Abstract
An equilibrium mixture of alternate quaternary structure assemblies can form a basis for allostery. The morpheein model of allostery is a concerted dissociative model that describes an equilibrium of alternate quaternary structure assemblies whose architectures are dictated by alternate conformations in the dissociated state. Kinetic and biophysical anomalies that suggest that the morpheein model of allostery applies for a given protein of interest are briefly described. Two methods are presented for evaluating proteins as potential morpheeins. One is a subunit interchange method that uses chromatography, dialysis, and mass spectroscopy to monitor changes in multimer composition. The other is a two-dimensional native gel electrophoresis method to monitor ligand-induced changes in an equilibrium of alternate multimeric assemblies.
Keywords: Allostery, morpheein, quaternary structure equilibrium, 2d native PAGE, subunit interchange
Section 1. Introduction of the morpheein concept
1.1 The morpheein model of allosteric regulation
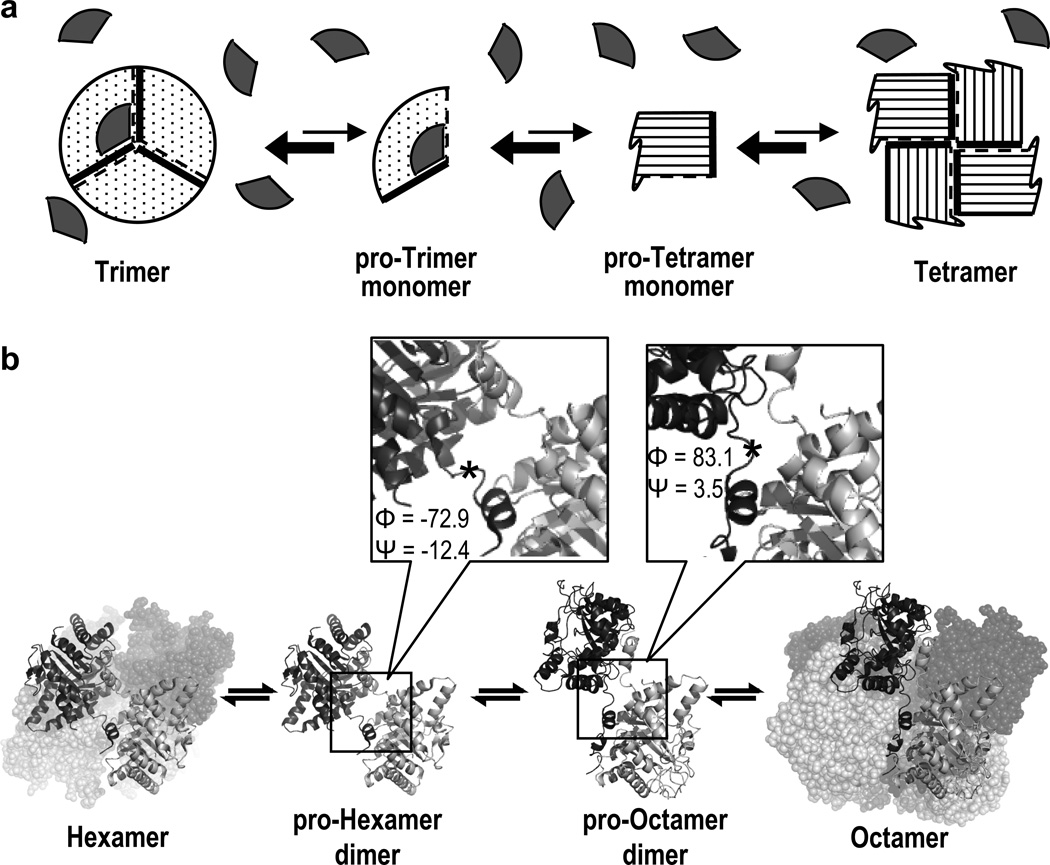
The morpheein model of allosteric regulation is a concerted dissociative model wherein the allosteric protein exists in a dynamic and reversible equilibrium of alternate oligomers, the interconversion of which requires a conformational change of composite subunits in the dissociated state prior to reassembly to an alternate oligomer (Fig. 1A) (1). In this model, one conformer of the protein allows assembly to one particular multimer, while an alternate conformer of the protein allows assembly to a structurally and functionally distinct alternate multimer. Small molecule binding that stabilizes one multimer over the other multimer will shift the quaternary structure equilibrium, altering the protein’s function toward that of the targeted multimer, thus providing a mechanism for allostery. In this model, the interconversion between conformers is fully reversible and does not require core structure refolding such as is seen in amyloidogenic proteins.
Figure 1. The morpheein model of allosteric regulation illustrated with a schematic example and with HsPBGS.
Panel A depicts a homo-oligomeric protein for which the available functionally distinct multimeric states are a trimer and a tetramer, and two conformations of a monomer, one that can trimerize and one that can tetramerize. The “rule of engagement” for multimeric assembly is the association of a thick line with a dashed line. An allosteric regulator molecule (depicted as a grey wedge) has the appropriate geometry to bind only to the pro-trimer and trimer forms and shift the equilibrium in that direction, thus allosterically stabilizing the trimer and promoting its function. Panel B illustrates the dynamic equilibria between the quaternary structure assemblies of HsPBGS. The two predominant forms in solution are the high activity octamer (PDB code 1E51) and the inactive hexamer (PDB code 1PV8); interconversion between these forms requires dissociation to a dimer, which can take on alternate conformations (4, 7). The interchange between dimer conformations involves alterations in a small number of Φ/Ψ angles that change the orientation of the αβ-barrel domains of each subunit with respect to each other. One significant change in Φ/Ψ angles is at residue 23 (*), highlighted in the magnified panels.
Proteins that can function in this way are called morpheeins. At present, the best-characterized morpheein is the enzyme porphobilinogen synthase (PBGS, EC 4.2.1.24), which catalyzes the first common step in tetrapyrrole biosynthesis. For PBGS, the functional distinction between alternate oligomers is the level of enzymatic activity; one oligomer is the “on” state and the other is the “off” state. This chapter covers two biophysical methods that can be used to address whether or not a particular protein functions as a morpheein; PBGS is used as an illustrative example. These methods are applicable to soluble proteins that migrate in native PAGE.
1.2 Atypical data suggestive of morpheeins
In general, one is taught to think about homo-oligomeric protein structure and function in terms of a single multimer or derivatives thereof (e.g. a tetramer that dissociates into its component dimers or monomers; a decamer that dissociates into its component pentamers, dimers, or monomers). This was the structural framework used to interpret data on PBGS for several decades. Most of this data was consistent with the idea that PBGS formed an octamer, as was our own laboratory data and emerging crystal structures (e.g. (2)). These octamers were considered to dissociate into their component tetramers, dimers, or monomers. However, select publications using a variety of biochemical and or biophysical methods suggested the possibility of a hexameric assembly.
Our subsequent detailed investigation into the root of these anomalous findings led to the realization that PBGS from many species can exist as an equilibrium of high activity octamers and low activity hexamers whose interconversion requires dissociation and occurs at the level of a conformationally flexible dimer (Fig. 1B) (3–6). For human PBGS (HsPBGS) the mole fraction of the dimers is ~0.5%, and the position of the equilibrium is pH dependent (7). At neutral pH, the protein is predominantly octameric and the octamer exhibits a high Vmax and a low Km. At basic pH, the equilibrium shifts toward the hexamer, which exhibits a low Vmax and a high Km. At basic pH, where there is a mix of octamers and hexamers, an experimental determination of Vmax and Km does not fit well to a single Michaelis-Menten equation. The data may conform to a fit reflecting negative cooperativity; however, the best fit (and the one that correctly reflects the behavior of PBGS in solution) is to a double hyperbolic equation reflecting a mixture of the two kinetically distinct (and slowly interconverting) multimeric states as we have described previously (8). Furthermore, small molecules have been demonstrated to bind to the protein in such a way as to shift this quaternary structure equilibrium, thus effecting function and establishing the equilibrium as a basis for allostery (4, 6, 9–11).
We have published extensively on physical and kinetic analyses of the PBGS morpheein equilibrium and do not reiterate these findings here (3, 7–8, 12). There are many kinetic and biophysical anomalies in the literature that suggest that various proteins may be morpheeins. While the purpose of this chapter is not to provide a comprehensive discussion of such anomalies, the following list provides a brief description of some characteristics that might suggest that a particular homo-oligomeric protein may use the morpheein model of allosteric regulation.
1.2.1. Biophysical indications of alternate quaternary structure stoichiometries
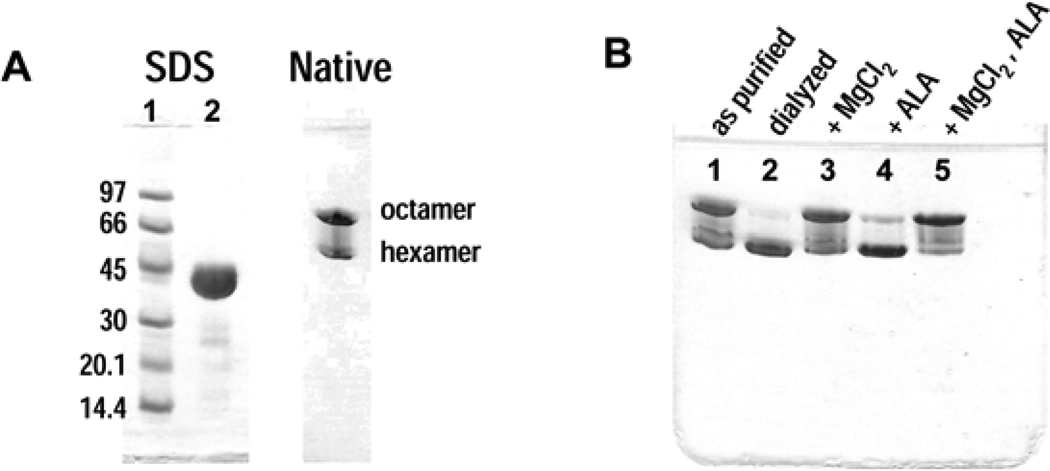
There are numerous examples of proteins for which different quaternary structure stoichiometries have been reported based on the use of different biophysical techniques for analysis (e.g. analytical ultracentrifugation, size exclusion chromatography, X-ray crystal structures). Historically, such data is typically reconciled by presuming that one oligomer is “correct” and the others are artifacts. However, as part of its normal function, a morpheein can experience different quaternary assemblies under different solution conditions, or in the presence of different ligands. Alternatively, a protein that is deemed homogeneous by SDS PAGE may be seen to separate into multiple forms using other methods such as native PAGE (Fig. 2A), or by various chromatographic methods (ion exchange, size exclusion, or hydrophobic). These alternate forms are often interpreted as being chemically distinct (e.g. having a covalent modification, or different disulfide linkages), but for morpheeins this interpretation is not necessarily valid. It is important to note that the use of tags for affinity purification will prevent the chromatographic identification of alternate morpheein forms that might otherwise separate. It is our experience that the addition of tags can also alter the quaternary structure equilibrium (13).
Figure 2. Electrophoretic analysis of pea PBGS.
A) SDS and native PAGE analysis of purified pea PBGS. SDS lane 1 – molecular weight markers; SDS lane 2 – pea PBGS (3 mg/mL) as purified (100 mM Tris-HCl, pH 8.5, 10 mM MgCl2); Native – pea PBGS (1 mg/mL) as purified. B) Native PAGE analysis of pea PBGS (1 mg/mL) at low ionic strength. lane 1 – as purified; lane 2 – dialyzed vs. 10 mM BTP pH 8.5; lane 3 – the sample shown in lane 2 with 10 mM MgCl2 added in the protein dilution step; lane 4 – the sample shown in lane 2 with 1 mM ALA added in the protein dilution step; lane 5 – the sample shown in lane 2 with 10 mM MgCl2 and 1 mM ALA added in the protein dilution step.
1.2.2. Kinetic indications of multiple protein forms
In the event that a component of the assay mix induces a dissociative quaternary structure transition from a low activity form to a high activity form, this can manifest as kinetic hysteresis, first defined by Frieden (14). An example of such a transition is seen for GDP-mannose dehydrogenase, which is a putative morpheein (1, 15). A dependence of the kinetic behavior of a protein on the order of addition of reaction components also suggests multiple protein forms may be in a metastable equilibrium (15–16). A protein concentration dependence to the specific activity of an enzyme can occur when a high activity oligomer of one stoichiometry is in equilibrium with a low activity oligomer of a different stoichiometry, and the protein concentration used in the assay is in the range of the dissociation constant for one or another oligomer (e.g. (17)).
1.2.3. Moonlighting functions (18)
Many proteins are being reported to have alternate and unrelated moonlighting functions. An outstanding example of a homo-oligomeric moonlighting protein is glyceraldehyde-3-phosphate dehydrogenase (GAPDH), which has been reported to have many alternate functions (e.g. (19–21)). Alternate GAPDH quaternary structure assemblies have been observed (22); while GAPDH functions as a homo-tetramer in glycolysis, it is possible that some of the moonlighting functions may arise from different oligomeric states. PBGS itself has two reported moonlighting functions, which have not yet been correlated with its alternate assemblies (23–24).
1.2.4. Inconsistent subunit-subunit interactions in different protein crystal structures
It is sometimes found that multi-domain proteins do not yield to protein crystal structure determination, while it is possible to obtain crystal structures from constructs containing a subset of the domains found in the mature protein. Subunit interactions apparent from these crystal structures are sometimes inconsistent with each other, though these discrepancies are rarely discussed in the literature. Although these inconsistencies may be attributed to crystal contact forces, it is possible that each is correct but represents a different multimeric form of the protein.
Any one of the aforementioned experimental anomalies can suggest that a particular protein may function as a morpheein, but none establish the dissociative allosteric mechanism illustrated in Figure 1. Here we describe two techniques, one of which established the dissociative mechanism of the PBGS quaternary structure equilibrium, and the other of which illustrates how one can monitor changes in quaternary structure as a result of allosteric modulators.
Section 2. Experiment 1 - Establishing subunit disproportionation during quaternary structure equilibration
2.1 Introduction to subunit disproportionation
A defining characteristic of the morpheein model of allostery is an obligatory dissociation to a lower order oligomer (or monomer) as part of oligomeric interconversion (Fig.1). This is distinct from a mechanism in which an octamer would simply lose one dimer to become a hexamer. In the case of PBGS, the transition through a dimeric intermediate implies that a given dimer can exchange between various PBGS hexamers and octamers throughout its “life” in a cell. The suite of techniques described below (which includes chromatography, dialysis, and mass spectroscopy) demonstrates a migration of subunits between the higher order oligomers.
Proteins that participate in a physiologically significant morpheein equilibrium must, by definition, sample the alternate assemblies under physiological conditions. Perturbations such as ligand binding, pH or ionic strength changes, or point mutations to a protein can alter the energy landscape to favor formation of one oligomer over another. A key tool in the disproportionation experiment is a mutant protein that alters the position of the quaternary structure equilibrium. Many single amino acid changes to HsPBGS alter the quaternary structure equilibrium (3, 9, 12). One naturally occurring mutation produces a variant, F12L, for which the equilibrium is dramatically shifted toward the low activity hexamer. This distinction between F12L and WT HsPBGS, for which the equilibrium favors the high activity octamer, makes F12L an invaluable experimental tool. The F12L mutation, where the site of mutation is distant from the enzyme active site yet the impact on catalysis is dramatic, can be considered within a frequently observed class of mutations that exert long-range effects. Many proteins have been characterized with mutations that have long-range effects, some of which may reflect alterations to an as-yet-uncharacterized equilibrium of quaternary structure assemblies. We propose that these mutants can serve the role that F12L serves in the described experiment.
2.2. Materials used to establish subunit disproportionation
2.2.1. WT+F12L hetero-oligomeric HsPBGS
A pET17b-derived plasmid coding for both WT and F12 HsPBGS was heterologously expressed in E. coli, and purified as described previously (3–4). Briefly, the purification involved ammonium sulfate fractionation followed by hydrophobic (phenyl Sepharose), anion exchange (Q-Sepharose) and size exclusion (Sephacryl S-300) chromatography. The purified protein was stored at −80 °C in buffer comprising 0.1 M potassium phosphate, pH 7, 10 mM β-mercaptoethanol, and 10 µM ZnCl2.
2.2.2. Analytical chromatography resin
Mono-Q anion exchange resin purchased from GE Healthcare equilibrated in 30 mM potassium phosphate, pH 7.0, 10 mM 2-mercaptoethanol, 10 µM ZnCl2.
2.2.3. Analytical chromatography buffer
30 mM potassium phosphate, pH 7.0, 10 mM 2-mercaptoethanol, 10 µM ZnCl2 with a gradient of 0.02–1.0 M KCl.
2.2.4. Trypsin
Sequencing grade, purchased from Promega.
2.2.5. Equilibrium dialysis buffer
0.1 M bis tris-propane (BTP) prepared in the absence or presence of the HsPBGS substrate, 5-aminolevulinic acid (ALA) (purchased as the hydrochloride from Sigma), and adjusted to pH 7.0 with KOH.
2.2.6. PAGE reagents
All SDS and native PAGE analyses were performed on a GE PhastSystem using 12.5% polyacrylamide gels with native (880 mM L-alanine, 250 mM tris [pH 8.8], made of 3% agarose IEF) or SDS (0.2 M tricine, 0.2 M tris, 0.55% SDS [pH 8.1], made of 2% agarose IEF) buffer strips purchased from GE Healthcare.
2.3 Methods used to establish subunit disproportionation
2.3.1. Initial isolation of WT+F12L HsPBGS hetero-hexamers and hetero-octamers
Purified WT+F12L hetero-oligomeric HsPBGS was subjected to anion exchange chromatography on Mono-Q resin. The column was equilibrated with analytical chromatography buffer (30 mM potassium phosphate, pH 7.0, 10 mM 2-mercaptoethanol, 10 µM ZnCl2), and a gradient of 0.02–1.0 M KCl was used to resolve the mixed oligomers into their components. The surface charge difference between hexameric and octameric HsPBGS is sufficient to achieve baseline separation on a Mono-Q column (3). The WT+F12L oligomers eluted from the Mono-Q column in two baseline-separated pools: Pool I eluted at a position consistent with that of homo-hexameric F12L; Pool II eluted at a position consistent with that of homo-octameric WT. Native PAGE analysis of the resolved pools confirmed that Pool I contained hexameric HsPBGS and Pool II contained octameric HsPBGS.
2.3.2. Analysis of the subunit composition of isolated WT+F12L HsPBGS hetero-hexamers and hetero-octamers
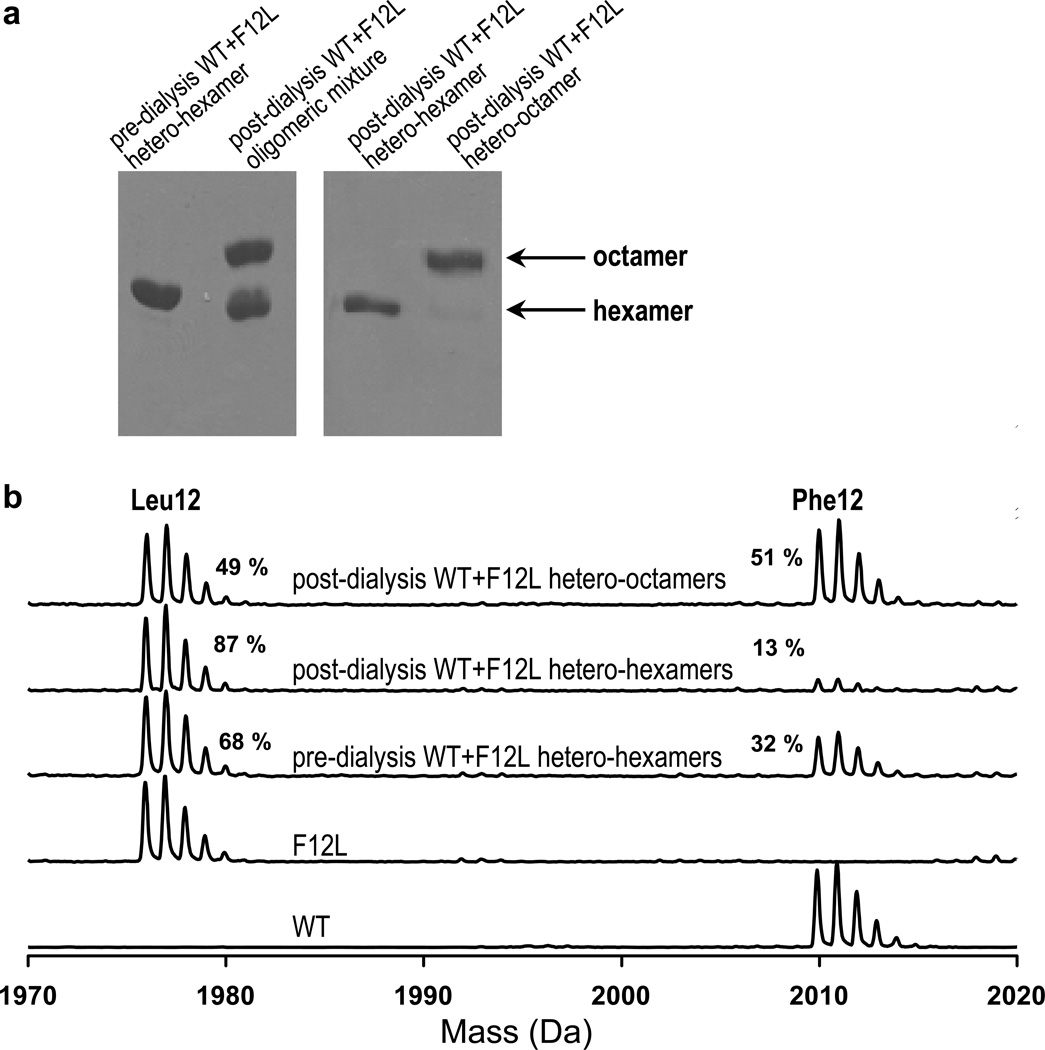
The pools containing hetero-hexameric or hetero-octameric HsPBGS were each concentrated to 1 mg/mL. The concentrated pools were dialyzed against 300 ml of 2 mM BTP-HCl buffer at pH 7.0 for 3 h to remove the phosphate from the Mono-Q buffer. Samples were subject to overnight trypsin digestion using a 1:20 (w/w) trypsin:protein ratio. The tryptic peptide mixtures were spotted on a gold plate with cyano-4-hydroxycinnamic acid matrix on top of that. The mass spectral data were collected using Reflex IV matrix-assisted laser desorption/ionization time-of-flight mass spectrometer (Bruker Inc.). Mass spectrometry of both pools, focusing on the N-terminal peptide of the tryptic digest, confirmed that each pool comprised hetero-oligomers (Fig.3). The hetero-hexamers (Pool I) contained ~70% Leu12 chains, while the hetero-octamers (Pool II) contained ~70% Phe12 chains.
Figure 3. Analysis of the disproportionation of subunits between HsPBGS oligomers.
A) Native PAGE analysis of WT+F12L HsPBGS oligomers before and after dialysis under turnover conditions demonstrates that substrate turnover facilitates oligomeric interconversion. Following dialysis to yield mixed hetero-oligomers, the hetero-hexamers were resolved from the hetero-octamers by anion exchange chromatography. B) Mass spectroscopy focusing on the N-terminal tryptic peptides of HsPBGS oligomers before and after substrate turnover conditions demonstrates subunit exchange during oligomeric interconversion.
2.3.3. Analysis of the stability of WT+F12L HsPBGS hetero-hexamers and hetero-octamers
The isolated hetero-oligomers appeared stable during storage. However, a time dependent activation of the hetero-hexamer (kinetic hysteresis) was observed during activity assay, suggesting that it could rearrange to the octamer during turnover. To address the dissociative mechanism of this phenomenon, hetero-hexamers were subjected to dialysis in the absence or presence of the substrate, ALA. The dialysis of HsPBGS proceeded for 48 hours at 37 °C, followed by analysis of the oligomeric composition by native PAGE. As shown previously, the hetero-hexamers remain hexameric in the absence of substrate, while the presence of substrate readily induces the formation of octamers (4). A general application of this method would use a ligand that has been demonstrated to shift the quaternary structure equilibrium of the protein being studied.
2.3.4. Demonstration of subunit disproportionation between HsPBGS hetero-oligomers
During dialysis in the presence of substrate WT+F12L hetero-hexamer is seen to convert to hetero-octamer (4). At approximately 50% conversion of hetero-hexamer to hetero-octamer the protein was subjected to anion exchange chromatography to resolve and isolate the resultant oligomers, as shown by native PAGE (Fig. 3A). Mass spectrometry of each oligomer (focusing again on the N-terminal tryptic peptide) revealed a dramatic disproportionation of chains between the oligomers (Fig. 3B). The pre-dialysis hetero-hexameric WT+F12L contained Leu12 and Phe12 chains in a ratio of 68:32. Following dialysis and chromatographic separation of the oligomers, the remaining hexamers contained Leu12 and Phe12 chains in a ratio of 87:13, while the newly assembled octamers contained Leu12 and Phe12 chains in a ration of 49:51. The observed disproportionation of chains is consistent with the oligomeric equilibria favoring the octamer for the WT, and the hexamer for F12L. The observed disproportionation of subunit chains definitively establishes a dissociative mechanism for oligomer interconversion, but does not define the quaternary structure of the dissociated form. If the oligomer did not dissociate during the transition, the ratio of chains in the oligomers would remain unchanged.
Section 3. Experiment 2 – Two-dimensional native PAGE analysis of the impact of allosteric modulators on oligomeric state
3.1 Allosteric modulators that shift an equilibrium of oligomeric states
As discussed above, the equilibrium between the alternate oligomers is freely reversible, and a protein that functions as a morpheein samples the available oligomers under physiological conditions. The impact of an allosteric ligand on a morpheein derives from ligand binding to one of the oligomeric forms in a manner that stabilizes it, thereby drawing the quaternary structure equilibrium towards that oligomer. An allosteric inhibitor (morphlock-1) for the Pisum sativum (green pea) PBGS (PsPBGS) has been characterized and found to inhibit PsPBGS by binding to and stabilizing the low activity hexamer, drawing the equilibrium towards that form (10). The equilibrium perturbing mechanism whereby the allosteric inhibitor binds to the hexamer is fundamentally different from a ligand induced “switch” mechanism where the ligand would bind to the octamer and induce a “switch” to the hexamer.
3.2 Introduction of two-dimensional native PAGE to assess oligomeric state changes
The quaternary structure equilibrium of PBGS is sufficiently dynamic that oligomeric interconversion can occur within the matrix of a native polyacrylamide gel (25). This phenomenon allowed us to monitor the impact of substrate or allosteric modifiers using a two-dimensional (2D) native PAGE technique. As detailed below for PsPBGS, the protein is resolved into its composite oligomers in the 1st dimension, the gel is subsequently incubated in a solution of the oligomer-perturbing ligand, and the results of the incubation are analyzed by a 2nd dimension native PAGE separation. The method described below has been optimized for the GE PhastSystem, whose cassette-style open-faced plastic-backed gels facilitate manipulations of the gels between dimensions. The method could be modified to run in a standard vertical slab gel apparatus for the 1st dimension, followed by a 2nd dimension on a flat-bed apparatus.
3.3. Materials used for two-dimensional native PAGE
3.3.1. Protein
Pisum sativum PBGS (PsPBGS) expressed and purified as previously described, stored at −80° C in buffer comprising 100 mM Tris HCl pH 8.5, 10 mM MgCl2 (17).
3.3.2. Dilution buffer
For PsPBGS, dilution of the protein to 1 mg/mL in 100 mM BTP-HCl at pH 8.5 yields a mixture of octamers and hexamers.
3.3.3. PAGE reagents
All native PAGE analyses were performed on a GE PhastSystem using 12.5% polyacrylamide gels with native (880 mM L-alanine, 250 mM tris [pH 8.8], made of 3% agarose IEF) (0.2 M tricine, 0.2 M tris, 0.55% SDS [pH 8.1], made of 2% agarose IEF) buffer strips purchased from GE.
3.3.4. Gel incubation buffer
100 mM BTP-HCl, pH 8.5. This buffer was also supplemented with 10 mM MgCl2 and 10 mM ALA to create assay turnover conditions for PsPBGS, or with 1 mM morphlock-1 (with a final DMSO concentration of 20%) to induce allosteric inhibition by stabilizing the inactive PsPBGS hexamer.
3.3.5. Activity assay buffer
100 mM BTP-HCl, 10 mM ALA, 10 mM MgCl2, pH 8.5.
3.3.6. STOP reagent
8 mL of 0.1 M BTP-HCl, pH 8.5 and 8 mL of 20% trichloroacetic acid.
3.3.7. Developing reagent
4 mL of 0.1 M BTP, pH 8.5 and 8 mL of modified Ehrlich’s reagent (20 g/L p-dimethylaminobenzaldehyde in 80% glacial acetic acid and 20% concentrated perchloric acid).
3.4 Methods for two-dimensional native PAGE
3.4.1. Preparation of the protein for electrophoresis
The starting sample conditions will vary for different proteins, and the nature of the oligomeric change being monitored. Generally the protein of interest should first be incubated in or dialyzed against a buffer in which the oligomeric equilibrium is pliable, and which yields an appropriate oligomeric distribution for the experiment. For example, when analyzing a hexamer-stabilizing inhibitor of PsPBGS, a starting sample that is predominantly octameric will yield the most dramatic results. Prior to the two-dimensional native PAGE experiment, appropriate initial conditions can be optimized by incubating the protein under various conditions (e.g. different pH, buffers, ligands), and examining the resultant oligomeric composition by one-dimensional native PAGE (as shown for PsPBGS in Fig. 2B).
Prior to electrophoresis, PsPBGS was diluted to 1 mg/mL in dilution buffer (100 mM BTP, pH 8.5). These conditions were chosen to yield a mixture of octamers and hexamers in the 1st dimension to illustrate the different effects of substrate and allosteric inhibitor on the protein in the 2nd dimension. One-dimensional native PAGE (Fig. 4A) confirms the desired oligomeric composition of the starting sample. The position of the PsPBGS quaternary structure equilibrium is influenced by ionic strength (higher ionic strength favors the octamer); protein concentration (higher protein concentration favors the octamer); the presence of an allosteric magnesium ion, which stabilizes the octamer by binding to an octamer-specific subunit-subunit interface; and the presence of substrate in conjunction with a catalytic magnesium ion, which also stabilizes the octamer (5).
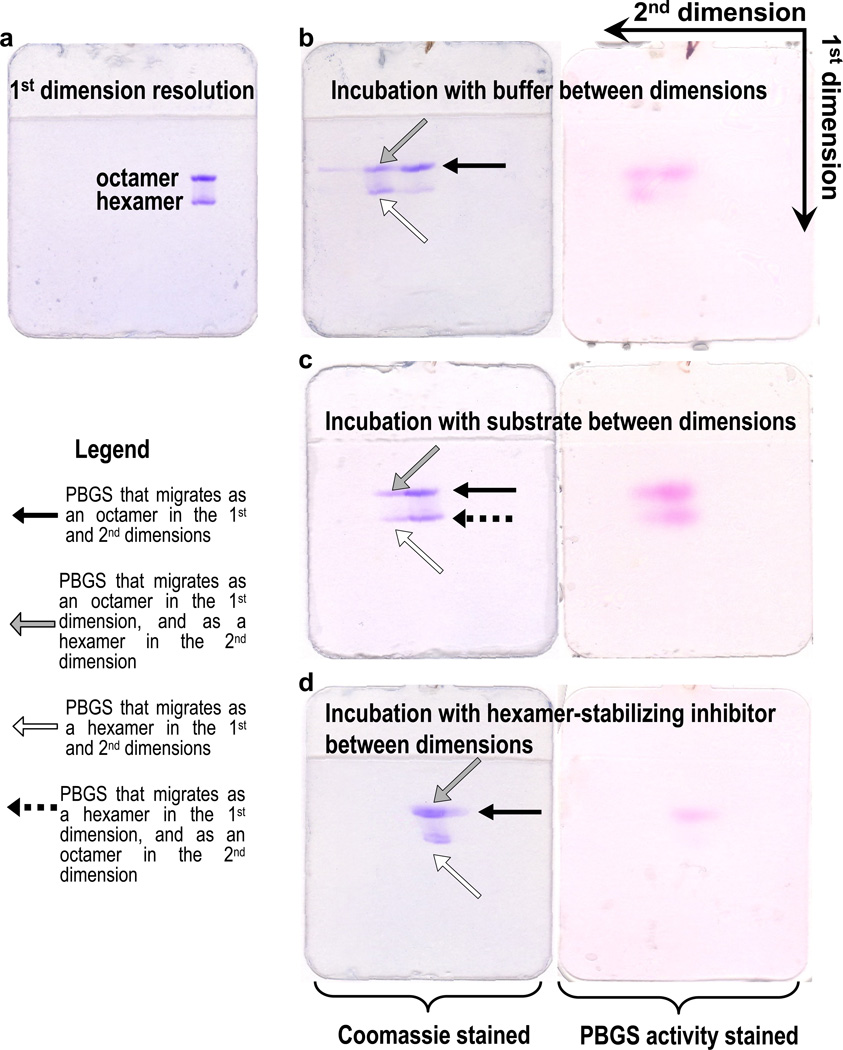
Figure 4. The impact of substrate or allosteric inhibitor binding on the oligomeric state of PBGS, as demonstrated by 2-dimensional native PAGE.
For all gels, a single 1 µl sample of 1mg/ml pea PBGS was applied in the next to the right-most sample well of a 12.5% PhastGel. A) PsPBGS resolves into ~70% octamer and ~30% hexamer after electrophoresis in the 1st dimension. The resolved gels were then incubated in following conditions for 20 min: B) 0.1 M BTP, pH 8.5 at 37°C, C) 0.1 M BTP, pH 8.5, 10 mM ALA, 10 mM MgCl2 (assay conditions) at 37 °C, D) 0.1 M BTP, pH 8.5, 1 mM morphlock-1, 20% DMSO. Following these incubations, the 2nd dimension separation was carried out immediately. All gels were then stained for PBGS activity (right panels) followed by Coomassie staining (left panels). The overall retarded protein mobility observed for the 2nd dimension separation for gel D versus gels B and C is due to the presence of DMSO in the incubation buffer on the gel matrix (control not shown).
3.4.2. Electrophoresis in the 1st dimension
A single 1 µL PsPBGS sample is applied to an 8 × 1 µL applicator comb in the 7th position from the left. It is critical to utilize the most narrow lane width possible, as wide bands will not meaningfully resolve in the 2nd dimension. The sample should be loaded in a lane near the edge of the gel to allow room for 2nd dimension migration, but not in the lane nearest the edge of the gel as protein in these lanes can run aberrantly. For the examples in Figure 4 B–D, electrophoresis in the 1st dimension was performed on a 12.5% polyacrylamide gel following a standard protocol. Different gel matrices could be selected based on the size of the oligomers of the protein of interest. The ideal matrix will cleanly resolve the alternate oligomers, while keeping both of them towards the center of the resolving gel (rather than abutting the stacking gel or the buffer front).
3.4.3. Incubation between dimensions of native PAGE
When the alternate oligomers have been resolved in the 1st dimension of native PAGE, the discrete effects of ligand binding on the oligomeric state of each can be examined by incubating the gel in a solution of that ligand. For the examples shown in Figure 4 B–D, the gels were incubated under three different conditions in Gel Incubation Buffer for 20 minutes at 37 °C with gentle shaking. These were 1) a control comprising buffer (100 mM BTP, pH 8.5), 2) assay turnover conditions (100 mM BTP pH 8.5, 10 mM MgCl2, and 10 mM ALA), and 3) in the presence of a hexamer stabilizing allosteric inhibitor (100 mM BTP pH 8.5, 1 mM morphlock-1, 20% DMSO). Following the incubation, gels were rinsed briefly in distilled water prior to native PAGE separation in the 2nd dimension.
3.4.4. Electrophoresis in the 2nd dimension
The 2nd dimension separation is performed perpendicular to the 1st dimension. For the GE PhastSystem, the gel is placed on the gel bed with a 90° clockwise rotation relative to the position for the 1st dimension. The gel should be carefully aligned to ensure that the top and bottom buffer strips contact the gel evenly, as further described by Samuel et al. (26). The duration of 2nd dimension electrophoresis will vary among different proteins. The conditions used for the 1st dimension are a good starting point for optimization.
3.4.5. Visualization of gel bands
Any common protein gel staining technique can be used to reveal the position of the bands following this 2D native PAGE protocol. In some cases, an activity assay can be performed prior to the protein visualization step. Analyzing the enzymatic activity associated with each provides an elegant demonstration of activation or inhibition of the protein by the ligand exposure between the two dimensions. This in-gel assay method is most suitable for enzyme assays that measure a colorimetric signal. In the illustrated case of PsPBGS, the formation of the product, porphobilinogen, can be monitored by the pink color that forms by reaction with modified Ehrlich’s reagent. Gels are incubated at 37 °C for 20 min in 15 mL of activity assay buffer (0.1 M BTP-HCl, 10 mM 5-aminolevulinic acid, 10 mM MgCl2, pH 8.5), and then transferred into 16 mL of STOP solution (8 mL of 0.1 M BTP, pH 8.5 and 8 mL of 20% trichloroacetic acid) for 1–2 min at room temperature. Gels are then transferred into 12 mL of developing reagent (4 mL of 0.1 M BTP, pH 8.5 and 8 mL of modified Ehrlich’s reagent) at room temperature for 4 min during which time a pink color develops. If the assay for a protein of interest has a colorimetric signal outside of the visible range, an appropriate scanner could be used for detection.
3.4.6. Interpretation of 2D native PAGE results
Electrophoresis in the 1st dimension resolves a morpheein into its component oligomers, a roughly equal mixture of octamers and hexamers in the illustrated case of PsPBGS. The incubation conditions between the 1st and 2nd dimension will have different effects on the octamers and hexamers. In the control experiment shown in Figure 4B, incubation of the gel in buffer between dimensions causes a portion of the protein that migrated as an octamer in the first dimension to migrate as a hexamer in the second dimension; the majority of the protein that migrated as a hexamer in the 1st dimension remains hexameric in the 2nd dimension. Activity is observed for all bands, which could derive from two possible interpretations: 1) the octameric and hexameric PsPBGS are both active forms of the protein, or 2) the inactive hexameric PsPBGS is converted to active octamers in the presence of substrate during the activity assay, and the observed activity is from the newly assembled octameric PsPBGS. The experiments illustrated in Figure 4C and 4D confirm the latter interpretation.
In the experiment illustrated in Figure 4C, incubation of the gel under assay turnover conditions between dimensions causes the majority of the protein in both the octameric and hexameric 1st dimension bands to migrate as octamer in the 2nd dimension. This experiment conclusively demonstrates that the substrate-mediated oligomeric interconversion can occur within the native PAGE matrix. Activity is again observed for all bands; the intensity of the activity staining roughly correlates with the Coomassie staining for protein.
In the experiment illustrated in Figure 4D, incubation of the gel in the presence of a hexamer stabilizing inhibitor (morphlock-1) between dimensions causes the majority of the protein from the 1st dimension octameric band to migrate as hexamer in the 2nd dimension. All of the 1st dimension hexameric band remains as hexamer. Activity staining reveals that the only active component is the octamer. The presence of the hexamer-stabilizing inhibitor is sufficient to prevent the interconversion in the presence of substrate during activity staining, and demonstrates that the activity observed in the hexamer bands of the gels in Figure 4B and 4C derives from PsPBGS that has converted to octamers.
Acknowledgments
This work was supported by the National Institutes of Health grants R01ES003654 (to E.K.J), R56AI077577 (to E.K.J), and CA006927 (to the Institute for Cancer Research). The authors thank our colleagues Drs. Gregory Adams, Mark Andrake, Erica Golemis and George D. Markham for helpful comments.
References
- 1.Jaffe EK. Morpheeins - a new structural paradigm for allosteric regulation. Trends in Biochemical Sciences. 2005;30:490–497. doi: 10.1016/j.tibs.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Erskine PT, Senior N, Awan S, Lambert R, Lewis G, Tickle LJ, Sarwar M, Spencer P, Thomas P, Warren MJ, ShoolinginJordan PM, Wood SP, Cooper JB. X-ray structure of 5-aminolaevulinate dehydratase, a hybrid aldolase. Nature Structural Biology. 1997;4:1025–1031. doi: 10.1038/nsb1297-1025. [DOI] [PubMed] [Google Scholar]
- 3.Breinig S, Kervinen J, Stith L, Wasson AS, Fairman R, Wlodawer A, Zdanov A, Jaffe EK. Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase. Nature Structural Biology. 2003;10:757–763. doi: 10.1038/nsb963. [DOI] [PubMed] [Google Scholar]
- 4.Tang L, Stith L, Jaffe EK. Substrate-induced interconversion of protein quaternary structure isoforms. Journal of Biological Chemistry. 2005;280:15786–15793. doi: 10.1074/jbc.M500218200. [DOI] [PubMed] [Google Scholar]
- 5.Kokona B, Rigotti DJ, Wasson AS, Lawrence SH, Jaffe EK, Fairman R. Probing the oligomeric assemblies of pea porphobilinogen synthase by analytical ultracentrifugation. Biochemistry. 2008;47:10649–10656. doi: 10.1021/bi801128d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez UD, Myachina F, Stith L, Jaffe EK. Advances in Computational Biology. 2010. Docking to large allosteric binding sites on protein surfaces. p in press, Springer in book series, Advances in Experimental Medicine and Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selwood T, Tang L, Lawrence SH, Anokhina Y, Jaffe EK. Kinetics and thermodynamics of the interchange of the morpheein forms of human porphobilinogen synthase. Biochemistry. 2008;47:3245–3257. doi: 10.1021/bi702113z. [DOI] [PubMed] [Google Scholar]
- 8.Lawrence SH, Jaffe EK. Expanding the Concepts in Protein Structure-Function Relationships and Enzyme Kinetics: Teaching using Morpheeins. Biochem Mol Biol Educ. 2008;36:274–283. doi: 10.1002/bmb.20211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang L, Breinig S, Stith L, Mischel A, Tannir J, Kokona B, Fairman R, Jaffe EK. Single amino acid mutations alter the distribution of human porphobilinogen synthase quaternary structure isoforms (morpheeins) Journal of Biological Chemistry. 2006;281:6682–6690. doi: 10.1074/jbc.M511134200. [DOI] [PubMed] [Google Scholar]
- 10.Lawrence SH, Ramirez UD, Tang L, Fazliyez F, Kundrat L, Markham GD, Jaffe EK. Shape shifting leads to small-molecule allosteric drug discovery. Chem Biol. 2008;15:586–596. doi: 10.1016/j.chembiol.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lawrence SH, Ramirez UD, Selwood T, Stith L, Jaffe EK. Allosteric inhibition of human porphobilinogen synthase. J Biol Chem. 2009;284:35807–35817. doi: 10.1074/jbc.M109.026294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaffe EK, Stith L. ALAD porphyria is a conformational disease. American Journal of Human Genetics. 2007;80:329–337. doi: 10.1086/511444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shanmugam D, Wu B, Ramirez U, Jaffe EK, Roos DS. Plastid-associated Porphobilinogen Synthase from Toxoplasma gondii - Kinetic and structural properties validate therapeutic potential. Journal of Biological Chemistry. 2010;285:22122–22131. doi: 10.1074/jbc.M110.107243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frieden C. Kinetic aspects of regulation of metabolic processes. The hysteretic enzyme concept. J Biol Chem. 1970;245:5788–5799. [PubMed] [Google Scholar]
- 15.Naught LE, Gilbert S, Imhoff R, Snook C, Beamer L, Tipton P. Allosterism and cooperativity in Pseudomonas aeruginosa GDP-mannose dehydrogenase. Biochemistry. 2002;41:9637–9645. doi: 10.1021/bi025862m. [DOI] [PubMed] [Google Scholar]
- 16.Jaffe EK, Bagla S, Michini PA. Reevaluation of a Sensitive Indicator of Early Lead-Exposure - Measurement of Porphobilinogen Synthase in Blood. Biological Trace Element Research. 1991;28:223–231. doi: 10.1007/BF02990469. [DOI] [PubMed] [Google Scholar]
- 17.Kervinen J, Dunbrack RL, Litwin S, Martins J, Scarrow RC, Volin M, Yeung AT, Yoon E, Jaffe EK. Porphobilinogen synthase from pea: Expression from an artificial gene, kinetic characterization, and novel implications for subunit interactions. Biochemistry. 2000;39:9018–9029. doi: 10.1021/bi000620c. [DOI] [PubMed] [Google Scholar]
- 18.Jeffery CJ. Moonlighting proteins--an update. Mol Biosyst. 2009;5:345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- 19.Karpel RL, Burchard AC. A basic isozyme of yeast glyceraldehyde-3-phosphate dehydrogenase with nucleic acid helix-destabilizing activity. Biochim Biophys Acta. 1981;654:256–267. doi: 10.1016/0005-2787(81)90180-5. [DOI] [PubMed] [Google Scholar]
- 20.Meyer-Siegler K, Mauro DJ, Seal G, Wurzer J, deRiel JK, Sirover MA. A human nuclear uracil DNA glycosylase is the 37-kDa subunit of glyceraldehyde-3-phosphate dehydrogenase. Proc Natl Acad Sci U S A. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim Biophys Acta. 2006;1762:502–509. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Ferreira-da-Silva F, Pereira PJ, Gales L, Roessle M, Svergun DI, Moradas-Ferreira P, Damas AM. The crystal and solution structures of glyceraldehyde-3-phosphate dehydrogenase reveal different quaternary structures. J Biol Chem. 2006;281:33433–33440. doi: 10.1074/jbc.M605267200. [DOI] [PubMed] [Google Scholar]
- 23.Guo GG, Gu M, Etlinger JD. 240-kDa proteasome inhibitor (CF-2) is identical to delta-aminolevulinic acid dehydratase. J Biol Chem. 1994;269:12399–12402. [PubMed] [Google Scholar]
- 24.Gross M, Hessefort S, Olin A. Purification of a 38-kDa protein from rabbit reticulocyte lysate which promotes protein renaturation by heat shock protein 70 and its identification as delta-aminolevulinic acid dehydratase and as a putative DnaJ protein. J Biol Chem. 1999;274:3125–3134. doi: 10.1074/jbc.274.5.3125. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe EK, Ali S, Mitchell LW, Taylor KM, Volin M, Markham GD. Characterization of the Role of the Stimulatory Magnesium of Escherichia-Coli Porphobilinogen Synthase. Biochemistry. 1995;34:244–251. doi: 10.1021/bi00001a029. [DOI] [PubMed] [Google Scholar]
- 26.Samuel M. "PHAST 2D," a two-dimensional electrophoretic technique on a single gel under native and denaturing conditions using pharmacia PhastSystem. Anal Biochem. 1995;224:457–459. doi: 10.1006/abio.1995.1072. [DOI] [PubMed] [Google Scholar]