RTF containing dental plaque bacteria was analyzed using Bz-l-Arg-NHPhNO2 (l-BApNA; PeptaNova, Sandhausen, Germany), a commercially available proteolytic substrate which is commonly used for the diagnosis of periodontal pathogens. For this purpose, 50 μl of suspension was incubated with 4 mM l-BApNA and 5 mM l-cysteine. Plates were incubated at 37°C for 60 min, and optical density at 405 nm (OD405) was measured using a microplate reader (Bio-Rad Laboratories, Veenendaal, The Netherlands). Paper points with an RF/min value of >5 or an OD405 of >1.2 were considered positive. Simultaneously, serving as the gold standard, gingival crevicular fluid eluted from the paper points from the same pockets was analyzed for the presence of a number of typical periodontal pathogens, including A. actinomycetemcomitans, P. intermedia, T. forsythia, Peptostreptococcus micros, Fusobacterium nucleatum, and P. gingivalis, using standard microbiological culture. Sensitivity is defined as the percentage of samples that are BikKam or l-BApNA positive as well as P. gingivalis culture positive. Specificity is defined as the percentage of samples which are both BikKam or l-BApNA negative and P. gingivalis culture negative (<10 CFU/ml).
RESULTS
Design of a FRET substrate library.
We developed a shotgun substrate library based on our study on d-amino-acid-containing peptides for the detection of Bacillus spp. (20). The library consisted of fluorogenic substrate peptides, each comprising two amino acids where (i) both amino acids were l-amino acids, (ii) the C-terminally located amino acid was a d-amino acid, and (iii) both amino acids were d-amino acids. All peptides were flanked with an amino hexanoic acid-linked FITC probe at the N terminus and a lysine-coupled KDbc quencher at the C terminus. The library consists of 115 peptides in total (see Table S1 in the supplemental material).
To test the library concept, culture supernatant of P. gingivalis was incubated with all FRET peptides present within the library. Of the 115 peptides, 8 were cleaved by the P. gingivalis culture supernatant. Strikingly, all P. gingivalis-positive substrates contained an arginine or a lysine residue (Table 2).
Specificity of the P. gingivalis substrates.
To examine the specificity of the P. gingivalis-positive substrates, the peptides were incubated with culture supernatants from eight different P. gingivalis strains and 12 other oral pathogens (Table 3). All eight substrates were cleaved by all of the P. gingivalis strains tested, except for BikKam9, which was not cleaved by the culture supernatant of P. gingivalis K2. Further, cleavage of the substrates varied in efficiency; BikKam9 and BikKam10 showed the lowest cleavage activity, whereas BikKam14 to BikKam16 showed the highest cleavage activity (Table 3). It was found that the majority of the substrates cleaved by P. gingivalis enzymes were not degraded by any of the other oral pathogens tested, the only exception being BikKam15, which was cleaved by proteases present in the culture supernatant of T. forsythia (Table 3).
Table 3.
Proteolytic activities of bacterial culture supernatants against P. gingivalis substratesa
| Strain or sample | Proteolytic activitya |
|||||||
|---|---|---|---|---|---|---|---|---|
| BikKam9 | BikKam10 | BikKam11 | BikKam12 | BikKam13 | BikKam14 | BikKam15 | BikKam16 | |
| P. gingivalis W50 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| P. gingivalis K− | + | + | ++ | ++ | ++ | +++ | +++ | +++ |
| P. gingivalis K1 | + | ++ | +++ | +++ | +++ | +++ | +++ | +++ |
| P. gingivalis K2 | − | + | + | ++ | ++ | +++ | ++ | +++ |
| P. gingivalis K3 | ++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ |
| P. gingivalis K4 | + | + | ++ | ++ | ++ | ++ | +++ | +++ |
| P. gingivalis K5 | ++ | ++ | ++ | +++ | +++ | +++ | +++ | +++ |
| P. gingivalis K6 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| P. gingivalis K7 | ++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ |
| A. naeslundii | − | − | − | − | − | − | − | − |
| A. odontolyticus | − | − | − | − | − | − | − | − |
| A. actinomycetemcomitans | − | − | − | − | − | − | − | − |
| F. nucleatum subsp. periodontium | − | − | − | − | − | − | − | − |
| F. nucleatum subsp. nucleatum | − | − | − | − | − | − | − | − |
| F. nucleatum subsp. polymorphum | − | − | − | − | − | − | − | − |
| P. micros | − | − | − | − | − | − | − | − |
| P. intermedia | − | − | − | − | − | − | − | − |
| P. nigrescens | − | − | − | − | − | − | − | − |
| T. forsythia | − | − | − | − | − | − | + | − |
| T. denticola | − | − | − | − | − | − | − | − |
| S. mitis I | − | − | − | − | − | − | − | − |
| S. oralis | − | − | − | − | − | − | − | − |
| Saliva | − | − | − | − | − | + | +++ | ++ |
Enzyme activity is defined in RF/min values as follows: <5 (−), no activity; 5 to 24 (+), low activity; 25 to 124 (++), moderate activity; >125 (+++), high activity.
To further evaluate the possibility of the use of these novel substrates in the diagnosis of periodontitis, saliva from seven healthy volunteers was incubated with the eight P. gingivalis substrates. As expected, the substrates which consisted of exclusively l-amino acids (BikKam14 to BikKam16) were cleaved by saliva whereas no cleavage activity was observed when the d-amino-acid-containing substrates were used (Table 3).
Mapping of the proteolytic characteristics of the P. gingivalis FRET substrates.
Of the substrates present in the library, only the peptides which contained arginine or lysine residues were cleaved by the culture supernatant of P. gingivalis. From this observation, we hypothesized that the enzymes responsible for the cleavage of the substrates could be the P. gingivalis-specific Arg-gingipain and Lys-gingipain peptidases. These gingipains are members of the cysteine peptidase family. Therefore, we verified in addition whether (i) the addition of l-cysteine and DTT could stimulate cleavage and (ii) inhibitors such as NEM and leupeptin could inhibit proteolytic activity.
The addition of DTT and l-cysteine resulted in the increased degradation of all substrates (Table 4). Especially the presence of l-cysteine led to a significant increase in activity of BikKam14 and BikKam16 cleavage (>10- to 28-fold compared to the control). In line, decreased proteolytic activity was observed when the reaction was performed in the presence of NEM or leupeptin. The cleavage activity of BikKam14 in the presence of leupeptin was unaffected (Table 4). No significant increase in substrate cleavage was observed when the gingipain stimulator glycyl-glycine was added to the reaction mixture (Table 4).
Table 4.
Effects of protease inhibitors and stimulators on the cleavage of FRET substrates by P. gingivalis W50a
| Compound | Relative activity (%) |
|||||||
|---|---|---|---|---|---|---|---|---|
| BikKam9 | BikKam10 | BikKam11 | BikKam12 | BikKam13 | BikKam14 | BikKam15 | BikKam16 | |
| None | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Leupeptin at (μM): | ||||||||
| 25 | 54 | 48 | 28 | 28 | 29 | 102 | 25 | 24 |
| 50 | 50 | 42 | 26 | 21 | 24 | 101 | 19 | 15 |
| 100 | 48 | 42 | 23 | 20 | 21 | 97 | 14 | 11 |
| NEM at (mM): | ||||||||
| 6,75 | 39 | 27 | 25 | 16 | 14 | 13 | 7 | 4 |
| 12,5 | 30 | 19 | 19 | 11 | 10 | 11 | 4 | 2 |
| 25 | 23 | 13 | 14 | 8 | 8 | 8 | 3 | 2 |
| DTT at (mM): | ||||||||
| 25 | 213 | 165 | 272 | 267 | 235 | 1,071 | 364 | 562 |
| 50 | 275 | 273 | 500 | 564 | 562 | 1,570 | 683 | 1,164 |
| 100 | 399 | 340 | 709 | 927 | 886 | 1,962 | 1,197 | 1,722 |
| l-Cysteine at (mM): | ||||||||
| 25 | 213 | 292 | 570 | 701 | 811 | 2,728 | 977 | 1,920 |
| 50 | 250 | 334 | 672 | 842 | 920 | 2,494 | 952 | 1,981 |
| 100 | 274 | 403 | 849 | 899 | 969 | 2,855 | 1,027 | 2,185 |
| Glycyl-glycine at (mM): | ||||||||
| 25 | 98 | 96 | 93 | 99 | 99 | 99 | 96 | 103 |
| 50 | 97 | 99 | 91 | 99 | 96 | 95 | 89 | 104 |
| 100 | 96 | 97 | 93 | 94 | 95 | 100 | 92 | 100 |
Inhibitors: leupeptin and NEM. Stimulators: DTT, l-cysteine, and glycyl-glycine.
Proteolytic activity of P. gingivalis gingipain knockout strains.
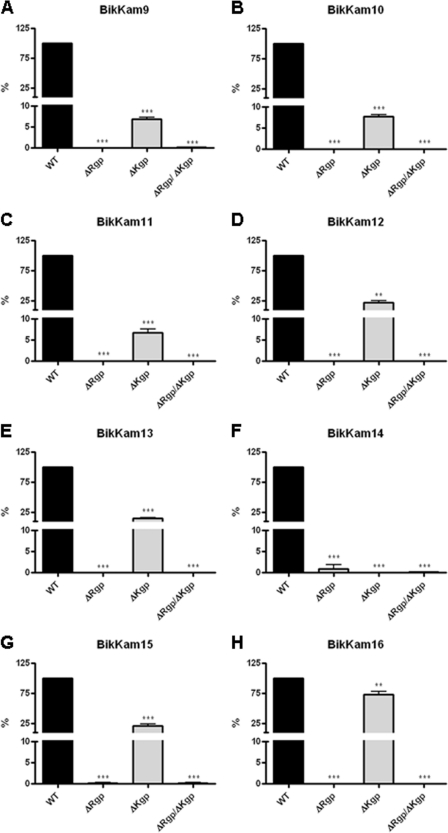
To further explore the role of gingipains in the cleavage of the artificial substrates, the activity of culture supernatants of P. gingivalis strains which lack the Arg-gingipains (ΔRgp), Lys-gingipain (ΔKgp), or both (ΔRgp/ΔKgp) was examined. In both ΔRgp and ΔRgp/ΔKgp supernatants, cleavage of the arginine containing substrates was absent (Fig. 1A to E, G, and H). The substrate which consists of only lysine residues, BikKam14, did show cleavage by ΔRgp, though it was with a much lower efficiency compared to the wild-type strain (Fig. 1F). As expected, there was no cleavage of BikKam14 by ΔKgp or ΔRgp/ΔKgp (Fig. 1F). Surprisingly, besides BikKam14, also decreased efficiency of cleavage by ΔKgp was observed on the other substrates (Fig. 1A to E, G, and H).
Fig 1.
Culture supernatant cleavage activity of gingipain knockout strains. Culture supernatants of P. gingivalis W50 knockout strains ΔRgp (KDP133), ΔKgp (K1A), and ΔKgp/ΔRgpA/ΔRgpB (KDP136) were incubated with each FRET substrate (A to H) at 37°C. As a positive control, wild-type strain W50 (WT) was used. Cleavage of the substrates was defined in RF/min. The enzyme activity of knockout strains was compared to P. gingivalis W50 activity using the unpaired, two-tailed Student t test. Results are expressed as mean ± standard error of the mean (n = 3). ***, P < 0.0001; **, P < 0.01.
Sensitivity of the P. gingivalis substrates.
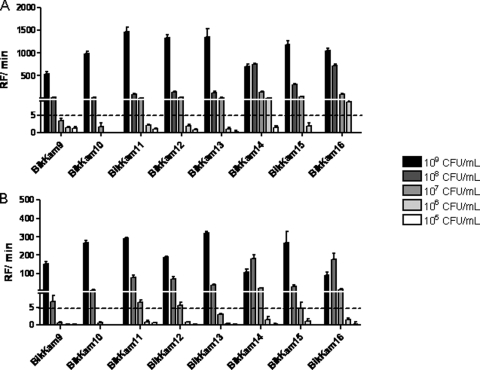
To study the sensitivity of the P. gingivalis-specific protease test, we analyzed serial dilutions of P. gingivalis culture using all eight substrates. BikKam9 and BikKam10 showed the lowest sensitivity, with a limit of detection of 108 CFU/ml (Fig. 2A). The minimal concentration of P. gingivalis that could be detected using BikKam11 to BikKam13 and BikKam15 was 107 CFU/ml (Fig. 2A). BikKam14 and BikKam15 showed the best sensitivity, with a limit of detection of 106 CFU/ml (Fig. 2A).
Fig 2.
In vitro sensitivities of P. gingivalis substrates. Serial dilutions of P. gingivalis W50 (A) and P. gingivalis-spiked paper points (B) were incubated with 16 μM each FRET substrate at 37°C. Enzyme activity was defined in RF/min. The cutoff of the assay was estimated at an RF/min value of 5. Results are expressed as mean ± standard error of the mean (n = 3).
To evaluate the possible use of paper points for the collection of clinical samples, paper points were spiked with serial dilutions of P. gingivalis. Subsequently, reactivity was tested using the BikKam substrates. It was found that the overall signal was lower than the diluted culture (Fig. 2B). This had a negative effect on the sensitivity of three of the BikKam substrates. The limit of detection of BikKam13 changed from 107 CFU/ml to 108 CFU/ml and the detection limit of BikKam14 and BikKam16 decreased from 106 CFU/ml to 107 CFU/ml (Fig. 2B). Despite the decrease in substrate cleavage, the sensitivity of the other substrates remained unaffected.
Screening of patient-derived paper points using P. gingivalis substrates.
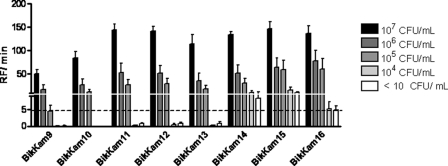
The clinical applicability of the novel protease assay was examined in situ by screening 72 paper points obtained from individuals suffering from periodontitis. As a gold standard, the presence of periodontal pathogens was quantified by routine culture (see Table S2 in the supplemental material). Twenty of the paper points were P. gingivalis culture positive, varying in P. gingivalis concentration from 107 to 104 CFU/ml. All eluates of the 72 paper points were analyzed using all eight substrates. P. gingivalis could be detected to a concentration of 106 to 107 CFU/ml using BikKam9 and BikKam10 and no false-positive activity was observed (Fig. 3). BikKam11 to BikKam13 showed a somewhat higher sensitivity; however, these substrates were also cleaved by 1 or 2 of the P. gingivalis culture-negative samples (Fig. 3). The only l-amino-acid-containing substrates (BikKam14 to BikKam16) yielded the highest sensitivity, but the specificity of these substrates was significant lower than that of BikKam9 to BikKam13 (Fig. 3).
Fig 3.
Screening of P. gingivalis substrates using patient-derived paper points. A total of 72 patient-derived paper points were incubated with 16 μM each FRET substrate. Fluorescence was measured for 90 min at 37°C. Enzyme activity was defined in RF/min. The cutoff of the assay was estimated at an RF/min value of 5. Results are expressed as mean ± standard error of the mean. 107 CFU/ml, n = 6; 106 CFU/ml, n = 6; 105 CFU/ml, n = 5; 104 CFU/ml, n = 3; <10 CFU/ml, n = 52.
Earlier, Schmidt and coworkers designed an assay for the detection of gingipain R activity based on the use of chromogenic substrates in which enzyme activity was measured using BANA (39). Therefore, the commercially available substrate l-BApNA was included in our paper point study and the results were compared to the results obtained using the BikKam substrates. l-BApNA showed a sensitivity for P. gingivalis (40%) lower than that of BikKam9 to BikKam13 (60 to 75%) (Table 5). Also the sensitivity of l-BApNA (85%) is lower than that of BikKam9 to BikKam13 (96 to 100%). Overall, the results obtained with our d-amino acid substrates correspond better to the microbiological culture diagnosis. Some of the paper points analyzed contained, besides P. gingivalis, other oral pathogens such as P. micros, P. intermedia, or F. nucleatum subsp. nucleatum. No significant correlation was observed between the false positives BikKam9 to BikKam13 and the presence of these other oral pathogens (see Table S2 in the supplemental material).
Table 5.
Validation of P. gingivalis substrates using patient-derived paper points
| Peptide | Sensitivitya (%) | Specificityb (%) |
|---|---|---|
| BikKam9 | 60 | 100 |
| BikKam10 | 60 | 100 |
| BikKam11 | 70 | 98 |
| BikKam12 | 75 | 96 |
| BikKam13 | 70 | 96 |
| BikKam14 | 95 | 64 |
| BikKam15 | 95 | 44 |
| BikKam16 | 95 | 73 |
| l-BApNA | 40 | 85 |
Percent BikKam or l-BApNA positive and culture positive.
Percent BikKam or l-BApNA negative and culture negative.
DISCUSSION
The presence of d-amino acids in fluorogenic protease substrates has proven to be crucial for their specificity. Recently we have successfully shown that substrates containing d-amino acids were specifically cleaved by bacterial proteases, as demonstrated for Bacillus spp. (20). No cleavage was observed in human secretions or fluids such as saliva or serum (20). To further explore this concept, 115 novel fluorogenic dipeptides were designed, each containing no, one, or two d-amino acids. Screening of this library using culture supernatant of several oral pathogens resulted in the identification of eight P. gingivalis-specific substrates. Five of these substrates indeed contained a d-amino acid (Table 2). Almost all of the substrates were cleaved by all of the P. gingivalis strains tested, the only exception being BikKam9, where no cleavage activity by P. gingivalis capsule serotype K2 was observed. The RF/min value of BikKam9 of this strain was, however, very close to the cutoff value of 5. All three substrates in which no d-amino acid was present (BikKam14 to BikKam16) were also cleaved by proteases present in saliva. Potentially, a diversity of hydrolases present in saliva can be responsible for this cleavage. Cathepsins, for instance, are known to recognize and cleave a wide range of peptide bonds, among which are Arg-Arg, Lys-Lys, and Phe-Arg (6, 10, 15, 22, 30, 32, 42). As well as the cathepsins, other salivary proteases, like alanine aminopeptidase and dipeptidyl peptidase IV, may play a role in the cleavage of these three substrates (2).
In addition to saliva, BikKam15 was also cleaved by the culture supernatant of T. forsythia. It is known that T. forsythia, similar to P. gingivalis, produces secretory proteases (40). One of these proteases, karilysin, has a high preference for substrates which contain a hydrophobic residue, such as phenylalanine, at the P1 position (21).
The sensitivity for P. gingivalis of the all-l-amino-acid-containing substrates (BikKam14 to BikKam16) was high. However, due to the presence of d-amino-acid-independent proteolytic activity in saliva, a low specificity was observed (Table 3 and 5). Also, using the clinical samples, a high number of false positives for these substrates were observed (Fig. 3 and Table 5). In parallel to BikKam14 to BikKam16, the commercially available compound l-BApNA also showed a relatively low specificity rate (85%). In contrast, for the d-amino-acid-containing substrates the specificity was high (96 to 100%), strengthening our earlier observation that the use of d-amino acids is crucial in the detection of bacterial proteases using fluorogenic substrates (20).
Despite the increased specificity, the presence of d-amino acids in FRET-peptides had a negative effect on sensitivity. The substrates which consisted exclusively of l-amino acids were able to detect lower P. gingivalis concentrations than the d-amino-acid-containing substrates in culture, as well as in patient samples (Table 3, 5). The sensitivity of the all l-amino acid substrates in diluted P. gingivalis culture was 106 to 107 CFU/ml, whereas for the d-amino-acid-containing substrates, the observed limit of detection was 10 times higher (107 to 108 CFU/ml). In general, using the patient samples, more efficient cleavage than that obtained with the diluted P. gingivalis culture was observed. Compared to culture, the limit of detection for all substrates was 10 times lower (Fig. 2A and Fig. 3). This discrepancy might be due to the fact that the pH of the paper point buffer (pH 7.5) is more optimal for gingipain activity than the pH we measured in our P. gingivalis cultures, pH 8.5 (data not shown) (12). Another explanation may be that the amount of gingipains in gingival crevicular fluid is higher, possibly due to the presence of host proteins which might trigger the production of these virulence factors.
Of the 115 substrates from the peptide library, only the substrates which contained an arginine or lysine residue were cleaved by P. gingivalis culture supernatants. Therefore, we hypothesized that P. gingivalis-specific Arg-gingipains and Lys-gingipain are responsible for substrate cleavage (34).
Previously it had been shown that the proteolytic activity of gingipains can be stimulated by the presence of DTT and l-cysteine and inhibited by NEM (5). The addition of these compounds influenced the proteolysis of all compounds, as expected, pointing toward the possible involvement of gingipains (Table 4). It was found that leupeptin inhibited the cleavage of all substrates, except for BikKam14, the only substrate in which no arginine residue is present (Table 4). This finding is in agreement with findings by Aduse-Opoku and coworkers, who observed inhibition of Arg-gingipain activity in the presence of leupeptin, whereas no effect on Lys-gingipain activity was observed (1). A literature search revealed that the amidolytic activity of both Arg- and Lys-gingipain on l-BApNA is stimulated by the addition of glycyl-glycine (5, 7). In the BikKam substrates, however, no amino group is present next to arginine. This can explain the observation that the addition of glycyl-glycine had no effect on the cleavage of our substrates (Table 4).
Experiments using P. gingivalis knockout strains further supported the hypothesis that cleavage of the BikKam substrates is mediated by the gingipains. Absence of these gingpains led to significantly reduced proteolytic activity (Fig. 1). In addition to the absence of Arg-gingipain in ΔRgp, it is known that ΔRgp strains secrete Lys-gingipain less efficiently (1, 41). This was confirmed by the decreased proteolytic activity of ΔRgp culture supernatant on the BikKam14 substrate (Fig. 1F). We were surprised to measure a reduced cleavage activity of ΔKgp on the arginine-containing substrates. We hypothesize that since the Dbc group is coupled to the substrate via a C-terminal lysine side chain, cleavage of the BikKam substrates by Lys-gingipain may take place next to the C-terminal KDbc residue. Further experiments are needed to confirm these assumptions. It has to be noted that all peptides present in the library contain a C-terminal KDbc group. Nevertheless, only the eight substrates described in this study were cleaved by P. gingivalis. This clearly points toward the essence of the complete amino acid sequence of the substrate for gingipain recognition and proteolysis. The results obtained strongly suggest a role for the gingipains in the degradation of the BikKam substrates. However, the precise chemical basis of BikKam cleavage remains to be elucidated.
The use of an enzyme based diagnostic tool based on the presence of P. gingivalis Arg-gingipain was previously described by Loesche and coworkers, who assayed plaque samples using l-BApNA (27, 28). They achieved a sensitivity similar to that of our test using BikKam11 to BikKam13, i.e., 6.6 × 107 ± 5.8 × 107 CFU/ml versus 107 CFU/ml, respectively. The specificity of the BikKam substrates, however, is significantly higher at 96 to 98% for the BikKams versus 85% for l-BApNA. The l-BApNA assay has been developed into a commercially available strip test which is used as a point-of-care test for P. gingivalis, T. denticola, and T. forsythia (BANA test strips; www.oratec.net). Our results imply that the specificity of this rapid and simple diagnostic tool for the detection of P. gingivalis can be improved by the replacement of l-BApNA by one of the d-amino-acid-containing substrates BikKam11, BikKam12, or BikKam13.
Regarding the applicability of these substrates as diagnostic tools for the detection of P. gingivalis, their sensitivities, ranging from 105 to 107 CFU/ml (Fig. 3), potentially match the range in which nonsurgical or surgical therapy is unsuccessful and antimicrobial treatment is needed (35). Consequently, we consider it tempting to hypothesize that the BikKam substrates would not only be helpful in identifying the presence of P. gingivalis in situ but also serve as indicators for antibiotic treatment.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the European Community Seventh Framework Program FP7/2007-2013, TEMPOtest-QC, under grant agreement 241742 and STW Valorisation Grant RapiDent 11535.
We thank M. Curtis, J. Potempa, and K. Nakayama for providing P. gingivalis knockout strains.
Footnotes
Published ahead of print 9 November 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Aduse-Opoku J, et al. 2000. Generation of Lys-gingipain protease activity in Porphyromonas gingivalis W50 is independent of Arg-gingipain protease activities. Microbiology 146(Pt. 8):1933–1940 [DOI] [PubMed] [Google Scholar]
- 2. Aemaimanan P, et al. 2009. Alanine aminopeptidase and dipeptidyl peptidase IV in saliva of chronic periodontitis patients. J. Periodontol. 80:1809–1814 [DOI] [PubMed] [Google Scholar]
- 3. Brunner J, et al. 2010. The core genome of the anaerobic oral pathogenic bacterium Porphyromonas gingivalis. BMC Microbiol. 10:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan HC, Wu CT, Welch KB, Loesche WJ. 2010. Periodontal disease activity measured by the benzoyl-dl-arginine-naphthylamide test is associated with preterm births. J. Periodontol. 81:982–991 [DOI] [PubMed] [Google Scholar]
- 5. Chen Z, Potempa J, Polanowski A, Wikstrom M, Travis J. 1992. Purification and characterization of a 50-kDa cysteine proteinase (gingipain) from Porphyromonas gingivalis. J. Biol. Chem. 267:18896–18901 [PubMed] [Google Scholar]
- 6. Choe Y, et al. 2006. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 281:12824–12832 [DOI] [PubMed] [Google Scholar]
- 7. Ciborowski P, Nishikata M, Allen RD, Lantz MS. 1994. Purification and characterization of two forms of a high-molecular-weight cysteine proteinase (porphypain) from Porphyromonas gingivalis. J. Bacteriol. 176:4549–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cullinan MP, Ford PJ, Seymour GJ. 2009. Periodontal disease and systemic health: current status. Aust. Dent. J. 54(Suppl. 1):S62–S69 [DOI] [PubMed] [Google Scholar]
- 9. Deo V, Bhongade ML. 2010. Pathogenesis of periodontitis: role of cytokines in host response. Dent. Today 29:60–66 [PubMed] [Google Scholar]
- 10. Duncan EM, et al. 2008. Cathepsin L proteolytically processes histone H3 during mouse embryonic stem cell differentiation. Cell 135:284–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dzink JL, Socransky SS, Haffajee AD. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316–323 [DOI] [PubMed] [Google Scholar]
- 12. Fujimura S, Hirai K, Shibata Y, Nakayama K, Nakamura T. 1998. Comparative properties of envelope-associated arginine-gingipains and lysine-gingipain of Porphyromonas gingivalis. FEMS Microbiol. Lett. 163:173–179 [DOI] [PubMed] [Google Scholar]
- 13. Haffajee AD, Socransky SS. 1994. Microbial etiological agents of destructive periodontal diseases. Periodontol. 2000 5:78–111 [DOI] [PubMed] [Google Scholar]
- 14. Hamlet SM. 2010. Quantitative analysis of periodontal pathogens by ELISA and real-time polymerase chain reaction. Methods Mol. Biol. 666:125–140 [DOI] [PubMed] [Google Scholar]
- 15. Higaki J, et al. 1996. Processing of beta-amyloid precursor protein by cathepsin D. J. Biol. Chem. 271:31885–31893 [DOI] [PubMed] [Google Scholar]
- 16. Holt SC, Kesavalu L, Walker S, Genco CA. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168–238 [DOI] [PubMed] [Google Scholar]
- 17. Holtfreter B, Kocher T, Hoffmann T, Desvarieux M, Micheelis W. 2010. Prevalence of periodontal disease and treatment demands based on a German dental survey (DMS IV). J. Clin. Periodontol. 37:211–219 [DOI] [PubMed] [Google Scholar]
- 18. Hugoson A, Sjodin B, Norderyd O. 2008. Trends over 30 years, 1973-2003, in the prevalence and severity of periodontal disease. J. Clin. Periodontol. 35:405–414 [DOI] [PubMed] [Google Scholar]
- 19. Jervøe-Storm PM, AlAhdab H, Koltzscher M, Fimmers R, Jepsen S. 2010. Quantification of periodontal pathogens by paper point sampling from the coronal and apical aspect of periodontal lesions by real-time PCR. Clin. Oral Investig. 14:533–541 [DOI] [PubMed] [Google Scholar]
- 20. Kaman WE, et al. 2011. Peptide-based fluorescence resonance energy transfer protease substrates for the detection and diagnosis of Bacillus species. Anal. Chem. 83:2511–2517 [DOI] [PubMed] [Google Scholar]
- 21. Karim AY, et al. 2010. A novel matrix metalloprotease-like enzyme (karilysin) of the periodontal pathogen Tannerella forsythia ATCC 43037. Biol. Chem. 391:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khouri HE, et al. 1991. A model to explain the pH-dependent specificity of cathepsin B-catalysed hydrolyses. Biochem. J. 275(Pt. 3):751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Laine ML, Appelmelk BJ, van Winkelhoff AJ. 1996. Novel polysaccharide capsular serotypes in Porphyromonas gingivalis. J. Periodontal Res. 31:278–284 [DOI] [PubMed] [Google Scholar]
- 24. Laine ML, Appelmelk BJ, van Winkelhoff AJ. 1997. Prevalence and distribution of six capsular serotypes of Porphyromonas gingivalis in periodontitis patients. J. Dent. Res. 76:1840–1844 [DOI] [PubMed] [Google Scholar]
- 25. Lamont RJ, Jenkinson HF. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loesche WJ. 1988. The role of spirochetes in periodontal disease. Adv. Dent. Res. 2:275–283 [DOI] [PubMed] [Google Scholar]
- 27. Loesche WJ, et al. 1990. Development of a diagnostic test for anaerobic periodontal infections based on plaque hydrolysis of benzoyl-dl-arginine-naphthylamide. J. Clin. Microbiol. 28:1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loesche WJ, Lopatin DE, Giordano J, Alcoforado G, Hujoel P. 1992. Comparison of the benzoyl-dl-arginine-naphthylamide (BANA) test, DNA probes, and immunological reagents for ability to detect anaerobic periodontal infections due to Porphyromonas gingivalis, Treponema denticola, and Bacteroides forsythus. J. Clin. Microbiol. 30:427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mattila PT, Niskanen MC, Vehkalahti MM, Nordblad A, Knuuttila ML. 2010. Prevalence and simultaneous occurrence of periodontitis and dental caries. J. Clin. Periodontol. 37:962–967 [DOI] [PubMed] [Google Scholar]
- 30. McDonald JK, Ellis S. 1975. On the substrate specificity of cathepsins B1 and B2 including a new fluorogenic substrate for cathepsin B1. Life Sci. 17:1269–1276 [DOI] [PubMed] [Google Scholar]
- 31. Moore WE. 1987. Microbiology of periodontal disease. J. Periodontal Res. 22:335–341 [DOI] [PubMed] [Google Scholar]
- 32. Nascimento FD, et al. 2011. Cysteine cathepsins in human carious dentin. J. Dent. Res. 90:506–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pisarewicz K, Mora D, Pflueger FC, Fields GB, Mari F. 2005. Polypeptide chains containing d-gamma-hydroxyvaline. J. Am. Chem. Soc. 127:6207–6215 [DOI] [PubMed] [Google Scholar]
- 34. Potempa J, Sroka A, Imamura T, Travis J. 2003. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 4:397–407 [DOI] [PubMed] [Google Scholar]
- 35. Preshaw PM. 2004. Antibiotics in the treatment of periodontitis. Dent. Update 31:448–450, 453, 454, 456 [DOI] [PubMed] [Google Scholar]
- 36. Rawlings ND, Barrett AJ, Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res. 38:D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robertson PB, et al. 1982. Collagenolytic activity associated with Bacteroides species and Actinobacillus actinomycetemcomitans. J. Periodontal Res. 17:275–283 [DOI] [PubMed] [Google Scholar]
- 38. Routsias JG, Goules JD, Goules A, Charalampakis G, Pikazis D. 2011. Autopathogenic correlation of periodontitis and rheumatoid arthritis. Rheumatology (Oxford) 50:1189–1193 [DOI] [PubMed] [Google Scholar]
- 39. Schmidt EF, Bretz WA, Hutchinson RA, Loesche WJ. 1988. Correlation of the hydrolysis of benzoyl-arginine naphthylamide (BANA) by plaque with clinical parameters and subgingival levels of spirochetes in periodontal patients. J. Dent. Res. 67:1505–1509 [DOI] [PubMed] [Google Scholar]
- 40. Sharma A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000 54:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shi Y, et al. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955–17960 [DOI] [PubMed] [Google Scholar]
- 42. Tersariol IL, et al. 2010. Cysteine cathepsins in human dentin-pulp complex. J. Endod. 36:475–481 [DOI] [PubMed] [Google Scholar]
- 43. van Heijenoort J. 2001. Formation of the glycan chains in the synthesis of bacterial peptidoglycan. Glycobiology 11:25R–36R [DOI] [PubMed] [Google Scholar]
- 44. van Steenbergen TJ, et al. 1993. Survival in transport media of Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis and Prevotella intermedia in human subgingival samples. Oral Microbiol. Immunol. 8:370–374 [DOI] [PubMed] [Google Scholar]
- 45. Williams RC. 1990. Periodontal disease. N. Engl. J. Med. 322:373–382 [DOI] [PubMed] [Google Scholar]
- 46. Xiang X, et al. 2010. An update on novel non-invasive approaches for periodontal diagnosis. J. Periodontol. 81:186–198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.