Abstract
Phenotypic and molecular methods were used to characterize the antibiotic resistance of 64 clinical isolates of Staphylococcus haemolyticus. By PCR of the mecA gene, 87% were found to be methicillin resistant. Approximately 55% harbored staphylococcal cassette chromosome mec element (SCCmec) type V, and only one SCCmec type IV. Many isolates (75%) displayed multiresistance, and pulsotype analysis showed a high diversity.
TEXT
Among coagulase-negative staphylococci (CoNS), Staphylococcus haemolyticus is the second most frequently isolated from human blood cultures (18) and has the highest level of antimicrobial resistance (3, 8). Methicillin resistance is conferred by the mecA gene, carried on the staphylococcal cassette chromosome mec element (SCCmec) (12). Eight types (I to VIII) of SCCmec have been assigned for Staphylococcus aureus (11), and SCCmec type V has already been found in CoNS, particularly in S. haemolyticus (13). The increase in the frequency of methicillin-resistant S. haemolyticus as the causal agent of hospital infections and the possibility of emergence of resistance to other antibiotics demand trustworthy characterization of the isolates and an investigation of clonal spreading within hospitals.
In the studies reported here, 64 clinical strains were isolated from patients at Hospital Naval Marcílio Dias, Rio de Janeiro, Brazil, between 2006 and 2008. The strains were isolated from the following clinical infections or sources in 31 males and 33 females: bacteremia (n = 45), skin (n = 2), urine (n = 13), and unknown source (n = 4). The isolates were identified at the hospital laboratory as S. haemolyticus by using the MicroScan WalkAway PC21 panel, and their identification was confirmed by specific PCR (17).
The resistance profiles of the strains for the main antibiotics used in Brazil were determined by disc diffusion tests according to CLSI guidelines (5). However, the mupirocin susceptibility testing was not preconized by CLSI, so the results for this antibiotic were interpreted as previously described (7, 9). The methicillin resistance was also evaluated by other phenotypic methods, such as the MIC for oxacillin (5), the MicroScan, and PCR of the mecA gene (6). The SCCmec type was determined in a multiplex PCR as previously described (14), except that the pair of primers mecI P2 and mecI P3 used as the internal control were replaced by MRS1 and MRS2 (6), which amplify a 154-bp fragment of the mecA gene.
Analysis of genetic relatedness and characterization of isolates using pulsed-field gel electrophoresis (PFGE) of genomic DNA digested with SmaI were carried out as previously described (20). Banding patterns were determined by visual inspection and by using Bionumerics software, version 6.0 (Applied Maths) using the Dice index and the unweighted-pair group method with arithmetic average. Similar PFGE genotypes were defined using a coefficient of similarity of up to 80%, and the subtypes were those with less than five-band variants, as recommended by van Belkun et al. (19).
As shown in Table 1, there was great variation in the antibiotic resistance profiles. In this collection, the strains were resistant to at least one of the antibiotics tested, and susceptibility to vancomycin was observed in all isolates. Moreover, 75% of the isolates were multiresistant, exhibiting resistance to more than three classes of antibiotics. It is important to emphasize that 88% of the strains were classified as methicillin resistant as defined by the cefoxitin disc diffusion assay. When other tests to evaluate methicillin resistance were performed and considering the detection of the mecA gene as definitive, seven discordant results were observed. One result was false negative for all phenotypic methods, and only one for the MicroScan. In fact, the detection of the mecA gene by PCR does not allow a functional characterization of this gene. For instance, some genetic mutations may prevent the production of active proteins. False-positive results were observed too. Five isolates with negative results for the mecA gene displayed controversial results with phenotypic methods. Three of them were classified as resistant only by the MicroScan, one by disc diffusion, and one by all phenotypic tests.
Table 1.
Antibiogram patterns of the 64 S. haemolyticus strains determined by the disc diffusion test
| Antimicrobial agent | No. (%) of S. haemolyticus isolates that were: |
||
|---|---|---|---|
| Susceptible | Intermediate | Resistant | |
| Penicillin | 3 (5) | 0 | 61 (95) |
| Ampicillin | 3 (5) | 0 | 61 (95) |
| Oxacillin | 8 (12) | 0 | 56 (88) |
| Cefoxitin | 8 (12) | 0 | 56 (88) |
| Gentamicin | 16 (25) | 1 (2) | 47 (73) |
| Ciprofloxacin | 18 (28) | 0 | 46 (72) |
| Erythromycin | 21 (33) | 2 (3) | 41 (64) |
| Trimethoprim-sulfamethoxazole | 27 (42) | 3 (5) | 34 (53) |
| Clindamycin | 34 (53) | 0 | 30 (47) |
| Chloramphenicol | 48 (75) | 0 | 16 (25) |
| Tetracycline | 50 (78) | 2 (3) | 12 (19) |
| Rifampin | 54 (84) | 3 (5) | 7 (11) |
| Mupirocin | 58 (90) | 1(2) | 5 (8) |
Among the 56 mecA-positive strains, only 32 (57%) could be assigned to known SCCmec types. Thirty-one isolates (55%) had SCCmec type V, and only one had SCCmec type IV. The SCCmec elements are common among S. haemolyticus isolates, and these microorganisms are considered to be potential SCCmec donors. Evidence has suggested horizontal transfer of SCCmec type V from methicillin-resistant S. haemolyticus to methicillin-susceptible S. aureus, resulting in the creation of a new methicillin-resistant S. aureus (MRSA) clone that could result in a potential outbreak (1). The remaining 24 strains (43%) were nontypeable by the method employed, which can be explained by the presence of novel structures or rearrangements and recombination of the SCCmec (4, 22). The emergence of new variants of the SCCmec element found in this study and the possibility of gene transfer will be further evaluated and characterized.
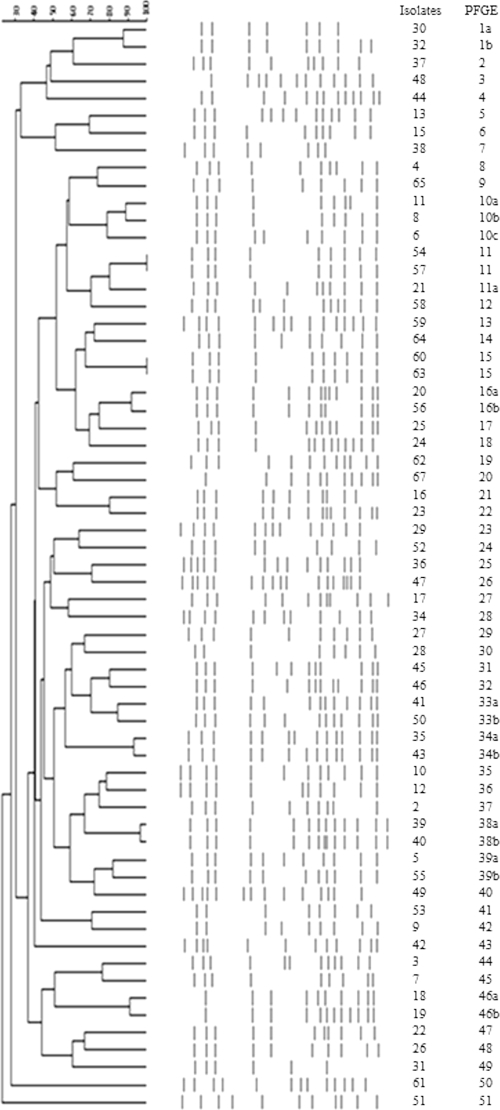
Based on PFGE clustering (Fig. 1), the 64 isolates were typed into 51 PFGE profiles (PFPs). Only two pairs of isolates (strains 54 and 57 and strains 60 and 63) exhibited identical PFPs. There was no single clone with a fixed pulsotype disseminated among these patients, although the strains with different PFPs often harbored the same SCCmec element, type V.
Fig 1.
Dendrogram from computer-assisted analysis of PFGE profiles obtained for 64 S. haemolyticus isolates.
S. haemolyticus is highly prevalent in the hospital environment, with a tendency to develop resistance to multiple antibiotics (15, 21). This was observed in this study, and the highest resistance rates were found for the β-lactams. There are several methods to detect methicillin resistance, and some of them were used here. As in other studies, some false-positive and false-negative results were found. In fact, a novel mecA homologue in S. aureus was described recently which was phenotypically resistant to methicillin but tested negative for the mecA gene. This gene was located in a novel SSCmec designated type XI (10). Resistance to oxacillin, without the mecA gene, may be due to either the overproduction or overexpression of penicillinase or alteration of other penicillin-binding proteins (2). With respect to the SCCmec typing, the results are comparable to the distribution of the SCCmec types among S. haemolyticus strains that appeared to be major reservoirs of type V (13, 16). Besides, the large genetic diversity among the samples, including those resistant to methicillin, highlights the possibility of horizontal spread of the SCCmec among the S. haemolyticus strains. The extreme plasticity of the S. haemolyticus genome was inferred through the complete genome sequencing of strain JCSC1435, which identified as many as 82 insertion sequences in its chromosome. This characteristic may result in frequent genomic rearrangements, phenotypic diversification, and acquisition of antibiotic resistance. This revealed how the medically important staphylococcal species diversify themselves to successfully colonize or infect the human host (18).
In conclusion, our tests showed a high prevalence of multiresistance among S. haemolyticus isolates. The phenotypic tests had good correlation with the genotypic characterization of methicillin resistance, but some discrepant results were observed. SCCmec type V was the most prevalent, although many strains were nontypeable. Despite the great genetic diversity, S. haemolyticus plays an important role as an efficient recipient and/or carrier of the SCCmec elements.
ACKNOWLEDGMENTS
This study was supported by Brazilian grants (from FAPERJ, CNPq, CAPES, and PRONEX).
Footnotes
Published ahead of print 5 October 2011
REFERENCES
- 1. Berglund C, Söderquist B. 2008. The origin of a methicillin-resistant Staphylococcus aureus isolate at a neonatal ward in Sweden—possible horizontal transfer of a staphylococcal cassette chromosome mec between methicillin-resistant Staphylococcus haemolyticus and Staphylococcus aureus. Clin. Microbiol. Infect. 14:1048–1056 [DOI] [PubMed] [Google Scholar]
- 2. Caierão J, et al. 2004. Evaluation of phenotypic methods for methicillin resistance characterization in coagulase-negative staphylococci (CNS). J. Med. Microbiol. 53:1195–1199 [DOI] [PubMed] [Google Scholar]
- 3. Chiew YF, et al. 2007. Detection of vancomycin heteroresistant Staphylococcus haemolyticus and vancomycin intermediate resistant Staphylococcus epidermidis by means of vancomycin screening agar. Pathology 39:375–377 [DOI] [PubMed] [Google Scholar]
- 4. Chung M, Dickinson G, De Lencastre H, Tomasz A. 2004. International clones of methicillin-resistant Staphylococcus aureus in two hospitals in Miami, Florida. J. Clin. Microbiol. 42:542–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. CLSI document M100-S20. CLSI, Wayne, PA [Google Scholar]
- 6. Del Vecchio VG, et al. 1995. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J. Clin. Microbiol. 33:2141–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Oliveira NE, Cardozo AP, Marques EA, dos Santos KRN, Giambiagi-deMarval M. 2007. Interpretative criteria to differentiate low and high level mupirocin resistance in Staphylococcus aureus. J. Med. Microbiol. 56:937–939 [DOI] [PubMed] [Google Scholar]
- 8. Froggatt JW, Johnston JL, Galetto DW, Archer GL. 1989. Antimicrobial resistance in nosocomial isolates of Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 33:460–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuchs PC, Jones RN, Barry AL. 1990. Interpretative criteria for disk diffusion susceptibility testing of mupirocin, a topical antibiotic. J. Clin. Microbiol. 28:608–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. García-Álvarez LG, et al. 2011. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11:595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ito T, Katayama Y, Hiramatsu K. 1999. Cloning and nucleotide sequence determination of the entire mec DNA of pre-methicillin-resistant Staphylococcus aureus N315. Antimicrob. Agents Chemother. 43:1449–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ito T, et al. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Milheiriço C, Oliveira DC, Lencastre H. 2007. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3374–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Monsen T, Karlsson C, Wiström J. 2005. Spread of clones of multidrug-resistant, coagulase-negative staphylococci within a university hospital. Infect. Control Hosp. Epidemiol. 26:76–80 [DOI] [PubMed] [Google Scholar]
- 16. Ruppé E, et al. 2009. Diversity of staphylococcal cassette chromosome mec structures in methicillin-resistant Staphylococcus epidermidis and Staphylococcus haemolyticus strains among outpatients from four countries. Antimicrob. Agents Chemother. 53:442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schuenck RP, Pereira EM, Iorio NL, dos Santos KRN. 2008. Multiplex PCR assay to identify methicillin-resistant Staphylococcus haemolyticus. FEMS Immunol. Med. Microbiol. 52:431–435 [DOI] [PubMed] [Google Scholar]
- 18. Takeuchi F, et al. 2005. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing staphylococcal species. J. Bacteriol. 187:7292–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. van Belkun A, et al. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl. 3):1–46 [DOI] [PubMed] [Google Scholar]
- 20. Vivoni AM, et al. 2005. Mupirocin for controlling methicillin-resistant Staphylococcus aureus: lessons from a decade of use at a university hospital. Infect. Control. Hosp. Epidemiol. 26:662–667 [DOI] [PubMed] [Google Scholar]
- 21. Yu MH, Chen YG, Yu YS, Chen CL, Li LJ. 2010. Antimicrobial resistance and molecular characterization of Staphylococcus haemolyticus in a Chinese hospital. Eur. J. Clin. Microbiol. Infect. Dis. 29:613–616 [DOI] [PubMed] [Google Scholar]
- 22. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. 2005. Novel multiplex PCR assay for characterization and concomitant sub-typing of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026–5033 [DOI] [PMC free article] [PubMed] [Google Scholar]