Abstract
From 2006 to 2009, 315 clinical methicillin-resistant Staphylococcus aureus (MRSA) isolates were collected from 5 hospitals across Israel. Most isolates (64%) were related to the global clones spa types t001-SCCmec-I (SCCmec-I stands for staphylococcal cassette chromosome mec type I) (n = 99; 31%), t002-SCCmec-II (n = 82; 26%), and t008-SCCmec-IV (n = 21; 7%), five of which were identified as MRSA strain USA-300. Seventeen strains unique to Israel were identified. SCCmec types IV and V were common among hospital-acquired isolates.
TEXT
Methicillin-resistant Staphylococcus aureus (MRSA) is an increasing problem throughout the world, both in hospitals and in the community. Several epidemic health care-associated MRSA (HA-MRSA) clones have emerged since the 1970s. In the last decade, five major pandemic clones, designated the Iberian, Brazilian, Hungarian, New York/Japan, and pediatric clones, have been identified, while other new or preexisting clones have emerged in certain areas (12). The recent worldwide spread of several community-associated MRSA (CA-MRSA) clones, and their dissemination into hospitals, has made the understanding of this epidemiology even more complex (10). In Israel, the proportion of MRSA among all S. aureus isolates in 2008 was 35%, similar to the proportion in Southern Europe and the United Kingdom (15) and lower than that in other Middle Eastern countries (4). There is little data on the molecular epidemiology of MRSA in Israel. The epidemiology of CA-MRSA was studied in the pediatric population of southern Israel, where 5.7% of infants were found to be colonized, mostly by a unique MRSA staphylococcal cassette chromosome mec type IV (SCCmec-IV) clonal complex (CC), 913 (1). However, data regarding the molecular epidemiology of community-acquired MRSA among adults, and data on HA-MRSA infections in Israel, are limited to small series or case reports (6, 29, 30).
In 2008, the National Center for Infection Control initiated a national survey of MRSA in Israel. The aims of the study were (i) to describe the molecular epidemiology of hospital-associated and community-associated MRSA infections in Israel and (ii) to determine whether internationally known MRSA strains (e.g., the USA-300 strain) have spread into Israel. Five general hospitals throughout Israel participated in the study: (i) Tel-Aviv Sourasky Medical Center (TA) in the Tel-Aviv area, (ii) Rambam Medical Center (RA) in northern Israel, (iii) Barzilai Medical Center (AS) in southern Israel, (iv) Meir Medical Center (ME) in the Sharon region of central Israel, and (v) Bikur Cholim (BC) in the Jerusalem area. Each laboratory was asked to submit prospectively collected MRSA isolates from blood samples or wounds, isolated within 72 h of admission to the hospital (CA-MRSA), or from 72 h on (HA-MRSA). MRSA isolates collected from 2006 to 2010 were eligible to be included in the study. Isolates were shipped and analyzed at the central study laboratory in Tel-Aviv, Israel.
Identification and susceptibility testing of MRSA isolates were done by using the VITEK2 (bioMérieux, Marcy l'Etoile, France) system; DNase testing and cefoxitin disk diffusion testing were performed according to Clinical and Laboratory Standards Institute guidelines (7). Genetic relatedness was determined by spa typing for all strains (18) and by pulsed-field gel electrophoresis (PFGE) for t008 isolates (27). spa types were determined with Ridom StaphType software version 2.2.1 (Ridom GmbH, Würzburg, Germany) and analyzed by the BURP algorithm, with the following parameters: spa types with fewer than five repeats were considered nongroupable, and spa types belonged to the same spa clonal complex if the cost was less than or equal to six (18). In addition, the corresponding multilocus sequence typing (MLST) clonal complex was assigned for each spa type based on the spa types in the Ridom StaphType database (http://spa.ridom.de/spatypes.shtml) and the study by Monecke et al. (24). The Panton-Valentine leukocidin (pvl) and arcA (t008 isolates only) genes were tested for by PCR (13, 27). SCCmec typing was assigned according to the mec and ccr complexes (36); subtyping of SCCmec-IV was done for t008 isolates (23).
A total of 315 MRSA isolates were collected from 2006 to 2009. A summary of the epidemiological features of the MRSA isolates in this study is presented in Table 1. Almost half of the isolates were collected at the Tel-Aviv Sourasky Medical Center (TASMC), Tel-Aviv, Israel. Eighty-six (51%) and 66 (45%) of the blood and wound cultures, respectively, were collected at least 72 hours after admission to the hospital.
Table 1.
Epidemiological characteristics of MRSA isolates in this study
| Center | No. of MRSA isolatesa |
Total no. of MRSA isolates | |||
|---|---|---|---|---|---|
| Blood |
Wound |
||||
| CA | HA | CA | HA | ||
| TA | 37 | 37 | 40 | 37 | 151 |
| RA | 21 | 34 | 27 | 16 | 98 |
| ME | 13 | 9 | 0 | 2 | 24 |
| AS | 11 | 4 | 13 | 12 | 40 |
| BC | 0 | 2 | 0 | 0 | 2 |
| Total (all) | 82 | 86 | 80 | 67 | 315 |
The number of community-associated (CA) MRSA isolates is the number of isolates cultured within 3 days of admission to the hospital. The number of health care-associated (HA) MRSA isolates is the number of isolates cultured from ≥3 days of admission to the hospital.
The molecular characteristics of MRSA strains according to spa and SCCmec types are presented in Table 2 and Fig. 1 (t008). The majority of isolates (229; 63%) belonged to 13 spa types which are related to spa CC-002 or MLST CC 5 (Table 2; see Fig. S1 in the supplemental material). The second largest was a group of 40 isolates (11%) and 11 spa types related to spa CC 008 or MLST CC 8 (Table 2 and Fig. S1). Except for five t008 isolates, all spa CC-008 isolates were negative by pvl testing.
Table 2.
Molecular and microbiological features of MRSA strains (other than spa-t008) in Israela
| SCCmec type and spa type | spa CCb | MLST CCc | No. of isolates | SCCmec type | No. of isolates that were: |
Comment(s)g | ||
|---|---|---|---|---|---|---|---|---|
| Clin Sd | CAe | Wf | ||||||
| SCCmec types I and III (n ≥ 2) | ||||||||
| 001 | 002 | 5 | 99 | Ih | 0 | 49 | 51 | Presumed Southern Germany clone (8); previously reported from Israel (6) |
| 002 | 002 | 5 | 82 | IIi | 8 | 44 | 33 | Presumed New York/Japan clone (8); previously reported from Israel (6) |
| 004 | 065/004 | 045 | 8 | II | 0 | 5 | 3 | Novel report* |
| 010 | 002 | 5 | 3 | II (2)h/I (1)i | 0 | 1 | 2 | Novel report* |
| 051 | 008 | 8 | 2 | I | 0 | 2 | 1 | Presumed Iberian clone (12) |
| 052 | 008 | 8 | 6 | I | 0 | 2 | 2 | Presumed Iberian clone (12) |
| 535 | E | 5 | 5 | II | 1 | 1 | 0 | Novel report* |
| 5212 | 008 | 2 | I | 0 | 0 | 1 | Novel report* | |
| SCCmec types IV/V | ||||||||
| 002 | 002 | 5 | 28 | V | 15 | 14 | 12 | Previously reported from hospitals in Israel (6) |
| 024 | 008 | 8 | 2 | IV | 2 | 0 | 1 | Presumed EMRSA-2/-6 (8); previously reported from Denmark (21) |
| 032 | S | 22 | 2 | IV | 1 | 1 | 0 | Presumed EMRSA-15; reported from both animals and humans in Germany (34) and the United Kingdom (25, 31) |
| 064 | 008 | 8 | 1 | IV | 1 | 1 | 1 | Previously isolated from domestic animals in Europe (20, 25, 32) |
| 065 | 065/004 | 45 | 8 | IV | 6 | 4 | 4 | Previously reported from Israel (1, 28, 29), presumed Berlin clone (12) |
| 105 | 002 | 5 | 1 | V | 1 | 0 | 1 | Novel report* |
| 127 | S | 1 | 1 | IV | 0 | 1 | 1 | Associated with pigs in Europe (16) |
| 159 | S | 121 | 1 | IV | 1 | 1 | 1 | Novel report* |
| 214 | 002 | 5 | 1 | V | 1 | 0 | 1 | Novel report* |
| 223 | 223/8610 | 22 | 1 | IV | 1 | 0 | 1 | Presumed EMRSA-15 (24) |
| 242 | 002 | 5 | 1 | V | 0 | 1 | 0 | Novel report* |
| 318 | 037 | 30 | 1 | IV | 0 | 0 | 1 | A pvl-positive strain, previously reported from Israel (1) and Shanghai (19) |
| 437 | S | 59 | 1 | V | 0 | 1 | 1 | A pvl-positive strain, commonly reported from Taiwan (5). |
| 509 | 002 | 5 | 1 | IV | 0 | 0 | 0 | Presumed “pediatric clone,” reported from France (9) |
| 570 | 002 | 5 | 1 | V | 1 | 0 | 1 | Novel report* |
| 723 | 008 | 8 | 1 | V | 0 | 0 | 0 | Novel report* |
| 796 | S | 7 | 1 | IV | 1 | 0 | 0 | Previously reported from China (22) |
| 991 | E | 913 | 4 | IV | 3 | 4 | 4 | A common colonizing strain in Bedouin children in Israel (1) |
| 1774 | 008 | 8 | 1 | IV | 1 | 0 | 1 | Previously reported from South Africa (26) |
| 1911 | 008 | 8 | 1 | IV | 1 | 1 | B | Novel report* |
| 2849 | 008 | 2 | IV/V | 2 | 1 | 2 | Novel report* | |
| 5160 | 008 | 1 | IV | 0 | 1 | 1 | Novel report* | |
| 6274 | 065/004 | 1 | IV | 0 | 1 | 0 | Novel report* | |
| 7544 | 002 | 1 | V | 0 | 0 | 1 | Novel report* | |
Data are presented for all strains excluding t008 strains (Fig. 1) and single-isolate, SCCmec type I to III spa types (n = 23). Additional spa-CC-002 strains presented in Fig. S1 in the supplemental material are t088, t105, t2358, and t5712.
spa CC, spa clonal complex. E, excluded from spa BURP analysis due to low (5>) repeat number; S, singleton on spa BURP analysis.
MLST CC, multilocus sequence typing clonal complex (presumed allocation was based on the Ridom spa database and Monecke et al. (24), unless specified otherwise in the comment).
Clin S, isolates tested that were susceptible to clindamycin.
CA, community associated (cultured within 3 days of admission to the hospital).
W, wound (cultured from wound or other nonblood sites).
An asterisk designates either a newly reported spa type or a new spa-SCCmec type combination.
Two isolates harbored SCCmec-II.
One isolate harbored SCCmec-I.
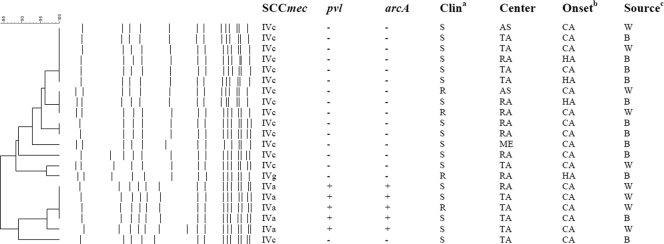
Fig 1.
Molecular features of spa-t008 strains in Israel. Molecular, microbiological, and epidemiological features of 21 spa-t008 isolates in Israel. The SCCmec type, presence (+) or absence (−) of the pvl and arcA genes, clindamycin drug resistance, center, onset, and source are shown in the columns to the right of the dendrogram and PFGE pattern. The Clina column shows the resistance (R) or susceptibility (S) of the strain to clindamycin. The Onsetb column shows whether the isolate was community associated (CA) (cultured within 3 days of admission to the hospital) or health care associated (HA) (cultured from ≥3 days of admission to the hospital). The Sourcec column shows whether the isolates were derived from blood cultures (B) or wound cultures (W).
The 21 t008 strains were divided into 2 major types (Fig. 1): (i) SCCmec-IVa, a pvl- or arcA-positive strain, identified as MRSA strain USA-300 by PFGE; (ii) SCCmec-IVc (1 isolate was SCCmec-IVg), a pvl- or arcA-negative strain, that possessed a PFGE pattern similar to that of a previously described Israeli t008 strain (30) presumably similar to the EMRSA-2/-6 clone (8). All 5 USA-300 isolates and 12 of 16 pvl-negative t008 isolates were categorized as CA based on the time of isolation.
Six novel MRSA spa types were found and nine isolates were typed with a unique combination of spa and SCCmec types (Table 2).
Almost all MRSA SCCmec-I to -III strains (Table 2) were resistant to clindamycin, whereas strains harboring SCCmec-IV/V were mostly susceptible (55/85). In contrast, most isolates were susceptible to rifampin (95%) and trimethoprim-sulfamethoxazole (98%); all strains were susceptible to vancomycin.
The present study is the first national survey of MRSA strains in Israel. The most common MRSA strains in our study were found to be related to common epidemic clones, such as the Southern Germany (t001/SCCmec-I), New York/Japan (t002/SCCmec-II), Berlin (t065/SCCmec-IV), EMRSA-2/-6 (t008/024/SCCmec-IV or pvl negative) and Iberian (t051/052/SCCmec-I) (8, 12) clones; these clones have been previously identified in Israel (1, 6). Interestingly, these clones were absent or rarely reported in a recent study from Lebanon (35).
Three pvl-producing strains were identified. The t318-SCCmec-IV strain (n = 1) has been previously reported from Israel (1) and other Middle Eastern countries (3, 14). t437-SCCmec-V, a common strain in Taiwan (5), has never been reported from our region, and USA-300 MRSA infections have never been reported from other countries in our region (3, 14, 35). Although we are not able to determine whether these USA-300 MRSA infections were acquired in Israel, these cases together with the recent report of a USA-300 MRSA infection in an Israeli child (17) suggest that this strain is likely to be present at a low level in our population.
The proportions of SCCmec types IV and V were 16% and 11%, respectively. The proportion of SCCmec-V is relatively high compared with other countries, as was also reported in a recent study from Israel (2). Many of the SCCmec-IV/V strains identified in our study are uniquely reported from Israel (Table 2). Four t991 SCCmec-IV isolates were cultured from wounds within 3 days of admission to the hospital, from AS, TA, and RA, indicating the presence of this clone in communities outside the Bedouin population in the Negev desert in Israel (1). t002-SCCmec-V, the most common SCCmec-IV/V-harboring strain in our study, is reported uniquely from Israel (6) and was isolated after at least 72 h of admission to the hospital in 14 out of 28 cases. Altogether, SCCmec-IV/V-harboring strains were isolated after at least 72 h of admission to the hospital in 35 of 85 cases, indicating that the SCCmec-IV/V types, cannot serve as a marker for community acquisition in Israel (11).
Our study has several limitations concerning the extent of representation of the MRSA population in Israel. First, our collection lacked a significant number of isolates from certain parts of the country, such as Jerusalem and the Galilee region. Second, despite the collection instructions, the proportion of wound and blood culture isolates was not even in all centers. Third, we are not able to determine how many of the isolates labeled as CA represented actual community-acquired infection, as many of the patients from whose specimens these isolates grew may have been recently hospitalized.
Our study delineates molecular epidemiological features that our MRSA isolates have in common with those in other parts of the world, while highlighting unique features of the Israeli isolates. With global transmission of MRSA (33), our local epidemiology may be changing due to the introduction of new strains.
Supplementary Material
Footnotes
Published ahead of print 26 October 2011
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1. Adler A, Givon-Lavi N, Moses AE, Block C, Dagan R. 2010. Carriage of community-associated methicillin-resistant Staphylococcus aureus in a cohort of infants in southern Israel: risk factors and molecular features. J. Clin. Microbiol. 48:531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alon D, Abd-Elkadir F, Chowers M, Paitan Y. 2011. MRSA SCCmec epidemiology in Israel: development and implementation of an MRSA SCCmec typing strategy. Eur. J. Clin. Microbiol. Infect. Dis. 30:1443–1452 [DOI] [PubMed] [Google Scholar]
- 3. Alp E, et al. 2009. MRSA genotypes in Turkey: persistence over 10 years of a single clone of ST239. J. Infect. 58:433–438 [DOI] [PubMed] [Google Scholar]
- 4. Borg MA, et al. 2007. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) in invasive isolates from southern and eastern Mediterranean countries. J. Antimicrob. Chemother. 60:1310–1315 [DOI] [PubMed] [Google Scholar]
- 5. Chen F-J, Hiramatsu K, Huang I-W, Wang C-H, Lauderdale T-LY. 2009. Panton-Valentine leukocidin (PVL)-positive methicillin-susceptible and resistant Staphylococcus aureus in Taiwan: identification of oxacillin-susceptible mecA-positive methicillin-resistant S. aureus. Diagn. Microbiol. Infect. Dis. 65:351–357 [DOI] [PubMed] [Google Scholar]
- 6. Chmelnitsky I, et al. 2008. SCCmec and spa types of methicillin-resistant Staphylococcus aureus strains in Israel. Detection of SCCmec type V. Eur. J. Clin. Microbiol. Infect. Dis. 27:385–390 [DOI] [PubMed] [Google Scholar]
- 7. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. Approved standard MS100-S20. Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 8. Cookson BD, et al. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 45:1830–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dauwalder O, et al. 2008. Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J. Clin. Microbiol. 46:3454–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. David MZ, Daum RS. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 23:616–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. David MZ, et al. 2008. What is community-associated methicillin-resistant Staphylococcus aureus? J. Infect. Dis. 197:1235–1243 [DOI] [PubMed] [Google Scholar]
- 12. Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13:222–235 [DOI] [PubMed] [Google Scholar]
- 13. Diep BA, et al. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 14. Enany S, Yaoita E, Yoshida Y, Enany M, Yamamoto T. 2010. Molecular characterization of Panton-Valentine leukocidin-positive community-acquired methicillin-resistant Staphylococcus aureus isolates in Egypt. Microbiol. Res. 165:152–162 [DOI] [PubMed] [Google Scholar]
- 15. European Antimicrobial Resistance Surveillance System (EARSS) 2009. EARSS annual report 2008. On-going surveillance of S. pneumoniae, S. aureus, E. coli, E. faecium, E. faecalis, K. pneumoniae, P. aeruginosa. Rijksinstituut voor Volksgezondheid en Milieu (RIVM), Bilthoven, The Netherlands. http://www.ecdc.europa.eu/en/activities/surveillance/EARS-Net/Documents/2008_EARSS_Annual_Report.pdf
- 16. Franco A, et al. 2011. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J. Antimicrob. Chemother. 66:1231–1235 [DOI] [PubMed] [Google Scholar]
- 17. Glikman D, Davidson S, Kudinsky R, Geffen Y, Sprecher H. 2010. First isolation of SCCmec IV- and Panton-Valentine leukocidin-positive, sequence type 8, community-associated methicillin-resistant Staphylococcus aureus in Israel. J. Clin. Microbiol. 48:3827–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hallin M, Deplano A, Denis O, De Mendonça R, De Ryck R, Struelens MJ. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45:127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han L-Z. 2010. Panton-Valentine leukocidin-positive MRSA, Shanghai. Emerg. Infect. Dis. 16:4–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huber H, Koller S, Giezendanner N, Stephan R, Zweifel C. 2010. Prevalence and characteristics of meticillin-resistant Staphylococcus aureus in humans in contact with farm animals, in livestock, and in food of animal origin, Switzerland, 2009. Euro Surveill. 15:1–4 [PubMed] [Google Scholar]
- 21. Larsen AR, et al. 2009. Emergence and characterization of community-associated methicillin-resistant Staphyloccocus aureus infections in Denmark, 1999 to 2006. J. Clin. Microbiol. 47:73–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y, et al. 2009. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob. Agents Chemother. 53:512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Milheiriço C, Oliveira DC, de Lencastre H. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: “SCCmec IV multiplex.” J. Antimicrob. Chemother. 60:42–48 [DOI] [PubMed] [Google Scholar]
- 24. Monecke S, et al. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moodley A, et al. 2006. spa typing of methicillin-resistant Staphylococcus aureus isolated from domestic animals and veterinary staff in the UK and Ireland. J. Antimicrob. Chemother. 58:1118–1123 [DOI] [PubMed] [Google Scholar]
- 26. Moodley A, Oosthuysen WF, DusÉ AG, Marais E. 2010. Molecular characterization of clinical methicillin-resistant Staphylococcus aureus isolates in South Africa. J. Clin. Microbiol. 48:4608–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mulvey MR, et al. 2001. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis J. Clin. Microbiol. 39:3481–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Regev-Yochay G, et al. 2006. Prevalence and genetic relatedness of community-acquired methicillin-resistant Staphylococcus aureus in Israel. Eur. J. Clin. Microbiol. Infect. Dis. 25:719–722 [DOI] [PubMed] [Google Scholar]
- 29. Regev-Yochay G, et al. 2005. Methicillin-resistant Staphylococcus aureus in neonatal intensive care unit. Emerg. Infect. Dis. 11:453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwaber MJ, et al. 2011. High prevalence of methicillin-resistant Staphylococcus aureus among residents and staff of long-term care facilities, involving joint and parallel evolution. Clin. Infect. Dis. 53:910–913 [DOI] [PubMed] [Google Scholar]
- 31. Soliman RS, Phillips G, Whitty P, Edwards DH. 2009. Distribution of meticillin-resistant Staphylococcus aureus spa types isolated from health-care workers and patients in a Scottish university teaching hospital. J. Med. Microbiol. 58:1190–1195 [DOI] [PubMed] [Google Scholar]
- 32. Stastkova Z, Karpiskova S, Karpiskova R. 2009. Occurrence of methicillin-resistant strains of Staphylococcus aureus at a goat breeding farm. Vet. Med. 54:419–426 [Google Scholar]
- 33. Stenhem M. 2010. Imported methicillin-resistant Staphylococcus aureus, Sweden. Emerg. Infect. Dis. 16:189–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Strommenger B, et al. 2006. Molecular characterization of methicillin-resistant Staphylococcus aureus strains from pet animals and their relationship to human isolates. J. Antimicrob. Chemother. 57:461–465 [DOI] [PubMed] [Google Scholar]
- 35. Tokajian ST, et al. 2010. Molecular characterization of Staphylococcus aureus in Lebanon. Epidemiol. Infect. 138:707–712 [DOI] [PubMed] [Google Scholar]
- 36. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.