Abstract
Recent studies revealed that micro RNA-10b (mir-10b) is highly expressed in metastatic breast cancer cells and positively regulates breast cancer cell migration and invasion through inhibition of HOXD10 target synthesis. In this study we designed anti-mir-10b molecules and combined them with poly L-lysine (PLL) to test the delivery effectiveness. An RNA molecule sequence exactly matching the mature mir-10b minor antisense showed strong inhibition when mixed with PLL in a wound-healing assay with human breast cell line MDA-MB-231. The resulting PLL-RNA nanoparticles delivered the anti-microRNA molecules into cytoplasm of breast cancer cells in a concentration-dependent manner that displayed sustainable effectiveness.
Keywords: microRNA-10b, breast cancer metastasis, nanoparticles
Introduction
The most important goal in treating breast cancer patients is to stop metastasis at the source; for example, by surgery to eradicate all cancer cells before they metastasize to distant organs. However, it has been widely reported that metastasis occurs even after surgery. Metastasis is a complex, multi-step process by which primary tumor cells invade adjacent tissue, enter the systemic circulation (intravasate), translocate through the vasculature, arrest in distant capillaries, extravasate into the surrounding tissue (parenchyma), and finally proliferate from microscopic growths (micrometastases) into macroscopic secondary tumors.1–3
Recently, it has become evident that, in addition to alterations in protein-encoding genes, abnormalities in non-coding genes can also contribute to cancer pathogenesis. In particular, a class of small cellular RNAs, termed microRNAs (miRNAs), acting as agents of the RNA interference pathway, can lead to silencing of their cognate target genes, doing so either by cleaving mRNA molecules or by inhibiting their translation.4,5 A recent screening of miRNAs toward inhibition of breast cancer metastasis using in vitro cultivated cells and animal models ended up with several anti-miRNA reagents.4,6,7 For example miRNA-10b (mir-10b), located on Hox gene clusters was identified for its role in positively regulating breast cancer cell migration and invasion by the transcription factor Twist.6 HOXD10 was reported to be a direct and functional target of mir-10b.4,6,8 RNA molecules of anti-mir-10b reduce the invasive properties of MDA-MB-231 breast cancer cells by ten fold.6 These studies established that anti-mir-10b RNA can be used directly to inhibit breast cancer metastasis. However, those foreign RNA molecules cannot be directly taken up by cells; several transfection techniques are applied to ensure successful uptake of these anti-mir RNA reagents into cultured cells or animal tissues.1,3,9 For example, cationic liposomes are widely used for DNA vectors and RNAi delivery, but with low efficiency of transfections. 10 Alternatively, synthetic polymers and nanoparticles have been applied in gene delivery in recent years because of their potential advantages of low immunogenicity.11,12 Poly (L-Lysine) (PLL) has been widely used as a gene delivery carrier because of its excellent nuclei acid condensation property and efficient protection of nuclei acid from the attack of nucleases.11,13–15 In this study, we used PLL (MW 40,000) to deliver anti-mir-RNA-10b molecules into the breast cancer cell line MDA-MB231. We found that unmodified RNA molecules have strong interactions with PLL, and the formed nanoparticles can deliver the RNA into breast cancer cytoplasm in a concentration-dependent manner with sustainable effectiveness.
Materials and Methods
Chemical and reagents
Custom ordered FITC labeled poly L-lysine of molecular weight 40k Da was purchased from Fisher Scientific Inc. 3-(4,5-dimethylthiazolyl-2)- 2,5-diphenyl tetrazolium bromide (MTT) was purchased from Invitrogen (catalog number V13154). Anti-mir-10b RNA major (sequence: 5′-CACAAAUUCGGUU-CUACAGGGUA-3′) and anti-mir-10b RNA minor (sequence: 5′-ACAGAUUCGAUU-CUAGGGGAAU-3′) were ordered from IDT (Coralville, IA). Negative RNA control (catalog AM4611) was ordered from Applied Biosystems (Austin, TX). The RNA cy5 labeling kit (catalog number MIR 3700) was ordered from Mirus Bio LLC (Madison, WI). Breast cancer cell MDA-MB-231 was purchased from ATCC (catalog number HTB-26). DMEM medium and FBS were purchased from ATCC (catalog 30-2008). Antibody for RHOC was purchased from Santa Cruz Biotechnology (catalog sc-26480). Antibody for beta-Actin was purchased from Abcam (catalog ab8229).
Cy5 labeling of RNA
Following the kit instructions, 1 mM RNA ( including major anti-micro-10b, minor anti-micro-10b, and negative RNA) was mixed with 2 mM Cy5 labeling reagent and incubated at room temperature (21 °C ± 1 °C) for 2 hours. The labeled RNA molecules were purified by standard procedures following instructions from the kit. The Cy5-labeled RNA molecules were aliquoted as 10 μM and stored at −80 °C before experiments.
Preparation and characterization of PLL-RNA nanoparticles
Anti-mir-10b RNA molecules and PLL were both dissolved into pure RNAse, DNAse-free water (Fisher Scientific, catalog number BP2484-100) and auto-claved at 121 °C for 30 min before experiments. Multiple tubes containing 0.5 mL of various concentration of FITC labeled PLL (2 μM to 2 mM) were added with a 0.5 mL of constant anti-mir-10b RNA (0.5 mM) solution, then vortexed for 5 min and incubated for 1 hour at room temperature. For the fluorescence measurement, excitation wavelength was at 485 nm, emission wavelength at 516 nm, slit widths at 5 nm, accumulation time of 0.4 sec. Multiple tubes containing 0.5 mL various concentrations (2 μM to 2 mM) of Cy5-labeled anti-mir-10b RNA was added with 0.5 mL fixed concentration of PLL (0.5 mM), then vortexed for 5 minutes and incubated for 1 hour at room temperature. The fluorescence was measured with a Fluoro Max-2 fluoresce photometer with the excitation at 630 nm, emission wavelength at 660 nm, slit widths at 5 nm, accumulation time of 0.4 sec.
Based on the titration of PLL to anti-mir-10b above, the 1:1 molar ratio was used to produce PLL-anti-mir-10b nanoparticles. 2 mM of anti-mir-10b was mixed with an equal volume of 2 mM PLL, and vortexed at room temperature for 5 min. The RNA loading were estimated by measuring the solution absorbance at 260 nm. During the reaction, RNA molecules bound to PLL and formed micelle, no detectable free RNA in the solution can be found by the solution absorbance at 260 nm. The final RNA concentration of stock of RNA-PLL was estimated as 1 mM. The particle size of anti-mir-10b-PLL was analyzed by dynamic light scattering (DLS).
DLS measurements of the nanoparticle size
The dynamic light scattering (DLS) instrument was home-built. A 532 nm laser was focused into a sample cell and the scattering light was collected by an optical fiber. A D532/10X filter (Chroma Technology, Brattleboro, VT) was used to exclude possible autofluorescence. To minimize the effect of after pulse, the scattering light output from the optical fiber was split by a cube beam splitter ( Thorlabs, Newton, NJ), and detected by a pair of SPCM-AQR-14 avalanche photodiodes (Perkin-Elmer Optoelectronics, Vaudreuil, Canada). The output was fed into a Flex01-05D multi-tau correlator (Correlator.com, Bridgewater, NJ), and the correlation function was calculated in real time. An NIST traceable particle size standard (Catalog 64010, Polysciences Inc. Warrington PA) was used to calibrate the setup.
Wound-healing assay
MDA-MB-231 cells were counted and plated 1 × 105/well in 24-well tissue culture plates to achieve 90% confluence. A vertical or horizontal wound was created using a 1000-μL pipette tip. The wounded cells were washed three times with PBS and DMEM medium with 10% FBS, and covered with either anti-mir-10b contained medium or control medium. The cells were then placed onto a Nikon Eclipse TE300 inverted microscope equipped with an environmental chamber (In Vivo Scientific, LLC) maintained at 37 °C for imaging. Random fields in each well were selected for imaging with a Nikon Plan Apo 4X/0.2 objective using a Retiga 1300 cooled CCD camera (QImaging) controlled by Volocity Acquisition (Perkin Elmer) software. The location of the imaged cells were recorded and saved with the Volocity software (Velocity Software, Inc.) allowing the same location to be imaged at 0 and 24 hours. Images were captured at designated times to assess wound closure. The inhibition effectiveness was estimated by the relative distance of wound closure.
Confocal microscope
To obtain 3-dimensional images of the cell uptake of RNA-nanoparticles, we used a Zeiss 710 laser scanning confocal microscope. A 20X NA 1.0 water-dipping objective was used. The FITC and Cy5 fluorophores were excited by the 488-nm and 633-nm lasers simultaneously. The emission band of 493–628 nm for the FITC-labeled PLL channel and 638–759 nm band for the Cy5-labeled RNA channel were recorded simultaneously.
In vitro toxicity assay
MDA-MB-231 cells were plated in a 96-well plate at a seeding density of 1×106 cells per well in 0.1 mL of growth medium consisting of DMEM plus 10% FBS. Cells were grown at 37 °C for 24 hours, and the medium was removed. Medium containing designed PLL-anti-mir-10b nanoparticles was incubated for an additional 24 hours, 100 μL of fresh medium containing 50 μg of MTT was added to each well, and cells were incubated for 4 hours. Lysis buffer (10 (W/v) SDS, 45% DMF, pH 4.7) was then added and incubated for 24 hours. Absorbance was measured at 570 nm in a microplate reader (Molecular Devices Corp.). Survival percentage was calculated as compared to no-treatment cells (100% survival).
Western blot
Cells were harvested in RIPA lysis buffer (150 mM NaCl, 10 mM Tris-HCl, pH 7.5, 1% NP40, 1% deoxycholate, 0.1% SDS, protease inhibitor cocktail (Roche)). Proteins from total cell lysate were analyzed by NuPAGE 4%–12% Bis-Tris gradient gel (Invitrogen), transferred to the PVDF membrane, blocked with 2% non-fat milk in PBS/Tween-20, blotted with the antibodies for RhoC 1:200 and beta-actin (1:10,000), and developed with anti-mouse IgGHRP 1:10,000 using the ECL kit from Invitrogen.
Statistics method
All wound healing experiments were repeated at least three times. The representative experiments were reported in the figures. For the Western blot, three repeated experiments were carried out. The ratio of intensity of the RhoC bands to the actin bands were quantified using the software of LumiAnalyst 3.1 (Roche). The significant difference of anti-mir-10b compared to controls was based on the t-test P value < 0.05 (n = 3).
Results
Design of anti-mir-10b RNA
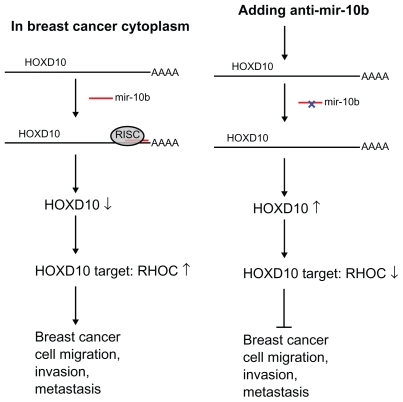
Theoretically RNA molecule that can complementarily form a complex with mature mir-10b will deplete the mir-10b, thus abolishing the mir- 10b-mediated breast cancer cell migration and metastasis (Fig. 1). According to the miRbase (http://www.miRbase.org), RNA sequence exactly complementary to mature hsa-mir-10b (major) is 5′-CACAAAUUCGGUUCUACAGGGUA-3′. The RNA sequence exactly complementary to mature hsa-mir-10b* (minor) is 5′-ACAGAU UCGAUUCUAGGGGAAU-3′. Based on the predicted secondary structure, the minor anti-mir-10b is more stable than major anti-mir-10b (Table S1). In our cell-based assays we decided to focus on the minor anti-mir-10b RNA molecule.
Figure 1.
Experiment design of using anti-mir-10b RNA molecule to inhibit breast cancer metastasis. Left panel: mir-10b in cytoplasm of breast cancer is overexpressed by Twist- induced mechanism.6,8,22 This leads to suppression of HOXD10, which the mir-10b binds to its UTR region and upregulates the HOXD10 target (ie, RHOC), thus resulting in breast cancer cell migration, invasion, and metastasis. Right panel: By adding anti-mir-10b RNA mediated by PLL nanoparticles, RISC (RNA Induced Silence Complex) is interrupted, and the HOXD10 target RhoC is downregulated, thus inhibiting the breast cancer cell migration, invasion, or metastasis.
Nanoparticle of PLL-anti-mir-10b
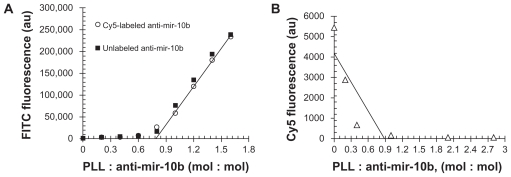
The polylysine main chain served the hydrophobic core, and the cationic side chain of lysine served as a hydrophilic surface corona that interacts with negatively charged RNA molecules. As we observed in our fluorescence titration, the fluorescence of PLL was quenched when PLL-anti-mir-10b formed complex. When the molar ratio of PLL: anti-mir-10b reached 0.9–1, the PLL fluorescence started linearly increased along with the amount of PLL increased (Fig. 2A). The Cy5 labeled anti-mir-10b and unlabeled anti-mir-10b showed same profile indicated the Cy5 labeling does not affect the binding of anti-mir-10b to PLL. When the PLL was titrated with different amount of fluorescence of Cy5 labeled anti-mir-10b, the fluorescence of Cy5 was measured. When the molar ratio of PLL : ant-mir-10b reached 0.9–1.0, the Cy5 fluorescence was completely quenched (Fig. 2B). These two titration experiments together suggested when molar ratio of PLL: anti-mir-10b was 0.9–1, the anti-mir-10b RNA formed complex with PLL. The DLS further suggested that 94% population of the solution formed nanoparticle micelle was ~200 nm (Fig. S1). This size was reported to be ideal to deliver the RNA molecule to transport through cell membrane.
Figure 2.
Characterization the complex of PLL-anti-mir-10b. (A) Constant anti-mir-10b RNA (Cy5 labeled (⋄), unlabeled (□)) were titrated with FITC-labeled PLL. (B) Constant PLL was titrated with Cy5-labeled anti-mir-10b RNA.
Cytotoxicity of PLL-micro-RNA
To identify an effective concentration of PLL-micro- RNA without inducing cell apoptosis, the cytotoxicity of these nanoparticles bearing anti-mir-10b RNA molecules toward MDA-MB-231 cells was examined using MTT assay (Fig. S2). There is essentially no cytotoxicity under the conditions tested (2–182 nM) according to MTT assay. In all of our experiments, 50 nM of anti-mir-10b was delivered by PLL-micro-RNA nanoparticles, and we did not observe either cytotoxicity or any abnormal cell morphology changes.
Inhibition of breast cancer invasion by wound-healing assay
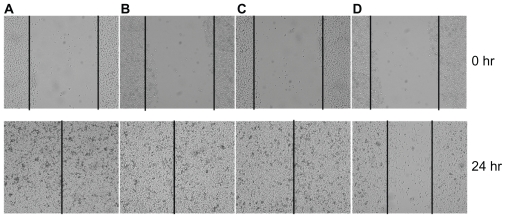
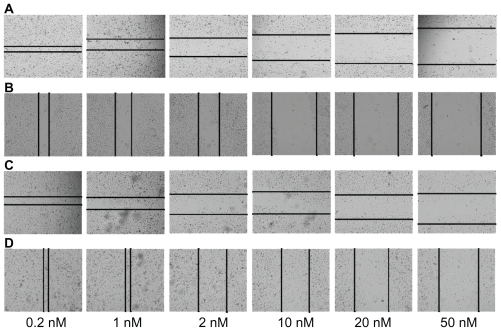
The wound-healing assay is widely used to present useful information for cell proliferations and cell migration. As previously reported, for MDA-MB-231, the cell invasion has a great corresponding relationship with cell migration assay and wound healing. The invasion properties of cancer cells were measured by the wound closure in these assays. In our study MDA-MB-231 cells were treated with different concentrations of anti-mir-10b combined with PLL, and the wound closure was analyzed. In the presence of no treatment or treatment with negative RNA molecules, the MDA-MB-231 cell can migrate into the scratched space in ~24 hours (Fig. 3). With PLL-anti-mir-10b-treatment, the wound was still open after 24 hours indicating that anti-mir-10b delivered by PLL interacted with mature mir-10b inhibiting the breast cancer cell migration or invasion. To further confirm the inhibition effectiveness, we loaded the cells with different concentrations of PLL-anti-mir-10b. The distance of the wound closure strongly correlates with concentration of PLL-anti-mir-10b (Fig. 4). That is the higher concentration of anti-mir-10b were delivered into the cell by PLL, the less chance of the cancer cells healed the wound.
Figure 3.
PLL-mediated anti-mir-10b inhibits breast cancer cell migration in wound-healing assay. 50 nM PLL alone (A), 50 nM anti-mir-10b (B), 50 nM negative control RNA molecules mixed with PLL (C), and 50 nM PLL-anti-mir-10b complex (D) were added into 1 × 105/well MDA-MB-231 cells for the wound-healing assay. The vertical lines indicate the wound formed at 0 hours (top panel) and 24 hours (bottom panel). Only in the D panel the wound healing is inhibited.
Figure 4.
PLL mediated anti-mir-10b inhibits breast cancer cell migration follows concentration-dependent manner and sustained effectiveness for 4 weeks. Different passages of MDA-MB-231 cells were carried on the wound-healing assay by adding different concentration (from 0.2 nM to 50 nM) of PLL-anti-mir-10b complex. (A) The first passage of MDA-MB-231 cells in the first week. The horizontal lines indicate the wound formed at 24 hours. (B) The second passage of MDA-MB-231 in the second week. The vertical lines indicate the wound formed at 24 hours. (C) The third passage of MDA-MB-231 in the third week. The horizontal lines indicate the wound formed at 24 hours. (D) The fourth passage of MDA-MB-231 in the fourth week. The vertical lines indicate the wound formed at 24 hours.
Breast cancer cell uptake of anti-mir-10b RNA into cytoplasm
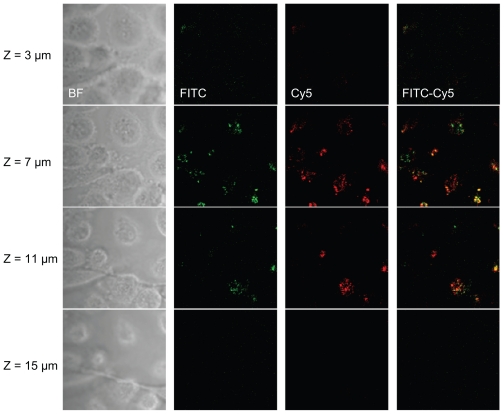
MicroRNAs are post-transcriptional regulators that bind to complementary sequences of target messenger RNA transcripts (mRNAs). The pre-miRNA hairpins are exported from the nucleus in a process involving the nucleocytoplasmic shuttle. In the cytoplasm, the pre-miRNA hairpin is cleaved by the Dicer. The miRNA maturation and regulation of target mRNA is mainly in the cytoplasm. To test if the anti-mir-10b molecules are delivered by nanoparticles into the cytoplasma, the uptake of PLL-anti-mir-10b nanoparticles was visualized by confocal microscope. In our experiments, the PLL was labeled by FITC, and anti-mir-10b molecule was labeled by Cy5. As shown in Figure 5, both FITC-labeled PLL and Cy5-labeled RNA were identified on the confocal microscope in same view in cytoplasm of MDA-MB-231 breast cancer cells. This strongly suggested that anti-mir-10b RNA molecules were delivered into cytoplasm by PLL-anti-mir-10b nanoparticles.
Figure 5.
PLL-mediated anti-mir-10b RNA cytoplasm delivery visualized by the confocal microscope. 50 nM PLL-anti-mir-10b complex were added into MDA-MB-231 cells. After 28 hours, the cell monolayers were washed with 1XPBS three times. Images were taken by Zeiss 710 laser scanning confocal microscope. FITC-labeled PLL (green), Cy5-labeled anti-mir-10b (red), and FITC-Cy5 merged view were visualized at 3 μm, 7 μm, 11 μm, and 15 μm Z-dimensions.
Sustained release of RNA by nanoparticles
PLL is a very stable drug delivery reagent, and has been proved to tolerate with extracellular and intracellular protease. But it is still a polypeptide material, it will be eventually degraded by the intracellular protease in several weeks. In our experiments, the denatured single chain RNA molecule was bound to polylysine nanoparticles. Therefore the releasing of anti-mir-10b must depend on how PLL is degraded. After the first 48-hour experiment, we detached the MDA-MB-231 cells and restarted the same experiments by making a perpendicular wound. In the second 48-hour experiment, the wound closure again showed concentration dependence as shown in Figure 4. We continued our assay for 4 weeks, continuously observing the sustained inhibition of cancer cell migration by this wound-healing assay. This observation suggested that PLL-anti-mir-10b particles may slowly release the small amount of RNA to inhibit the mature miR-10b that was overexpressed in breast cancer cells.
Downregulation of RhoC by PLL-anti-mir-10b
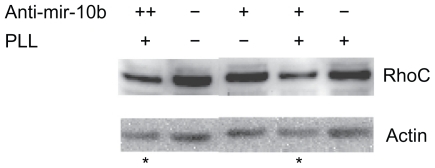
To confirm that the delivered RNA molecules can interfere with the regulation of miR-10b in MDA-MD-231, we analyzed the downstream target of miR-10b by Western blot experiments. In the absence of PLL or anti-mir-10b, the cancer cells expressed RhoC at basal level (Fig. 6). When anti-mir-10b RNA (50 nM) was added without PLL, it did not change the RhoC level. When PLL (50 nM) was present along with anti-mir-10b molecules (50 nM), the RhoC was dramatically reduced (Fig. 6). When we increased the concentration of anti-miR-10b (100 nM), the RhoC was further suppressed (Fig. 6).
Figure 6.
RhoC is suppressed by PLL-delivered cytoplasm anti-mir-10b RNA molecules. Western blot analysis of RhoC level with anti-mir-10b delivered by PLL. With presenting of 50 nM anti-mir-10b along with the constant PLL, the level of RhoC was significantly (P < 0.05, n = 3) decreased compared to its basal expression without adding PLL and/ or anti-mir-10b (denoted with star). When 100 nM of anti-mir-10b added (++), the RhoC was further suppressed (denoted with star).
Discussions
To deliver the anti-miR-10b RNA molecule into breast cancer cells to inhibit the cancer metastasis, we developed a novel RNA-PLL complex. The nanoparticles size was about 200 nm. This size was reported to be ideal to deliver the RNA molecule to transport through cell membrane.16 The wound-healing assay is widely used to present useful information for cell proliferations and cell migrations.6,17,18 So we tested this nanoparticles in the wound healing assays of breast cancer MDA-MB-231. This nanoparticle complex proved highly effective because of the slow release of trace amount of anti-mir-10b for up to 4 weeks in our cell assays with no toxicity. Our data has shown that a single dose of PLL-anti-mir-10b (50 nM) was sustained in the cells after multiple passages and thus has retained the anti-metastasis function up to 4 weeks. Although we are targeting miR-10b in this study, this platform could potentially be used with any other anti-miRNA.
As we observed in this study, microRNA molecule (anti-mir-10b) was bound and protected by PLL, and PLL-anti-mir-10b nanoparticle was capable to deliver microRNA molecule into breast cancer cytoplasm effectively. The molecular mechanism of PLL delivery is due to the strong interactions between the RNA backbone and the cationic polylysine. The formed nanoparticles protected the microRNA intact from extracellular or intracellular nucleases. In the cytoplasm of human cancer cells, the pre-miRNA hairpin is cleaved by the Dicer. This endoribonuclease interacts with the 3′ end of the hairpin and cuts away the loop joining the 3′ and 5′ arms, yielding an imperfect miRNA: miRNA duplex. Since the PLL nanoparticle was tightly bound to the anti-miR-10b, and protected it from RNAse degradation, it may need more energetic efforts to release those anti-miR-10b molecules into cytoplasm. This makes the selectivity of PLL-RNA with perfectly matched mature mir-10b in the cytoplasm. Regardless of the 4-week sustainable effectiveness, the slow degradation of PLL in the cell may be a possible mechanism as reported previously.19–21 Considering the cell proliferation, the initial amount of delivered nanoparticles in the cells must be diluted over 4 weeks of passaging. Perhaps the slow degradation of PLL made the delivered anti-mir-10b molecules still was able to bind to its target mir-10b even the amount of anti-10b has been diluted. More detailed pharmacokinetics study is needed to evaluate the effectiveness and sustainability of this nanoparticle delivery system in cells in future.
The wound-healing assay is widely used to present useful information for cell proliferations and cell migrations.6,17,18 Although the MDA-MB-231 is a model breast cancer cell line for metastasis studies, the PLL-microRNA-10b may also work with other type of metastasis breast cancer cells. And the wound-healing assay is the simplest test for cell proliferation and migrations, the inhibition of wound healing may not completely reflect the cancer metastasis. Boyden-chamber type cell migration assay and 3-D culture tests may offer opportunities to systematically study the micro-RNA-10b mediated breast cancer metastasis. And this PLL-microRNA nanoparticles may thus offer bright applications for prevent the breast cancer metastasis.
In conclusion, in this study, we showed that a single addition of a trace amount of polylysine-anti-mir-10b nanoparticle complex into breast cancer cells resulted in sustained inhibit the wound-healing for up to 4 weeks in breast cancer cells, and the HOXD10 target RhoC is downregulated. No detectable toxicity was associated with the delivery of nanoparticles at the tested concentrations. These findings may stimulate more studies on the development of targets and drugs that can be further tailored toward specific abnormalities in breast cancer.
Supplementary Materials
Table S1.
Thermo stability of anti-mir-10bs.
| Name | Anti-mir-10b (major) | Anti-mir-10b (minor) |
|---|---|---|
| Sequence | 5′-CACAAAUUCGGUUCUACAGGGUA-3′ | 5′-ACAGAUUCGAUUCUAGGGGAAU-3 |
| ΔG | 0.6 kcal · mole−1 | −3.68 kcal · mole−1 |
| TM | 14.1 °C | 56.9 °C |
| ΔH | −15.8 kcal · mole−1 | −38.3 kcal · mole−1 |
| ΔS | −55 cal · K−1 mole−1 | −116 cal · K−1 mole−1 |
Note: The results are based on the free online software OligoAnalyzer 3.1 from IDTDNA (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/).
Nanoparticle size was estimated by the dynamic light scattering.
Note: The PLL-anti-mir-10b nanoparticle solution was analyzed by DLS. The main peak with an average size of 260nm contained 94% of the nanoparticles
Cytotoxicity of PLL-anti-mir-10b.
Note: MTT cell proliferation method was used to estimate the cytotoxicity of different concentration of anti-mir-10b.
Acknowledgments
We thank Julie Wiley for proofreading the manuscript. We also thank Dr. Sudhakar Chintharlapalli, and Dr. Chunghong Gong for helpful discussion. This research is supported by the US Department of Defense Breast Cancer Postdoctoral Fellowship W81XWH-10-1-0031, and by the NIH National Institute of General Medical Sciences (grant number R01GM080987). A part of our experiments were performed at the Environmental Molecular Science Laboratory (EMSL), a national scientific user facility sponsored by DOE. PNNL is operated by Battelle for DOE under contract DE-AC05-76RL01830.
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Allen BJ. Reviews on Recent Clinical Trials. 2008;3:185. doi: 10.2174/157488708785700339. [DOI] [PubMed] [Google Scholar]
- 2.Escalona S, Blasco JA, Reza MM, Andradas E, Gomez N. Medical oncology (Northwood, London, England) 27:114. doi: 10.1007/s12032-009-9182-3. [DOI] [PubMed] [Google Scholar]
- 3.Pantel K, Alix-Panabieres C, Riethdorf S. Nature Reviews. 2009;6:339. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- 4.Ma L, Weinberg RA. Cell Cycle (Georgetown, Tex) 2008;7:570. doi: 10.4161/cc.7.5.5547. [DOI] [PubMed] [Google Scholar]
- 5.Negrini M, Calin GA. Breast Cancer Res. 2008;10:203. doi: 10.1186/bcr1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma L, Teruya-Feldstein J, Weinberg RA. Nature. 2007;449:682. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 7.Sasayama T, Nishihara M, Kondoh T, Hosoda K, Kohmura E. International Journal of Cancer. 2009;125:1407. doi: 10.1002/ijc.24522. [DOI] [PubMed] [Google Scholar]
- 8.Bourguignon LY, Wong G, Earle C, Krueger K, Spevak CC. The Journal of Biological Chemistry. doi: 10.1074/jbc.M110.162305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mamounas M, Leavitt M, Yu M, Wong-Staal F. Gene Therapy. 1995;2:429. [PubMed] [Google Scholar]
- 10.Felgner PL, Gadek TR, Holm M, et al. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:7413. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi YH, Liu F, Choi JS, Kim SW, Park JS. Human Gene Therapy. 1999;10:2657. doi: 10.1089/10430349950016690. [DOI] [PubMed] [Google Scholar]
- 12.Toncheva V, Wolfert MA, Dash PR, et al. Biochimica et Biophysica Acta. 1998;1380:354. doi: 10.1016/s0304-4165(98)00004-x. [DOI] [PubMed] [Google Scholar]
- 13.Choi JS, Lee EJ, Choi YH, Jeong YJ, Park JS. Bioconjugate Chemistry. 1999;10:62. doi: 10.1021/bc9800668. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli UK. Proceedings of the National Academy of Sciences of the United States of America. 1975;72:4288. doi: 10.1073/pnas.72.11.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Jeong JH, Park TG. J Control Release. 2002;79:283. doi: 10.1016/s0168-3659(02)00002-0. [DOI] [PubMed] [Google Scholar]
- 16.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Biochem J. 2004;377:159. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng L, Jiang G, Mei H, et al. BMC Cancer. 10:33. doi: 10.1186/1471-2407-10-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu Z, Willmarth NE, Zhou J, et al. Proceedings of the National Academy of Sciences of the United States of America. 107:8231. doi: 10.1073/pnas.1002080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong J, Kim BS, Char K, Hammond PT. Biomacromolecules. 2011;12:2975. doi: 10.1021/bm200566k. [DOI] [PubMed] [Google Scholar]
- 20.Singh HD, Wang G, Uludag H, Unsworth LD. Acta Biomater. 2010;6:4277. doi: 10.1016/j.actbio.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 21.Abbasi M, Lavasanifar A, Berthiaume LG, Weinfeld M, Uludag H. Cancer. 2010;116:5544. doi: 10.1002/cncr.25321. [DOI] [PubMed] [Google Scholar]
- 22.Li G, Wu Z, Peng Y, et al. Cancer Letters. 299:29. doi: 10.1016/j.canlet.2010.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Thermo stability of anti-mir-10bs.
| Name | Anti-mir-10b (major) | Anti-mir-10b (minor) |
|---|---|---|
| Sequence | 5′-CACAAAUUCGGUUCUACAGGGUA-3′ | 5′-ACAGAUUCGAUUCUAGGGGAAU-3 |
| ΔG | 0.6 kcal · mole−1 | −3.68 kcal · mole−1 |
| TM | 14.1 °C | 56.9 °C |
| ΔH | −15.8 kcal · mole−1 | −38.3 kcal · mole−1 |
| ΔS | −55 cal · K−1 mole−1 | −116 cal · K−1 mole−1 |
Note: The results are based on the free online software OligoAnalyzer 3.1 from IDTDNA (http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/).
Nanoparticle size was estimated by the dynamic light scattering.
Note: The PLL-anti-mir-10b nanoparticle solution was analyzed by DLS. The main peak with an average size of 260nm contained 94% of the nanoparticles
Cytotoxicity of PLL-anti-mir-10b.
Note: MTT cell proliferation method was used to estimate the cytotoxicity of different concentration of anti-mir-10b.