Abstract
The CapitalBio Mycobacterium identification microarray system is a rapid system for the detection of Mycobacterium tuberculosis. The performance of this system was assessed with 24 reference strains, 486 Mycobacterium tuberculosis clinical isolates, and 40 clinical samples and then compared to the “gold standard” of DNA sequencing. The CapitalBio Mycobacterium identification microarray system showed highly concordant identification results of 100% and 98.4% for Mycobacterium tuberculosis complex (MTC) and nontuberculous mycobacteria (NTM), respectively. The sensitivity and specificity of the CapitalBio Mycobacterium identification array for identification of Mycobacterium tuberculosis isolates were 99.6% and 100%, respectively, for direct detection and identification of clinical samples, and the overall sensitivity was 52.5%. It was 100% for sputum, 16.7% for pleural fluid, and 10% for bronchoalveolar lavage fluid, respectively. The total assay was completed in 6 h, including DNA extraction, PCR, and hybridization. The results of this study confirm the utility of this system for the rapid identification of mycobacteria and suggest that the CapitalBio Mycobacterium identification array is a molecular diagnostic technique with high sensitivity and specificity that has the capacity to quickly identify most mycobacteria.
INTRODUCTION
Members of the genus Mycobacterium are important pathogens for lung diseases, posing an important public health problem worldwide. The genus Mycobacterium includes bacteria that belong to the Mycobacterium tuberculosis complex (MTC), including M. tuberculosis, M. bovis, M. africanum, M. canetti, and M. microti, as well as collective nontuberculous mycobacteria (NTM). In recent years, the spread of AIDS and the increase in the size of the immunocompromised population have contributed to the expansion of infections caused by NTM. As treatment and infection control measures vary according to the etiologic species, early, rapid, and accurate identification of mycobacteria is urgently needed to shorten the culture duration of tuberculosis (TB).
The low rate of identification of mycobacteria is partially due to the use of traditional methods, such as growth characteristics, colony morphology, and biochemical tests, which are cumbersome and time consuming, taking 4 to 8 weeks for growth in culture, and do not satisfy the demands of rapid diagnosis and clinical treatment. Recently, the development of newer diagnostic methods has yielded promising tools for the identification of mycobacteria (1, 3–6, 7–10, 12–15, 18), leading to considerable improvements in both the speed and accuracy of identification. Among these molecular diagnostic methods, DNA microarray assays have undergone substantial development, as this method allows for simultaneous analyses of thousands of genes in a short time period and is useful for phylogenetic analysis and species identification.
The CapitalBio Corporation in Beijing recently developed a new DNA microarray assay, the CapitalBio Mycobacterium identification assay. It is an extremely powerful detection system based on reverse molecular hybridization, which couples PCR technology with hybridization of the resulting amplification products on microarrays. Generally, multiple DNA probes with known identities are fixed on a glass surface for molecular hybridization with DNA samples. Here, we report a method that includes the labeling of single-stranded DNA (ssDNA) from a target gene from a Mycobacterium species in clinical isolates, subsequent hybridization of the labeled DNA to species-specific oligonucleotide probes on a microarray, and detection of the label's fluorescence. The total procedure for determination of mycobacterial species takes only 6 h, and this microarray system is designed to detect and identify a large number of strains in a single reaction.
This study was designed to evaluate the performance of the CapitalBio Mycobacterium identification microarray system for the identification of clinical isolates of Mycobacterium species. The efficacy of the biochip for the identification of clinical isolates was confirmed by DNA sequencing.
MATERIALS AND METHODS
Bacterial strains and clinical samples.
M. tuberculosis strain H37Rv was provided by the Chinese Institute for the Control of Pharmaceutical and Biologic Products. Nineteen strains of the following nontuberculous Mycobacterium reference strains were used: M. chelonae, M. abscessus, M. fortuitum, M. intracellulare, M. avium, M. kansasii, M. gordonae, M. terrae, M. smegmatis, M. szulgai, M. malmoense, M. nonchromogenicum, M. scrofulaceum, M. xenopi, M. aurum, M. marinum, M. ulcerans, M. gilvum, and M. phlei. In addition, four more nonmycobacterial strains were used: Staphylococcus aureus, Enterococcus faecalis, Escherichia coli, and Pseudomonas aeruginosa. All reference strains were acquired from various international collections, and they covered a wide spectrum of mycobacterial species. A total of 486 isolates were obtained from sputum, pleural fluid, and bronchoalveolar lavage fluid samples after 8 weeks of culture with Lowenstein-Jensen slants at 37°C, and all strains were initially identified by biochemical tests. Moreover, 40 clinical samples, including 18 sputum, 12 pleural fluid, and 10 bronchoalveolar lavage fluid samples, from tuberculosis patients at Shanghai Pulmonary Hospital from 2008 to 2009 were used.
CapitalBio Mycobacterium identification array.
The CapitalBio Mycobacterium identification array assay (CapitalBio Corp.) was performed according to the manufacturer's instructions. Briefly, genomic DNA was isolated from bacteria cultured on Lowenstein-Jensen medium. The bacterial culture was then suspended in 80 μl lysis buffer, vortexed on a CapitalBio Extractor at 36°C for 5 min, incubated at 94°C for 5 min, and centrifuged at 5,000 rpm for 1 min at room temperature. Supernatant (2 μl) that contained the extracted DNA was transferred to a PCR tube containing 18 μl of the amplification reagent. Negative and positive controls were included in each experiment to exclude the possibilities of contamination and failure of amplification. The asymmetric PCR was performed in two amplification rounds, with an initial activation step at 37°C for 10 min and then DNA denaturation at 94°C for 10 min, followed by a first round of exponential amplification of 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 40 s, a second round of linear amplification of 10 cycles of 94°C for 30 s and 72°C for 60 s, and a final extension step at 72°C for 7 min.
In a hybridization tube, 6 μl of each amplified DNA sample and 9 μl of prewarmed CapitalBio hybridization buffer were heated to 95°C for 5 min and cooled on ice for 3 min. Next, 13.5 μl of this hybridization mixture was transferred onto the gene microarray, and then the sealed hybridization plate was incubated at 50°C for 2 h in a thermostat-controlled water bath. Using a thermostated shaker, wash buffer I and wash buffer II were prewarmed at room temperature and then shaken at 80 to 100 rpm for 3 min at room temperature. The hybridization chips were analyzed using a LuxScan 10K confocal laser scanner (CapitalBio Corp.), and the fluorescence intensities were quantified using dedicated software called the Mycobacteria Identification Array Test System (CapitalBio Corp.); the hybridization results were printed automatically.
For clinical samples, 0.5-ml portions of samples were used for detection and identification with the same procedure as for isolates. The other portions of the samples were cultured in Lowenstein-Jensen medium for DNA sequencing.
DNA sequencing.
PCR products from 486 clinical isolates and 40 positive cultures from clinical samples were sequenced to assess the CapitalBio Mycobacterium identification array. A fragment of the 16S rRNA gene was amplified with primers (5′-TGGCTCAGGACGAAC-3′ and 5′-GGCTTGCGCCCATTGTGC-3′), and sequences were determined by double-end sequencing with an automated model 377 DNA sequencer (Applied Biosystems, Foster City, CA) with fluorescently labeled dideoxynucleotide terminators (ABI Prism BigDye Terminator cycle sequencing ready reaction kit; Applied Biosystems).
RESULTS
CapitalBio Mycobacterium identification array results. (i) Reference strains.
Each of the 20 mycobacterial reference strains reacted with the Mycobacterium genus-specific-probe internal control (IC), and they hybridized specifically with each complex- or species-specific probe, respectively. For example, M. intracellulare only hybridized with the species-specific probe of M. intracellulare, and it did not hybridize with any other of the species-specific probes on the multiplex probe array. The four nonmycobacterial strains (S. aureus, E. faecalis, E. coli, and P. aeruginosa) were classified as negative controls and were not recognized by the microarray.
(ii) Clinical isolates.
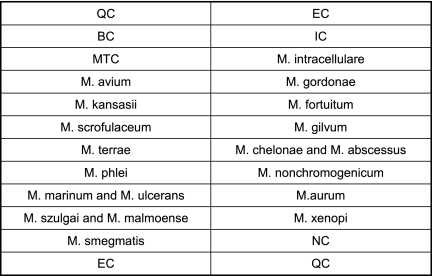
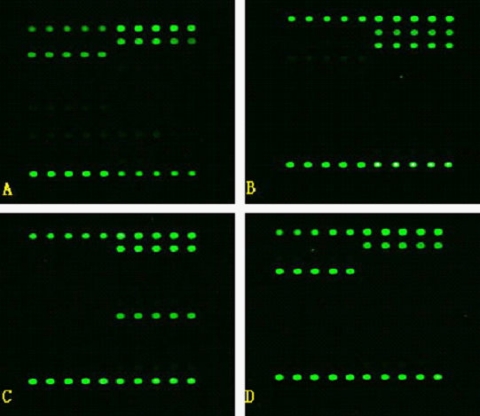
A total of 486 clinical isolates were tested with the newly developed probe array. The CapitalBio Mycobacterium identification array showed highly concordant results of 100% and 98.4% identification of MTC and NTM, respectively. Of the clinical isolates that reacted with the genus-specific Mycobacterium probe IC, 358 isolates also hybridized correctly with the MTC-specific probe and 128 isolates were identified as NTM. Of the 128 NTM isolates, 126 (98.4%) were identified to the species level or the group level. Two NTM isolates (M. parascrofulaceum and M. monacense) were not identified to the species level. For one specimen, culture methods identified the isolate as MTC; interestingly, the scanning image obtained on the microarray showed a composite of the patterns specific for M. avium and M. intracellulare, suggesting that this patient had a dual infection. The probes printed onto the microarray are shown in Fig. 1, and actual scanning images obtained from the microarray are shown in Fig. 2.
Fig 1.
The array of probes on the microarray included 17 species-specific probes and 5 quality control probes. Each probe and the control probes have 5 spots. QC, surface chemistry quality control; EC, hybridization positive external control; BC, blank control; NC, negative control; IC, Mycobacterium genus-specific internal control.
Fig 2.
Representative examples of hybridization patterns on the probe array. Green fluorescence reveals positive hybridization to the probe. Microarray analysis was performed after amplification of DNAs from cultured clinical specimens with primer pairs 16S1 and 16S2 and IS1 and IS2, immobilization of specific probes on a glass slide, and hybridization on the glass slide with fluorescence-5-labeled DNA of clinical isolates of M. tuberculosis (A), M. intracellulare (B), M. chelonae (C), and M. avium (D).
(iii) Clinical samples.
The 40 clinical samples, which included 18 sputum, 12 pleural fluid, and 10 bronchoalveolar lavage fluid samples, were collected from suspected tuberculosis cases, and tested by using the CapitalBio Mycobacterium identification array. All 18 sputum samples were successfully detected and identified by the array, but only 2 pleural fluid and 1 bronchoalveolar lavage fluid sample were detected and positively identified. The identification results of 21 positive samples were all MTB.
Comparison of CapitalBio Mycobacterium identification array results with DNA sequencing results.
DNA sequencing, which is considered the “gold standard” for the identification of mycobacteria, is the most dependable technology for detecting differences in DNA sequences (9). The sequences of the PCR products from 486 isolates were determined by standard DNA sequencing. The identification of all strains was completed by comparison of the nucleotide sequence with a library of the Ribosomal Differentiation of Medical Microsystems database (RIDOM). The CapitalBio Mycobacterium identification array system displayed a sensitivity of 99.6% and a specificity of 100% compared with the results of DNA sequencing (Table 1). The DNA microarray results conformed to the sequencing data, with the exception of two species that only hybridized with the IC probe; therefore, only 0.4% of the isolates (2/486) were not identified to species level. DNA sequencing revealed that these strains were M. parascrofulaceum and M. monacense. The sensitivity of the CapitalBio Mycobacterium identification array was 52.5% for detection and identification in clinical samples. It was 100% for sputum, 16.7% for pleural fluid, and 10% for bronchoalveolar lavage fluid, respectively (Table 2).
Table 1.
Comparison of mycobacterial identification by the CapitalBio mycobacterial identification array and nucleic acid sequencing
| Species of identification | Identification (no. of strains) using: |
|
|---|---|---|
| CapitalBio Mycobacterium identification array | 16S rRNA gene sequencing | |
| MTC | MTC (358) | MTC (358) |
| M. intracellulare | M. intracellulare (32) | M. intracellulare (32) |
| M. avium | M. avium (15) | M. avium (15) |
| M. gordonae | M. gordonae (2) | M. gordonae (2) |
| M. kansasii | M. kansasii (11) | M. kansasii (11) |
| M. fortuitum | M. fortuitum (4) | M. fortuitum (4) |
| M. scrofulaceum | M. scrofulaceum (1) | M. scrofulaceum (1) |
| M. gilvum | M. gilvum (0) | M. gilvum (0) |
| M. terrae | M. terrae (2) | M. terrae (2) |
| M. chelonae-M. abscessus | M. chelonae-M. abscessus (47) | M. chelonae-M. abscessus (47) |
| M. phlei | M. phlei (0) | M. phlei (0) |
| M. nonchromogenicum | M. nonchromogenicum (1) | M. nonchromogenicum (1) |
| M. marinum | M. marinum/ulcerans (4)a | M. marinum (3) |
| M. ulcerans | M. ulcerans/marinum (4)a | M. ulcerans (1) |
| M. szulgai | M. szulgai/malmoense (3)a | M. szulgai (2) |
| M. malmoense | M. malmoense/szulgai (3)a | M. malmoense (1) |
| M. smegmatis | M. smegmatis (4) | M. smegmatis (4) |
| M. aurum | M. aurum (0) | M. aurum (0) |
| M. xenopi | M. xenopi (0) | M. xenopi (0) |
| M. para-scrofulaceum | Mycobacterium spp. (1) | M. para-scrofulaceum (1) |
| M. monacense | Mycobacterium spp. (1) | M. monacense (1) |
M. marinum/M. ulcerans and M. szulgai/M. malmoense cannot be identified to the species level by the CapitalBio Mycobacterium identification array. The 16S rRNA sequencing showed that 4 strains identified as M. marinum/M. ulcerans were 3 strains of M. marinum and 1 strain of M. ulcerans and that 3 strains identified as M. szulgai/M. malmoense were 2 strains of M. szulgai and 1 strain of M. malmoense.
Table 2.
Comparison of mycobacterial identification of isolates in clinical samples using the CapitalBio Mycobacterium identification array and nucleic acid sequencing
| Sample type | No. of samples | No. of isolates identified by: |
Concordance rate (%) | |
|---|---|---|---|---|
| CapitalBio Mycobacterium identification array in samples | 16S rRNA sequencing in Lowenstein-Jensen cultures | |||
| Sputum | 18 | 18 | 18 | 100 |
| Pleural fluid | 12 | 2 | 12 | 16.7 |
| Bronchoalveolar lavage fluid | 10 | 1 | 10 | 10 |
| Total | 40 | 21 | 40 | 52.5 |
DISCUSSION
Mycobacterial species are usually identified by time-consuming culture methods. Classical detection methods have some disadvantages: first, the number of organisms in a clinical sample is not always large enough for detection by microscopic methods; second, long periods are required for culture; and third, not all acid-fast bacilli are M. tuberculosis, and therefore, an identification test must be used to differentiate Mycobacterium species. However, rapid molecular genetic diagnostic tests, including PCR amplification, DNA probe technology, and pyrosequencing, have recently been developed for this purpose. In addition, microarrays are a valuable tool for the specific detection of microorganisms directly from clinical samples. Compared with the results of 16S rRNA gene sequencing, the CapitalBio Mycobacterium identification array was able to identify up to 17 Mycobacterium species. The sensitivity and specificity of the CapitalBio Mycobacterium identification microarray system were 99.6% and 100%, respectively, when used with clinical isolates from respiratory specimens. Three other commercial detection systems, namely, the Amplicor Mycobacterium tuberculosis test (Roche Diagnostic Systems), the Amplified Mycobacterium tuberculosis direct test (Gen-Probe), and the LCx Mycobacterium tuberculosis assay (Abbott Laboratories), yielded sensitivities of 75%, 65.2%, and 79.2%, respectively, with clinical isolates from respiratory specimens (3). The sensitivity of the biochip was apparently higher.
Another commercial method with characteristics similar to those of the CapitalBio Mycobacterium identification array is the INNO-LiPA mycobacteria assay. The 17 most frequently isolated mycobacterial species can be identified by both systems. Tortoli et al. reported that the overall sensitivity and specificity of the INNO-LiPA mycobacteria were 100% and 94.4%, respectively (17). Therefore, the CapitalBio Mycobacterium identification array was more specific than the INNO-LiPA mycobacteria. All 17 Mycobacterium strains that were tested were correctly identified, with no cross-hybridization, in our study. However, some limits of specificity of the M. fortuitum probe have been reported for the INNO-LiPA assay (specifically, cross-reactivity with M. senegalense and M. mageritense) (17). The INNO-LiPA assay is a line probe assay. Because of the space limitations of the strip, relatively few probes can be printed on the membrane, and it is difficult to present more or alternative probes for further species identification on a single assay strip. In contrast, the biochip can readily accommodate a large number of probes and the information content could easily increase to resolve more species. The INNO-LiPA assays are sold as CE marked or FDA approved. Considering the prevalence of mycobacterial species found in clinical samples in China, the biochip can readily meet the requirements of clinical diagnosis.
Microarrays on the slides were analyzed by using a LuxScan 10K confocal laser scanner, and the fluorescence intensities were quantified by using dedicated software called Mycobacteria Identification Array Test System. The results are printed automatically, which makes the identification more objective and accurate, and the data can be saved directly in computers, making daily work more convenient. For both the INNO-LiPA TB assay and the GenoType assay, the operator must manually paste the strip onto an evaluation sheet and read the results by eye using an interpretation chart, which could potentially introduce operator error.
One of the unique features of the CapitalBio Mycobacterium identification array is that the kit uses a novel asymmetric PCR method, and sufficient quantities of ssDNA are produced during the PCR. Therefore, the kit enables the performance of hybridization immediately after PCR amplification (21). Most importantly, the microarray assay can detect dual TB infections, such as those caused by M. avium and M. intracellulare or M. avium and M. kansasii, at the same time. In our study, the biochip identified one dual infection caused by M. avium and M. intracellulare.
As a new method applied in the clinical laboratory, the CapitalBio Mycobacterium identification array directly detected and identified Mycobacterium isolates from 40 clinical samples, including sputum, pleural fluid and bronchoalveolar lavage fluid samples. Isolates were successfully detected by the biochip system in 21 of the 40 samples. DNA sequencing of the 16S rRNA gene was performed for further identification. The overall concordance rate between the array and the DNA sequencing results was 52.5%. It was 100% for sputum, 16.7% for pleural fluid, and 10% for bronchoalveolar lavage fluid, respectively. Zhu et al. (22) used this system to analyze sputum samples, and the concordance rate between the biochip assay and the DNA sequencing results was 100%. The detection rate in sputum in our evaluation was consistent with the results of Zhu et al. The results showed that the CapitalBio Mycobacterium identification array can be used directly for detection and identification in sputum, but the detection rate is much lower in pleural fluid and bronchoalveolar lavage fluid. This microarray assay allows the fast and exact identification of Mycobacterium tuberculosis directly from sputum samples.
It is has been proven that early treatment could improve the quality of life and survival rate of patients with tuberculosis (2, 11, 19). Therefore, it is particularly important to develop an economical, simple, and convenient diagnostic method that can diagnose tuberculosis at an early stage. The CapitalBio Mycobacterium identification array, from PCR amplification to interpretation of the results, can be completed in 6 h. Regarding the kit's costs, the price for detection of mycobacteria by the biochip is about $30 per sample (May 2010), which is similar to the cost of the GenoType CM strip. Although the cost of the kit is higher than that of culture-based methods, the biochip requires a shorter time period before the results are available, which can reduce patient hospital charges due to the rapid initiation of therapy.
Although two highly variable regions (A and B) of the 16S rRNA gene could identify 50 species of Mycobacterium to the species level, some Mycobacterium species still could not be identified with a single probe because of intraspecies heterogeneity of 16S rRNA (20). As reported before, some Mycobacterium species shared the same 16S rRNA gene sequence (M. kansasii and M. gastri) or very similar 16S rRNA gene sequences (e.g., M. malmoense and M. szulgai) (16), which was well substantiated by our evaluation. The CapitalBio Mycobacterium identification array cannot differentiate the following organisms: species in the Mycobacterium tuberculosis complex (including M. tuberculosis, M. bovis, M. africanum, and M. microti), M. marinum and M. ulcerans, and M. szulgai and M. malmoense. Because they have high degrees of similarity, they can be identified to the group level but not to the species level. In our evaluation, four strains of M. marinum and M. ulcerans and three strains of M. szulgai and M. malmoense could only be identified to the group level. 16S rRNA sequencing was required to identify these strains, including three strains of M. marinum, one strain of M. ulcerans, two strains of M. szulgai, and one strain of M. malmoense. Moreover, two uncommon clinical isolates (M. parascrofulaceum and M. monacense) were not identified to the species level. Due to the lack of species-specific probes for M. parascrofulaceum and M. monacense on the microarray, these clinical isolates failed to be identified to the species level.
In summary, the resurgence of tuberculosis has highlighted the urgent need for sensitive, reliable, and fast methods for the laboratory detection of mycobacteria. This study demonstrates that the CapitalBio Mycobacterium identification array is a reliable and rapid approach for the identification of the most frequently isolated mycobacteria. This approach could be integrated into the workflow of a routine laboratory and, thus, enhance the level of diagnostic capability of each tuberculosis laboratory.
ACKNOWLEDGMENTS
This work was supported by the National Great Research Program of China (2008zx10003-009). We thank the TB team of the CapitalBio Corporation for technical assistance.
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Abass NA, Suleiman KM, Jalii IE. 2010. Differentiation of clinical Mycobacterium tuberculosis complex isolates by their GyrB polymorphism. Indian J. Med. Microbiol. 28:26–29 [DOI] [PubMed] [Google Scholar]
- 2. Alwood K, et al. 1994. Effectiveness of supervised, intermittent therapy for tuberculosis in HIV-infected patients. AIDS 8:1103–1108 [DOI] [PubMed] [Google Scholar]
- 3. Brown TJ, Power EG, French GL. 1999. Evaluation of three commercial detection systems for Mycobacterium tuberculosis where clinical diagnosis is difficult. J. Clin. Pathol. 52:193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eing BR, Becker A, Sohns A, Ringelmann R. 1998. Comparison of Roche Cobas Amplicor Mycobacterium tuberculosis assay with in-house PCR and culture for detection of M. tuberculosis. J. Clin. Microbiol. 36:2023–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang TS, et al. 2003. Rapid detection of pulmonary tuberculosis using the BD ProbeTEC ET Mycobacterium tuberculosis Complex Direct Detection Assay (DTB). Diagn. Microbiol. Infect. Dis. 46:29–33 [DOI] [PubMed] [Google Scholar]
- 6. Jan IS, et al. 1998. Evaluation of an automatic polymerase chain reaction assay for identification of Mycobacterium tuberculosis in respiratory specimens. J. Formos. Med. Assoc. 97:204–209 [PubMed] [Google Scholar]
- 7. Maugein J, Fourche J, Vacher S, Grimond C, Bebear C. 2002. Evaluation of the BD ProbeTec ET DTB assay (1) for direct detection of Mycobacterium tuberculosis complex from clinical samples. Diagn. Microbiol. Infect. Dis. 44:151–155 [DOI] [PubMed] [Google Scholar]
- 8. Mazzarelli G, et al. 2003. Evaluation of the BD ProbeTec ET system for direct detection of Mycobacterium tuberculosis in pulmonary and extrapulmonary samples: a multicenter study. J. Clin. Microbiol. 41:1779–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neonakis IK, Gitti Z, Krambovitis E, Spandidos DA. 2008. Molecular diagnostic tools in mycobacteriology. J. Microbiol. Methods 75:1–11 [DOI] [PubMed] [Google Scholar]
- 10. O'Sullivan CE, Miller DR, Schneider PS, Roberts GD. 2002. Evaluation of Gen-Probe amplified Mycobacterium tuberculosis direct test by using respiratory and nonrespiratory specimens in a tertiary care center laboratory. J. Clin. Microbiol. 40:1723–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pablos-Mendez A, Sterling TR, Frieden TR. 1996. The relationship between delayed or incomplete treatment and all-cause mortality in patients with tuberculosis. JAMA 276:1223–1228 [DOI] [PubMed] [Google Scholar]
- 12. Piersimoni C, et al. 1998. Comparative evaluation of the new Gen-Probe Mycobacterium tuberculosis amplified direct test and the semiautomated Abbott LCx Mycobacterium tuberculosis assay for direct detection of Mycobacterium tuberculosis complex in respiratory and extrapulmonary specimens. J. Clin. Microbiol. 36:3601–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Piersimoni C, et al. 2002. Performance assessment of two commercial amplification assays for direct detection of Mycobacterium tuberculosis complex from respiratory and extrapulmonary specimens. J. Clin. Microbiol. 40:4138–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reischl U, Lehn N, Wolf H, Naumann L. 1998. Clinical evaluation of the automated COBAS AMPLICOR MTB assay for testing respiratory and nonrespiratory specimens. J. Clin. Microbiol. 36:2853–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rimek D, Tyagi S, Kappe R. 2002. Performance of an IS6110-based PCR assay and the COBAS AMPLICOR MTB PCR system for detection of Mycobacterium tuberculosis complex DNA in human lymph node samples. J. Clin. Microbiol. 40:3089–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suffys PN, et al. 2001. Rapid identification of mycobacteria to the species level using INNO-LiPA Mycobacteria, a reverse hybridization assay. J. Clin. Microbiol. 39:4477–4482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tortoli E, Mariottini A, Mazzarelli G. 2003. Evaluation of INNO-LiPA MYCOBACTERIAv2: improved reverse hybridization multiple DNA probe assay for mycobacterial identification. J. Clin. Microbiol. 41:4418–4420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang JY, et al. 2004. Performance assessment of a nested-PCR assay (the RAPID BAP-MTB) and the BD ProbeTec ET system for detection of Mycobacterium tuberculosis in clinical specimens. J. Clin. Microbiol. 42:4599–4603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang JY, et al. 2005. Mycobacterium tuberculosis inducing disseminated intravascular coagulation. Thromb. Haemost. 93:729–734 [DOI] [PubMed] [Google Scholar]
- 20. Yamada-Noda M, et al. 2007. Mycobacterium species identification—a new approach via dnaJ gene sequencing. Syst. Appl. Microbiol. 30:453–462 [DOI] [PubMed] [Google Scholar]
- 21. Zhu LX, et al. 2007. Multiplex asymmetric PCR-based oligonucleotide microarray for detection of drug resistance genes containing single mutations in Enterobacteriaceae. Antimicrob. Agents Chemother. 51:3707–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu LX, et al. 2010. Biochip system for rapid and accurate identification of mycobacterial species from isolates and sputum. J. Clin. Microbiol. 48:3654–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]