Abstract
Diagnostic tests for respiratory viral infections have traditionally been performed on nasopharyngeal swabs or washings. Reverse transcriptase PCR (RT-PCR) is rapid, sensitive, and specific for viral infection diagnosis but is rarely applied to sputum samples. Thus, we evaluated the diagnostic yield of RT-PCR for detection of nine virus types by the use of nose and throat swabs (NTS) and sputum samples from patients admitted to the hospital with acute respiratory tract illnesses. Adults hospitalized with acute respiratory tract illnesses were recruited during the winters of 2008 and 2009. At enrollment, combined nose and throat swabs and sputum samples were collected for RT-PCR for detection of nine common respiratory virus types. A total of 532 subjects admitted for 556 respiratory illnesses were enrolled. A total of 189 virus strains were identified. The diagnostic yields for detection of any virus were 23% (126/556) for NTS RT-PCR and 36% (146/404) for sputum RT-PCR. A total of 83 (44%) of 189 viral detections were positive by both methods, 43 (23%) were positive by NTS alone, and 63 (33%) were positive only with sputum samples. The inclusion of RT-PCR performed with sputum samples significantly increased the diagnostic yield for respiratory viral infections in adults. Further studies designed to adapt the use of sputum samples for commercial RT-PCR respiratory virus assays are needed.
INTRODUCTION
The diagnosis of respiratory viral infections has traditionally been performed by viral culture or antigen testing of nasopharyngeal swabs or washings (26). Although nasal washes offer a better yield than swabs for viral infection diagnosis, this procedure is poorly tolerated and rarely used with hospitalized adults (11, 27). In addition, nasopharyngeal sampling using thin flexible applicators can be technically difficult to perform and nasal swabs with rigid cotton applicators have been found to be more acceptable to ill adult patients (17). Compounding the difficulty of viral infection diagnosis in adults is the insensitivity of viral culture and antigen testing in this population (6, 9, 25). New sensitive and specific molecular methods such as reverse transcriptase PCR (RT-PCR) have greatly improved the accuracy of viral testing and, because results are available within hours, can directly affect patient care (7, 14, 28). Lower-airway secretions collected in the form of bronchoalveolar lavage (BAL) fluid have been recognized as important for viral infection diagnosis for immunocompromised patients and persons infected with severe acute respiratory syndrome (SARS) coronavirus and H1N1 2009 influenza virus (2, 8, 19, 20). Although adequate sputum samples are sufficiently representative of the lower airways and can be collected by noninvasive means, they are rarely used for viral culture or antigen testing (16). This is likely due to the viscous consistency of sputum, which makes samples difficult to process. Toxic effects on cell culture and overgrowth of bacteria also limit its use for traditional viral tests. However, these issues should not preclude testing sputum for the presence of viral RNA by RT-PCR. Thus, we evaluated the diagnostic yield of RT-PCR for nine virus types in nose and throat swabs (NTS) and sputum samples from patients admitted to the hospital with acute respiratory tract illnesses.
MATERIALS AND METHODS
Patient population.
Adults over 21 years of age admitted to Rochester General Hospital (RGH) with a diagnosis at admission that was potentially compatible with acute respiratory tract infection (community-acquired pneumonia [CAP], acute exacerbation of chronic obstructive pulmonary disease [COPD], acute bronchitis, asthma, upper respiratory tract infection, influenza, viral syndrome, respiratory failure, and congestive heart failure [CHF] precipitated by infection) were recruited from 1 November to 30 May during the 2008 to 2009 and 2009 to 2010 seasons. Patients were screened within 24 h of admission, each subject or a legal guardian provided written informed consent, and the institutional review boards of the University of Rochester and Rochester General Hospital approved the study.
Illness evaluation.
At enrollment, demographic, clinical, and laboratory information was collected. Nose and throat swabs (NTS) were collected by sequentially sampling each naris with a single sterile cotton swab, sampling the throat with a second swab, and placing the two swabs in a single tube containing 3 ml of viral transport media. The nasal sample was obtained by inserting the swab approximately 1 in. into the nasal cavity and rubbing firmly in a circular motion on the nasal septum and lateral wall of the nasal cavity for 5 s each. An attempt was made to collect an adequate sputum sample as soon as possible once a patient was entered into the study. When patients had a productive cough but were unable to expectorate, sputum was induced with normal saline solution and bronchodilators.
Laboratory methods. (i) Reverse transcriptase PCR (RT-PCR) assays.
For sample extraction, sputum was diluted with an equal volume of distilled water and subjected to a vortex procedure for 1 to 5 min, depending on the sputum viscosity prior to extraction. RNA was extracted from 250 μl of NTS or sputum samples by the use of a commercial phenol-chloroform preparation (LS Stat; Tel Test, Friendswood, TX).
(ii) Real-time RT-PCR.
For each virus, extracted RNA was converted to cDNA by the use of RT, a conserved forward primer, and nucleotides (deoxynucleoside triphosphates [dNTPs]). The product was then treated with uracil N-glyconase (UNG). The treated cDNA was placed in a PCR tube with a conserved reverse primer, dNTPs (with uracil replacing thymidine), Taq polymerase, and a fluorescent labeled probe. Reactions were run using conditions specific for each of the primer-probe combinations. Relative fluorescent units (RFU) were monitored during each cycle at 490 μm (6-carboxyfluorescein [FAM]) and 580 μm (Texas Red). The cycle threshold (CT) was determined when the RFU exceeded 10× the standard deviation of the baseline RFU values. Primer and probe sequences are as follows: for influenza A virus (M gene), the forward primer sequence was 5′ GACCRATCCTGTCACCTCTGAC 3′, the reverse primer sequence was 5′ AGGGCATTYTGGACAAAKCGTCTA 3′, and the probe sequence was 5′ TGCAGTCCTCGCTCACTGGGCACG 3′; for influenza B virus (NS1 gene), the forward primer sequence was 5′ TCCTCAAYTCACTCTTCGAGCG 3′, the reverse primer sequence was 5′ CGGTGCTCTTGACCAAATTGG 3′, and the probe sequence was 5′ CCAATTCGAGCAGCTGAAACTGCGGTG 3′; for respiratory syncytial virus (RSV A) (F gene), the forward primer sequence was 5′ CACCCTGTTGGAAAC 3′, the reverse primer sequence was 5′ CTCTGTCAGTTCTTG 3′, and the probe sequence was 5′ ATGTTGGACCCTTCTTTTGTGTTGGTTGTA 3′; for RSV B (F gene), the forward primer sequence was 5′ CACCTTGCTGGAAAT 3′, the reverse primer sequence was 5′ CTCTGTCAGTTCTTG 3′, and the probe sequence was 5′ ATATTTGATCCTTCTTTGATGTTGGTGGTG 3′; for human metapneumovirus (hMPV) (N gene), the forward primer sequence was 5′ TCTCTTCAAGGGATTCA 3′, the reverse primer sequence was 5′ AATGATGAAGGTGTCAC 3′, and the probe sequence was 5′ TCATACAAGCATGCTATATTAAAAGAGTCTCAGTA 3′; for coronavirus OC43 (pol gene), the forward primer sequence was 5′ AATACCTTTCTTGGCTCGAGTAAT 3′, the reverse primer sequence was 5′ GTGGATTCTGCTCAAG 3′, and the probe sequence was 5′ TACAGAATGCGCTGTTTCTGCAGTCTGTGAAT 3′; for coronavirus 229E (pol gene), the forward primer sequence was 5′ AATACCTTTCTTTGC TCTAGTAAT 3′, the reverse primer sequence was 5′ GTGGATTCTGCTCAAG 3′, and the probe sequence was 5′ ACAGGCATGAGCAGTATCTGATGTCTGTGCGA 3′; for parainfluenza virus 1 (PIV1) (N gene), the forward primer sequence was 5′ CAAGTTTGGATAGTGTTGG 3′, the reverse primer sequence was 5′ AAGACCAGGGCAC 3′, and the probe sequence was 5′ CGGTTCCATCCTGTCTGAATGCTTC 3′; for PIV2 (N gene), the forward primer sequence was 5′ GACAGTGATGGTGAG 3′, the reverse primer sequence was 5′ GGTTGTTTGGTTGTCCA 3′, and the probe sequence was 5′ CTCTCCTCTATTCTCTCTGATAGCTTGATC3′; and for PIV3 (N gene), the forward primer sequence was 5′ TCAGAGACATCTTTCCAC 3′, the reverse primer sequence was 5′ ATTGTTCTGGCTCTTCA 3′, and the probe sequence was 5′ CTATTGCCATCTCTATGGCTGATCC 3′.
RESULTS
During two winters from 2008 to 2010, 532 subjects admitted for 556 respiratory illnesses were enrolled. The subjects averaged 65 years of age, and a high percentage had chronic underlying diseases such as diabetes (37%), congestive heart failure (29%), and COPD (40%). The leading primary admission diagnoses were COPD exacerbation (28%), pneumonia (25%), bronchitis (16%), asthma (15%), and CHF (8%).
An NTS sample was obtained for all 556 illnesses, and 404 (73%) sputum samples were available for analysis. A viral infection was identified in 183 of 556 (33%) illnesses. Of the 183 illnesses associated with viral infection, a single virus was identified in 177 cases and 6 were associated with two viruses. Thus, a total of 189 viruses were identified, as shown in Table 1.
Table 1.
Distribution of viral infection diagnoses in 556 respiratory illnesses
| Virus | No. of positive results by any test |
|---|---|
| Influenza A | 43 |
| Influenza B | 4 |
| Respiratory syncytial virus (RSV) | 43 |
| Parainfluenza (PIV)1 | 3 |
| Parainfluenza (PIV) 2 | 4 |
| Parainfluenza (PIV) 3 | 12 |
| Human metapneumovirus (hMPV) | 30 |
| Coronavirus OC43 | 38 |
| Coronavirus 229E | 12 |
| Total | 189 |
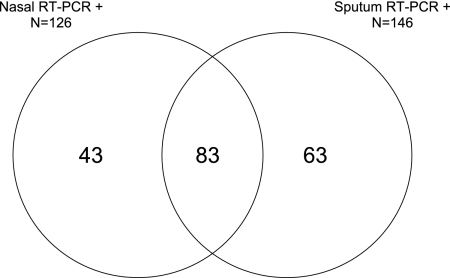
The diagnostic yields for any virus detected were 23% (126/556) for NTS RT-PCR and 36% (146/404) for sputum RT-PCR. Of 189 viral detections, 83 (44%) were positive by both methods, 43 (23%) were positive by NTS alone, and 63 (33%) were positive only by sputum RT-PCR (Fig. 1). The distributions of positive samples were similar for individual virus types, ranging from 45% to 75% of NTS tested and 86% to 100% of sputum samples tested (Table 2). The majority of those positive by NTS alone (33/43 [77%]) did not have sputum available for testing. In contrast, when sputum was available, it was rarely RT-PCR negative for those with identified viral infections (10/156 [6%]). Importantly, RT-PCR was positive only for sputum from 14 of 43 (33%) influenza A virus infections and from 2 of 4 (50%) influenza B virus infections. The diagnostic yields of NTS and sputum RT-PCR were not significantly different for seasonal compared to pandemic influenza (data not shown).
Fig 1.
Distribution and overlap of positive diagnostic tests for viral infection diagnosis. Each section of the diagram indicates the number of distinct viral infection diagnoses that were positive for each of the viral assays (NTS RT-PCR and sputum RT-PCR).
Table 2.
Numbers and percentages of positive RT-PCR results by sample for each virus type
| Assay | No. of positive results/total no. of samples tested (%)a |
|||||||
|---|---|---|---|---|---|---|---|---|
| Flu A (n = 43) | Flu B (n = 4) | RSV (n = 43) | PIV (n = 19) | hMPV (n = 30) | OC43 (n = 38) | 229E (n = 12) | Total (n = 189) | |
| NTS PCR | 29/43 (67) | 2/4 (50) | 34/43 (79) | 15/19 (79) | 21/30 (70) | 17/38 (45) | 8/12 (67) | 126/189 (67) |
| Sputum PCR | 34/36 (94) | 3/3 (100) | 31/32 (97) | 16/17 (89) | 24/27 (89) | 32/34 (94) | 6/7 (86) | 146/156 (94) |
Flu A, influenza A virus; Flu B, influenza B virus; OC43, coronavirus OC43; 229E, coronavirus 229E.
Overall, patient and clinical characteristics determined for illnesses associated with a positive NTS RT-PCR result were similar to those determined for illnesses associated with negative NTS and positive sputum RT-PCR, although upper respiratory symptoms (nasal congestion, sore throat, or hoarseness) and a duration of symptoms of less than 1 week prior to evaluation were more common in the NTS RT-PCR-positive group (Table 3). There was a trend toward greater inhaled steroid use and concomitant bacterial infection in the group with solely sputum-positive test results compared to those with positive NTS RT-PCR results, but rates of smoking and chronic obstructive pulmonary disease (COPD) were not different.
Table 3.
Clinical and illness characteristics of patients with RT-PCR-positive diagnoses
| Characteristic | RT-PCR (NTS positive) (n = 124)a | RT-PCR (sputum positive only) (n = 59) | P value |
|---|---|---|---|
| Age (mean ± SD) | 66.4 ± 17.3 | 62.0 ± 17.9 | 0.11 |
| No. (%) of women | 72 (58) | 38 (64) | 0.43 |
| No. (%) with COPD | 43 (35) | 25 (42) | 0.33 |
| No. (%) of smokers | 82 (66) | 43 (73) | 0.40 |
| No. (%) administered systemic corticosteroids | 13 (10) | 8 (14) | 0.62 |
| No. (%) administered inhaled corticosteroids | 39 (31) | 27 (46) | 0.07 |
| No. (%) with upper respiratory tract symptom(s) | 106 (85) | 40 (68) | 0.01 |
| Dyspnea | 115 (93) | 54 (92) | 0.77 |
| Sputum production | 100 (81) | 49 (83) | 0.84 |
| Pneumonia | 32 (26) | 14 (24) | 0.86 |
| Bacterial infection | 21 (17) | 17 (29) | 0.08 |
| No. (%) under intensive care | 12 (10) | 8 (14) | 0.45 |
| No. (%) deceased | 2 (0.02) | 2 (0.03) | 0.60 |
| Duration of symptoms (mean ± SD) | 5.1 ± 4.5 | 5.7 ± 5.2 | 0.42 |
| Duration of symptoms > 7 days | 22 (18) | 20 (34) | 0.02 |
n, number of illnesses. Some illnesses were associated with more than one virus. The NTS-positive group included subjects with positive and negative sputum RT-PCR results.
DISCUSSION
Viral infection diagnostics have been revolutionized by the development of RT-PCR, and the substantial burden of these ubiquitous pathogens is now being increasingly recognized (1, 5, 15, 28, 29). However, some fundamental issues, including the utility of swabs versus washes, nose versus throat versus nasopharyngeal samples, and flocked versus regular swabs, remain regarding the determination of the optimal specimens for viral testing (24). Throughout those debates, the use of sputum has rarely been considered and none of the currently available commercial RT-PCR assays for respiratory virus detection are approved for sputum testing (10). The observation that viruses can be detected in sputum by RT-PCR has been previously reported from studies of patients with COPD and asthma (3, 12, 13, 22, 23, 30). However, the use of sputum as a diagnostic tool for patient care has likely been ignored because of difficulties in procuring adequate samples and because of its highly viscous nature, which makes processing difficult. The fact that sputum was obtained in only 73% of cases highlights the difficulties presented by sputum collection. Yet our data show that these difficulties are not insurmountable and that the added diagnostic yield of sputum can be substantial if it can be collected.
The association of a positive NTS RT-PCR result with the presence of upper respiratory tract symptoms and a shorter duration of illness prior to evaluation suggests that our findings reflect a progression of virus from the upper to the lower airways rather than sampling issues or random detection of low levels of RNA. It is possible that the decreased sensitivity of NTS RT-PCR in hospitalized patients is due in part to drying of the nasal mucosa secondary to nasal prong oxygen administration.
The accurate diagnosis of influenza is immediately useful for appropriate isolation and treatment of hospitalized patients. During the H1N1 2009 influenza pandemic, a number of investigators reported the utility of RT-PCR testing using lower respiratory tract samples from patients with negative nasopharyngeal swabs (4, 21). In most instances, BAL fluid or endotracheal secretions were tested. Since the majority of influenza patients do not require intubation or bronchoscopy, our data suggest that sputum is a reasonable alternative. The identification of specific viral infections other than influenza virus infections is also important for the institution of appropriate infection control policies for immunocompromised patients, particularly in hospital settings. Lastly, accurate detection of all viruses is important for epidemiologic research and vaccine and therapeutic trials.
Our data indicate that NTS and sputum testing yield complementary results. The most cost-effective method of viral testing would likely involve combining samples rather than performing separate RT-PCR assays. It might be possible to use the viral transport media from the NTS swab to dilute the sputum sample. Alternatively, a swab might be dipped in the sputum sample and combined in the viral transport media with the NTS. In acute care settings, simple procedures are most likely to be implemented; thus, further studies of combined sample testing and specimen processing are needed.
Our study has several limitations. Quantitative RT-PCR data would have been useful to help address the question of active replication in the lower airways versus low residual levels of RNA from a prior infection. We did not test for rhinovirus or adenoviruses, and it is possible that testing for these viruses would have provided different results for the yields of NTS and sputum RT-PCR. Finally, the use of flocked swabs for collection of nasal secretions might have increased the yield from NTS specimens, resulting in fewer significant differences between NTS and sputum samples (18).
In conclusion, we found that inclusion of RT-PCR using sputum samples significantly increased the diagnostic yield for respiratory viral infections. Our data indicate that RT-PCR testing of sputum has merit and should be considered in studies of respiratory illnesses. As RT-PCR assays are validated and become commercially available, better methods to process and test sputum would be worthwhile.
ACKNOWLEDGMENTS
We thank Patricia Hennessey and Mary Criddle for specimen collection and Jamie Biear for sample processing.
A. R. Falsey has served as a consultant for sanofipasteur, GSK Biologics, Medimmune, AstraZeneca, and Novartis. E. E. Walsh has served as a consultant for Novartis, Alnylam, AstraZeneca, Medimmune, and Boehringer Ingelheim. M. A. Formica has no conflicts to declare.
This study was supported by the NIAID (grant 1R01AI079446-01).
Footnotes
Published ahead of print 16 November 2011
REFERENCES
- 1. Beckham JD, et al. 2005. Respiratory viral infections in patients with chronic obstructive pulmonary disease. J. Infect. 50:322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blyth CC, Iredell JR, Dwyer DE. 2009. Rapid-test sensitivity for novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 361:2493 doi:10.1056/NEJMc0909049 [DOI] [PubMed] [Google Scholar]
- 3. Borg I, et al. 2003. Evaluation of quantitative real-time PCR for the detection of respiratory syncytial virus in pulmonary diseases. Eur. Respir. J. 21:944–951 [DOI] [PubMed] [Google Scholar]
- 4. Cao B, et al. 2009. Clinical features of the initial cases of 2009 pandemic influenza A (H1N1) virus infection in China. N. Engl. J. Med. 361:2507–2517 doi:10.1056/NEJMoa0906612 [DOI] [PubMed] [Google Scholar]
- 5. Caram LB, et al. 2009. Respiratory syncytial virus outbreak in a long-term care facility detected using reverse transcriptase polymerase chain reaction: an argument for real-time detection methods. J. Am. Geriatr. Soc. 57:482–485 doi:10.1111/j.1532–5415.2008.02153.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Casiano-Colón AE, Hulbert BB, Mayer TK, Walsh EE, Falsey AR. 2003. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. J. Clin. Virol. 28:169–174 doi:S1386653203000027 [pii]. [DOI] [PubMed] [Google Scholar]
- 7. Dae Serres G, et al. 2009. Importance of viral and bacterial infections in chronic obstructive pulmonary disease exacerbations. J. Clin. Virol. 46:129–133 doi:10.1016/j.jcv.2009.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Englund JA, et al. 1996. Rapid diagnosis of respiratory syncytial virus infections in immunocompromised adults. J. Clin. Microbiol. 34:1649–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Falsey AR, McCann RM, Hall WJ, Criddle MM. 1996. Evaluation of four methods for the diagnosis of respiratory syncytial virus infection in older adults. J. Am. Geriatr. Soc. 44:71–73 [DOI] [PubMed] [Google Scholar]
- 10. Ginocchio CC. 2011. Strengths and weaknesses of FDA-approved/cleared diagnostic devices for the molecular detection of respiratory pathogens. Clin. Infect. Dis. 52(Suppl 4):S312–S325 doi:10.1093/cid/cir046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gooskens J, Swaan CM, Claas EC, Kroes AC. 2008. Rapid molecular detection of influenza outbreaks in nursing homes. J. Clin. Virol. 41:7–12 doi:10.1016/j.jcv.2007.10.023 [DOI] [PubMed] [Google Scholar]
- 12. Harju TH, et al. 2006. Pathogenic bacteria and viruses in induced sputum or pharyngeal secretions of adults with stable asthma. Thorax 61:579–584 doi:10.1136/thx.2005.056291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Horn ME, Reed SE, Taylor P. 1979. Role of viruses and bacteria in acute wheezy bronchitis in childhood: a study of sputum. Arch. Dis. Child. 54:587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kherad O, Rutschmann OT. 2010. Viral infections as a cause of chronic obstructive pulmonary disease (COPD) exacerbation. Praxis (Bern 1994) 99:235–240 doi:10.1024/1661–8157/a000034. (In German.) [DOI] [PubMed] [Google Scholar]
- 15. Kim YJ, Boeckh M, Englund JA. 2007. Community respiratory virus infections in immunocompromised patients: hematopoietic stem cell and solid organ transplant recipients, and individuals with human immunodeficiency virus infection. Semin. Respir. Crit. Care Med. 28:222–242 doi:10.1055/s-2007-976494 [DOI] [PubMed] [Google Scholar]
- 16. Kimball AM, et al. 1983. Isolation of respiratory syncytial and influenza viruses from the sputum of patients hospitalized with pneumonia. J. Infect. Dis. 147:181–184 [DOI] [PubMed] [Google Scholar]
- 17. Lieberman D, et al. 2009. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling. J. Clin. Microbiol. 47:3439–3443 doi:10.1128/JCM.00886-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moore C, Corden S, Sinha J, Jones R. 2008. Dry cotton or flocked respiratory swabs as a simple collection technique for the molecular detection of respiratory viruses using real-time NASBA. J. Virol. Methods 153:84–89 doi:10.1016/j.jviromet.2008.08.001 [DOI] [PubMed] [Google Scholar]
- 19. Mulrennan S, et al. 2010. Pandemic influenza (H1N1) 2009 pneumonia: CURB-65 score for predicting severity and nasopharyngeal sampling for diagnosis are unreliable. PLoS One 5:e12849 doi:10.1371/journal.pone.0012849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peiris JSM, Yuen KY, Osterhaus AD, Stohr K. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431–2441 [DOI] [PubMed] [Google Scholar]
- 21. Perez-Padilla R, et al. 2009. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N. Engl. J. Med. 361:680–689 doi:10.1056/NEJMoa0904252 [DOI] [PubMed] [Google Scholar]
- 22. Rohde G, et al. 2003. Respiratory viruses in exacerbations of chronic obstructive pulmonary disease requiring hospitalisation: a case-control study. Thorax 58:37–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simpson JL, Moric I, Wark P, Johnston L, Gibson PG. 2003. Use of induced sputum for the diagnosis of influenza and infections in asthma: a comparison of diagnostic methods. J. Clin. Virol. 26:339–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singh K, Vasoo S, Stevens J, Schreckenberger P, Trenholme G. 2010. Pitfalls in diagnosis of pandemic (novel) A/H1N1 2009 influenza. J. Clin. Microbiol. 48:1501–1503 doi:10.1128/JCM.02483-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steininger C, Kundi M, Aberle SW, Aberle JH, Popow-Kraupp T. 2002. Effectiveness of reverse transcription-PCR, virus isolation, and enzyme-linked immunosorbent assay for diagnosis of influenza A virus infection in different age groups. J. Clin. Microbiol. 40:2051–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Storch GA. 2000. Diagnostic virology. Clin. Infect. Dis. 31:739–751 [DOI] [PubMed] [Google Scholar]
- 27. Talbot HK, Falsey AR. 2010. The diagnosis of viral respiratory disease in older adults. Clin. Infect. Dis. 50:747–751 doi:10.1086/650486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Templeton KE, et al. 2005. Improved diagnosis of the etiology of community-acquired pneumonia with real-time polymerase chain reaction. Clin. Infect. Dis. 41:345–351 doi:10.1086/431588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weinberg GA, et al. 2004. Superiority of reverse-transcription polymerase chain reaction to conventional viral culture in the diagnosis of acute respiratory tract infections in children. J. Infect. Dis. 189:706–710 doi:10.1086/381456 [DOI] [PubMed] [Google Scholar]
- 30. Xiang X, et al. 2002. Comparison of three methods for respiratory virus detection between induced sputum and nasopharyngeal aspirate specimens in acute asthma. J. Virol. Methods 101:127–133 [DOI] [PubMed] [Google Scholar]