Background: Rhizobium LPS has four GalA residues.
Results: RgtDE add GalA to the lipid A and synthesize Dod-P-GalA. RgtABCDE mutants are affected in DOC and PmxB sensitivity.
Conclusion: Sequence of GalA addition to LPS is RgtDABC. Lipid A GalA provides membrane stability and PmxB resistance.
Significance: GalA negative charges are required for membrane stability and implicated for interaction with plant host antimicrobial peptides.
Keywords: Antimicrobial Peptides, Glycosyltransferases, Host Defense, Lipopolysaccharide (LPS), Symbiosis, Rhizobium leguminosarum, Galacturonic Acid, Nitrogen Fixation
Abstract
Rhizobium lipopolysaccharide (LPS) contains four terminally linked galacturonic acid (GalA) residues; one attached to the lipid A and three attached to the core oligosaccharide moiety. Attachment of the GalA residues requires the lipid donor dodecaprenyl-phosphate GalA (Dod-P-GalA), which is synthesized by the GalA transferase RgtE reported here. The galacturonosyl transferases RgtA, -B, and -C utilize Dod-P-GalA to attach GalAs on the LPS core region, and RgtD attaches GalA to the lipid A 4′ position. As reported here, the functions of the rgtD and rgtE genes were determined via insertion mutagenesis and structural characterization of the mutant lipid A. The rgtE− mutant lacked Dod-P-GalA as determined by mass spectrometry of total lipid extracts and the inability of rgtE− mutant membranes to provide the substrate for heterologously expressed RgtA activity. In addition, we created single mutations in each of the rgtA, -B, -C, -D, and -E genes to study the biological function of the GalA residues. The structures of the core oligosaccharide region from each of the rgt mutants were elucidated by glycosyl linkage analysis. Each mutant was assayed for its sensitivity to sodium deoxycholate and to the antimicrobial cationic peptide, polymyxin B (PmxB). The rgt mutants were more sensitive than the parent strain to deoxycholate by varying degrees. However, the rgtA, -B, and -C mutants were more resistant to PmxB, whereas the rgtD and E mutants were less resistant in comparison to the parent strain.
Introduction
The Gram-negative, nitrogen-fixing, endosymbiotic bacterium Rhizobium leguminosarum contains four terminally linked galacturonic acid (GalA)2 residues on its lipopolysaccharide (LPS) (1, 2). Three of the GalA residues are attached to the core region, whereas one GalA is attached to the lipid A (Fig. 1). The lipid donor dodecaprenyl-phosphate GalA (Dod-P-GalA) is required for the attachment of all four GalA residues to the LPS as reported by Kanjilal-Kolar et al. (3) and reported here. Three galacturonic acid transferase (GalAT) enzymes RgtA, -B, and -C were previously shown in an in vitro assay to transfer GalA from Dod-P-GalA to the synthetic substrate Man-Kdo2-[4′-32P]lipid IVA (3, 4). These results demonstrated that the GalATs RgtA and RgtB attach GalA to the branching Kdo likely at the 4 and 5 positions. However, it was unknown to which position each enzyme attaches the GalA residue. The RgtC enzyme attaches GalA to the Man residue at the 4 position. In addition, sequential addition of GalA was observed in vitro where by the RgtB and RgtC enzymes were active only in the presence of RgtA, and the RgtC enzyme was only active in the presence of both RgtA and RgtB (3, 4).
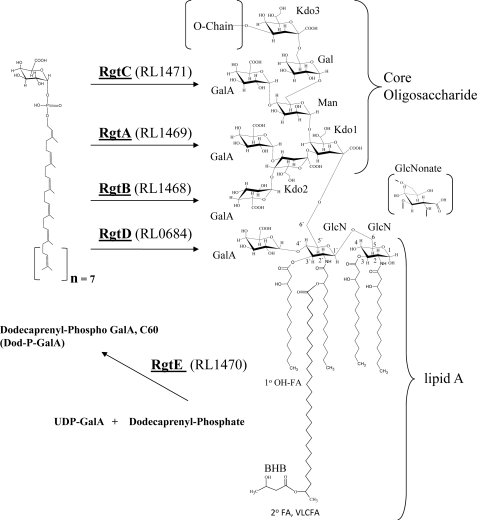
FIGURE 1.
The general structure of the R. leguminosarum bv viciae 3841 LPS core and lipid A regions. GlcN, 2-aminoglucosamine; Kdo, 3-deoxy-d-manno-2-octulosonic acid; 1o OH-FA, primary β-hydroxy fatty acids attached to the GlcN backbone of the lipid A; 2o FA, secondary very long chain fatty acid (VLCFA), 27OHC28, attached to the 3′ 1o OH-FA to form an acyloxyacyl moiety; BHB, β-hydroxybutyryl group. The proximal GlcN can be oxidized to GlcNonate by the outermembrane oxidase LpxQ. Position 27 of the very long chain fatty acid can contain a hydroxyl or β-hydroxybutyryl group. As described in this study, RgtE is required for the synthesis of the lipid donor Dod-P-GalA which can then be utilized by the galacturonosyl transferases RgtA, -B, -C, and -D in the transfer of GalA to lipid A and Core where indicated.
Rhizobia, like several Gram-negative bacterial species, synthesize the lipid A precursor molecule Kdo2-lipid IVa. However, the 1 and 4′ phosphates are subsequently removed, respectively, by phosphatases LpxE and LpxF (5, 6), and in turn GalA is added to the 4′ position. The proximal lipid A GlcN can be oxidized by the outer membrane monoxygenase LpxQ to form a negatively charged 2-aminogluconate (GlcNonate) residue (7). Before this report, the putative lipid A-4′-GalAT, RgtD, and the Dod-P-GalAT, RgtE, were unknown. In this report we describe the function of the rgtD and rgtE genes by single gene mutagenesis. We also describe the additional preparation of single gene knock outs in rgtA, -B, and -C of R. leguminosarum bv. viciae 3841, a nitrogen-fixing endosymbiont of pea. In this study the effect of each mutation on membrane stability, LPS structure, and LPS synthesis is described.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
For a list of bacterial strains and plasmids used in this work, see Table 1. Escherichia coli strains were grown at 37 °C using Luria Broth (LB). Rhizobium leguminosarum strains were grown on Tryptone-yeast extract (TY) with 10 mm CaCl2 or minimal (Y) medium at 30 °C as described previously containing 5% sucrose where needed (8). Antibiotics were used in the following concentrations; tetracyclin (15 μg/ml), kanamycin, Km (50 μg/ml), gentamicin (R. leguminosarum, 30 μg/ml, E. coli 15 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strains and Plasmid | Characteristics | Source |
|---|---|---|
| Strains | ||
| E. coli | ||
| Top10 | F- mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 | Invitrogen |
| Δ(araleu) 7697 galU galK rpsL (Strr) endA1 nupG | ||
| R. leguminosarum bv. Viciae | ||
| 3841 | Strain 300 Strr, Nod+, Fix+ | (38) |
| EL203 (rgtA−) | Strain 3841 rgtA::accC1, Gmr Nod+ | This study |
| EL204 (rgtB−) | Strain 3841 rgtB::accC1, Gmr Nod+ | This study |
| EL205 (rgtC−) | Strain 3841 rgtC::accC1, Gmr Nod+ | This study |
| EL206 (rgtD−) | Strain 3841 rgtD::accC1, Gmr Nod+ | This study |
| EL202 (rgtE−) | Strain 3841 rgtE::accC1, Gmr, Nod+, Fix+ | This study |
| EL203 (rgtA−)/pRK-RgtA | Strain EL203 (rgtA−) complemented with plasmid pRK-RgtA (4), GmrTcr | This study |
| EL204 (rgtB−)/pRK-RgtB | Strain EL204 (rgtB−) complemented with plasmid pRK-RgtB (4), Gmr Tcr | This study |
| EL205 (rgtC−)/pRK-RgtC | Strain EL205 (rgtC−) complemented with plasmid pRK-RgtC (4), Gmr Tcr | This study |
| EL202 (rgtE−)/pMKGE | Strain EL202 (rgtE−) complemented with plasmid pMKGE (4), Gmr Tcr | This study |
| Plasmids | ||
| pUC18 | Cloning vector, Amr | Fermentas |
| pMS255 | Source of aacC1 Gm cassette, Gmr | (9) |
| pRK2013 | Mobilizing plasmid for pEX-Tc, Col E1 replicon, Kanr | (25) |
| pEX18-Tc | Suicide vector, allows positive selection for integration, Tcr | (10) |
| pRgtA-KO | pEX18 containing rgtA::aacC1 insert used to engineer EL203 (rgtA−), Gmr, Tcr | This study |
| pRgtB-KO | pEX18 containing rgtB::aacC1 insert used to engineer EL204 (rgtB−), Gmr, Tcr | This study |
| pRgtC-KO | pEX18 containing rgtC::aacC1 insert used to engineer EL205 (rgtC−), Gmr, Tcr | This study |
| pRgtD-KO | pEX18 containing rgtD::aacC1 insert used to engineer EL206 (rgtD−), Gmr, Tcr | This study |
| pRgtE-KO | pEX18 containing rgtE::aacC1 insert used to engineer EL202 (rgtE−), Gmr, Tcr | This study |
Insertion Mutagenesis of Galacturonosyl Transferase Genes
The GalAT genes, rgtA, -B, -C, -D, and -E, were individually mutated by inserting the gentamicin resistant gene aacC1 (9) near the center of each gene, thereby disrupting gene function. Plasmid extractions, gel extractions, and PCR/enzyme cleanup was performed using Qiagen mini-prep kits. Restriction enzymes were purchased from Promega, Inc. (Madison, WI). For each mutagenesis procedure, ∼1-kb fragments containing either half of the open reading frame and upstream DNA or half of the open reading frame and downstream DNA were amplified (iProofTM High Fidelity polymerase, Bio-Rad) from the R. leguminosarum bv. viciae 3841 genome. The primers were constructed containing restriction enzyme sites for cloning of each fragment (Table 2). Each PCR fragment was cloned into plasmid pUC18 by digestion of pUC18 and PCR products with respective restriction enzymes that recognize the engineered primers followed by ligation (Table 2). The resulting plasmids created, pRgtAUp, pRgtADwn, pRgtBUp, pRgtBDwn, pRgtCUp, pRgtCDwn, pRgtDUp, pRgtDDwn, pRgtEUp, and pRgtEDwn were transformed into E. coli. The cloned upstream fragments were released by restriction digest using the same enzymes that were used to clone them and separated by gel electrophoresis followed by gel extraction. The isolated upstream fragments were subcloned into the plasmids containing the downstream fragments by double restriction digestion and ligation using the enymes; pRgtADwn (BamHI, EcoRI), pRgtBDwn (BamHI, XbaI), pRgtCDwn (SmaI, EcoRI), pRgtDDwn (BamHI, XbaI), and pRgtEDwn (BamHI, EcoRI). The resulting plasmids contained upstream DNA, the open reading frames with an engineered single cloning site near the center,and downstream DNA (pRgtAUpDwn, pRgtBUpDwn, pRgtCUpDwn, pRgtDUpDwn, and pRgtEUpDwn) and were transformed into E. coli. The gentamicin-resistant gene cassette aacC1 (1 kb) was released from plasmid pM255 (9) by restriction digest and isolated by gel electrophoresis followed by gel extraction. The digested aacC1 fragments were inserted into plasmids containing up and downstream DNA by single restriction digestion and ligation using the enzymes pRgtAUpDwn (BamHI), pRgtBUpDwn (BamHI), pRgtCUpDwn (SmaI), pRgtDUpDwn (BamHI), and pRgtEUpDwn (BamHI). The resulting plasmids (pRgtA-KO, pRgtB-KO, pRgtC-KO, pRgtD-KO, pRgtE-KO) were transformed into E. coli. The inserts from each plasmid were released by restriction digestion followed by gel electrophoresis and gel extraction using the enzymes pRgtA-KO (XbaI, EcoRI), pRgtB-KO (XbaI, EcoRI), pRgtC-KO (EcoRI, HindIII), pRgtD-KO (SacI, XbaI), and pRgtE-KO (XbaI, EcoRI). The fragments were cloned into the suicide vector pEX18-tetracyclin (10), which harbors the sacB lethal gene to create plasmids pEX/RgtA-KO, pEX/RgtB-KO, pEX/RgtC-KO, pEX/RgtD-KO, and pEX/RgtE-KO, which were transformed into E. coli.
TABLE 2.
Primer list
Underlined regions indicate the engineered endonuclease restriction sites. Italic letters indicate regions homologous to R. leguminosarum bv. viciae 3841 genomic DNA. Fwd, forward; Rev, reverse.
| Gene | Primer |
|---|---|
| RgtAUp | Fwd, ACGTGGATCCGCAGGTGAAGCTGATGG |
| RgtAUp | Rev, ACGTGAATTCGCAGCACCAGCAGGAAG |
| RgtADwn | Fwd, ACGTTCTAGACTGGGTGCCCGACAATC |
| RgtADwn | Rev, ACGTGGATCCGGATCTTGCGCAGGAAC |
| RgtBUp | Fwd, ACGTTCTAGATTTCGCGCTGATGCCTG |
| RgtBUp | Rev, ACGTGGATCCCTTCTCCAGTGTGCGTG |
| RgtBDwn | Fwd, ACGTGGATCCGCTCTCGCCATCATCA |
| RgtBDwn | Rev, ACGTGAATTCTGGCCCGCGTCTATTG |
| RgtCUp | Fwd, ACGTGAATTCGCATGCATCGGCAGCA |
| RgtCUp | Rev, ACGTCCCGGGTCGGCAGTCATTCGC |
| RgtCDwn | Fwd, ACGTCCCGGGTTTTTCGCGCGCTCT |
| RgtCDwn | Rev, ACGTAAGCTTGTGCCGATGTCGTTGC |
| RgtDUp | Fwd, ACGTTCTAGAGAGATCCCTGAGGCCTG |
| RgtDUp | Rev, ACGTGGATCCGCCGCACCGTCGTATTG |
| RgtDDwn | Fwd, ACGTGGATCCTCAGAGGCAGGAGCGAC |
| RgtDDwn | Rev, ACGTGAGCTCAGCGCCTTCAGCAGCAC |
| RgtEUp | Fwd, ACGTTCTAGACTGGCTGCCCGACAATC |
| RgtEUp | Rev, ACGTGGATCCGGATCTTGCGCAGGAAC |
| RgtEDwn | Fwd, ACGTGGATCCGCAGGTGAAGCTGATGG |
| RgtEDwn | Rev, ACGTGAATTCGCAGCACCAGCAGGAAG |
A triparental mating was conducted to move each pEX/Rgt-KO into 3841 via conjugal transfer. The donor strain E. coli Top 10 harboring the pEX/Rgt-KO, the helper strain E. coli carrying the helper plasmid pRK2013 (20), and the recipient strain 3841 were cultured overnight in 100 ml of media. The E. coli strains were grown in LB (37 °C) broth, and the R. leguminosarum bv. viciae 3841 was grown in TY broth (30 °C). Two ml each of cultured donor and helper were centrifuged at 4000 × g for 10 min. The cell pellets were each resuspended in 4 ml of TY medium and centrifuged at 4000 × g for 10 min. The supernatant was discarded until there was just enough left to make a resuspension. The entire suspension (∼500 μl) was spotted on a nitrocellulose membrane previously laid on a TY agar plate and kept at 30 °C overnight. The resulting paste was washed off the membrane with 4 ml of sterile deionized water. To select for double recombinants, various amounts of the mating mix were plated onto minimal Y agar plates containing 30 μg/ml gentamicin and 5% sucrose. Colonies were picked and streaked on TY/gentamicin plates. Mutant colonies were isolated using a PCR screen and given the strain names EL202 (rgtE−), EL203 (rgtA−), EL204 (rgtB−), EL205 (rgtC−), and EL206 (rgtD−). The EL202 (rgtE−), EL203 (rgtA−), 204 (rgtB−), and 205 (rgtC−) mutants were, respectively, complemented with the previously described plasmids pMKGE, pRK-RgtA, pRK-RgtB, and pRK-RgtC by triparental mating (3, 4).
Isolation of Lipopolysaccharide
Briefly, bacteria were grown to mid-late log phase, pelleted, and washed with 0.9% sodium chloride followed by two washes with deionized water. The pelleted bacteria were subjected to hot phenol-water extraction as described previously (11). Water layer material was dialyzed in 12–14-kDa molecular mass cutoff dialysis bags against deionized 10 liter of deionized water for 5 days, each day exchanging the water. The sample was lyophilized and subjected to DNase, RNase, and protease treatment as described previously (12) and dialyzed in 12–14-kDa molecular mass cutoff bags for 3 days and lyophilized.
DOC-PAGE Analysis
Gel electrophoresis in the presence of DOC was performed as previously described (13). Briefly, 1 μg of extracted LPS material was loaded onto an 18% acrylamide gel, and current was applied (30 mA (constant), 400 V) in the presence of DOC buffer. The gel was subjected to Alcian blue/silver staining as previously described (14).
Glycosyl and Fatty Acid Composition Analysis of Lipopolysaccharide
The composition of LPS was determined by gas chromatography/mass spectrometry (GC/MS) of trimethylsilyl (TMS) methyl glycosides and methyl ester fatty acids as described previously (15, 16). Isolated LPS, 300 μg per sample, was subjected to methanolysis (80 °C, 16–18 h) followed by N-acetylation (100 °C, 1 h) and trimethylsylation (80 °C, 30 min). The TMS derivatives were dissolved in hexane and injected into a Hewlett Packard 5890 Series II GC/MS analyzer fitted with a 30-m EC-1 column (Alltech).
Lipid A Isolation and Analysis
Lipid A was isolated by the standard mild acid hydrolysis procedure (17). Briefly, ∼5 mg of isolated LPS was treated with 1% acetic acid (100 °C, 1 h) to cleave the labile Kdo ketosidic linkage between the core moiety and the lipid A. The lipid A/core-polysaccharide mixture was partitioned three times by chloroform:methanol:water (2:2:1.8, v:v:v) extraction. The lipid A-containing chloroform layers were pooled and dried under a stream of air.
The lipid A preparations were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS). Isolated lipid A was dissolved in a 3:1 (v:v) chloroform:methanol mixture, mixed 1:1 (v:v) with 2,4,6-trihydroxyacetophenone (THAP), matrix and 0.5 μl was spotted on a MALDI plate. Spectra were acquired by a MALDI-TOF analyzer (Applied Biosystems 5800) in the positive reflectron mode operating at a 20-kV extraction voltage scanning 800–3000 mass units.
Linkage Analysis of the Kdo and Uronosyl Residues in LPS
The Kdo and uronic acid linkages in the intact LPS samples were identified by preparing the partially methylated carboxyl-reduced alditol acetate (PMAA) derivatives following the procedures described previously for Rhizobium etli LPS (2, 18) with additional modifications. The LPS samples were first subjected to permethylation using the sodium hydroxide procedure (19), modified to promote recovery of carboxymethylated Kdo residues as described for sialic acid containing glycoconjugates (20).
The purpose of these modified procedures is to preserve the Kdo carboxymethyl esters during the aqueous work-up after methylation, thus minimizing base-catalyzed elimination from the Kdo and subsequent loss of linkage. This modified work-up consists of 1) cooling the methylation reaction mixture and adding excess chloroform (2 ml), 2) low speed centrifugation to remove excess precipitated NaOH and NaI, and 3) acidification of the chloroform supernatant by washing with dilute 5% aqueous acetic acid (3 ml). After low speed centrifugation, the chloroform layer containing the permethylated LPS was washed nine more times with water and then dried under nitrogen stream.
The resulting permethylated LPS was then subjected to reduction of the Kdo and uronosyl carboxymethyl groups with lithium triethylborodeuteride (Superdeuteride, Aldrich) in THF (2 h at room temperature), mild acid hydrolysis (0.1 m trifluoroacetic acid, 100 °C, 30 min) to cleave the Kdo ketosidic linkages, reduction of Kdo residues at C-2 carbonyl group (using NaBD4 in water/ethanol), normal hydrolysis (2 m trifluoroacetic acid, 121 °C, 2 h) to cleave the remaining uronosyl and neutral sugar linkages, reduction of the newly formed aldehydo sugars (with NaBD4 in 50 mm NH4OH), and acetylation of the resulting partially methylated alditols to yield the PMAA derivatives. Acetylation was performed in acetonitrile-pyridine-acetic anhydride containing 4-N,N′-dimethylaminopyridine as the catalyst for 4 h at room temperature as described (21). The PMAA derivatives were analyzed by GC/MS (electron impact) using a 30-m SP-2330 capillary column (Supelco) programmed from 80 to 250 °C.
Preparation of E. coli Membranes
Cultures, 1 liter, of E. coli harboring plasmids pET23a or pRgtA were grown to mid-logarithmic phase and harvested by centrifugation at 4000 × g and 4 °C for 20 min. The cell pellets were resuspended in prechilled 50 mm HEPES, pH 7.5. Membranes were prepared as previously described (4).
Extraction and Purification of Total Lipids from rgtE− Mutant
Cultures, 1 liter each, of the 3841 parent, rgtE− mutant, and complemented strains were grown to mid/late logarithmic phase and harvested by centrifugation at 4000 × g for 15 min and washed 1 time with phosphate-buffered saline. Lipids were extracted from the pellets as described previously (3).
Analysis of Membrane Lipids by Combined Liquid Chromatography-Electrospray Ionization Mass Spectrometry (LC-ESI/MS)
Normal phase LC-ESI/MS of lipids was performed using an Agilent 1200 Quaternary LC system coupled to a QSTAR XL quadrupole time-of-flight tandem mass spectrometer (Applied Biosystems, Foster City, CA). Data were collected in the negative mode. An Ascentis® Si HPLC column (5 μm, 25 cm × 2.1 mm) was used. Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v/v). Mobile phase B consisted of chloroform/methanol/water/aqueous ammonium hydroxide (600:340:50:5, v/v/v/v). Mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (450:450:95:5, v/v/v/v). The elution program consisted of the following; 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% mobile phase C over 3 min and held at 100% C for 3 min and finally returned to 100% A over 0.5 min and held at 100% A for 5 min. The total LC flow rate was 300 μl/min. The post-column splitter diverted ∼10% of the LC flow to the ESI source of the Q-Star XL mass spectrometer, with MS settings as follows: IS (ion spray) = −4500 V, CUR (curtain gas) = 20 p.s.i., GS (source gas) = 20 p.s.i., DP (declustering potential) = −55 V, and FP (focusing potential) = −150 V. Nitrogen was used as the collision gas for MS/MS experiments. Data acquisition and analysis were performed using the instrument's Analyst QS software.
In Vitro Reconstitution of GalAT Activity
The activity assay methods were adapted from Kanjilal-Kolar et al. (3, 4). Briefly, the standard reaction mixture contained 50 mm MES, pH 6.5, 0.05% Nonidet P-40, 2 mm MgCl2, and 2.5 μm Kdo2-1-dephospho-[4′-32P]lipid IVA (106 cpm/nmol). When mentioned, total extracted lipid was added to the reaction at a final concentration of 0.25 mg/ml, and purified donor was added at a final concentration of 5 μm. Washed membranes were used as the enzyme source at a final concentration of 0.25 mg/ml. The reaction mixtures were kept at 30 °C for 10 or 30 min. Reactions were stopped by spotting 4 μl on a Silica Gel 60 plate. Plates were developed in a solvent system containing chloroform, pyridine, 88% formic acid, and water (30:70:16:10, v/v/v/v), dried, and exposed overnight on a PhosphorImager screen. Product was detected using PhosphorImager (Storm 840, Amersham Biosciences).
Polymyxin B and DOC Resistance Assay
Polymyxin B gradient plates were made by pouring at an angle 25 ml of TY agar media containing 50 μg/ml polymyxin B (PmxB) in a square 100 × 15-mm Petri dish. After the agar solidified, they were placed level, 40 ml of TY agar media was poured on top, and the plates were allowed to cool. The PmxB was allowed to diffuse throughout the plates overnight. A loop full of bacteria grown for 4 days on TY agar media was dissolved in 1 ml of sterile phosphate-buffered saline, pH 7.4. Then 10 μl of bacterial suspension was spotted on the gradient plate and streaked from highest to lowest PmxB concentration. The plates were incubated for 4 days at 30 °C. Sodium deoxycholate gradient plates were made as described above using TY containing 2 mm DOC. Bacteria were prepared and streaked onto the gradient plates and incubated as described above.
Plant Growth and Inoculation
Surface-sterilized pea seeds, variety Early Alaska (Bunton Sead Co., Inc.) were germinated in the dark at room temperature for 3 days on 0.8% water agar plates and transferred to sterile Erlenmeyer flasks (1 seed per flask) containing a foam top and 300 ml of nitrogen-free 1.5% agar plant media (Fahraeus media). The Erlenmeyer flasks were surrounded with brown paper bags and placed in a Conviron growth chamber. Approximately 10 days post-germination, plants were inoculated with bacteria (1 ml of late log/early stationary phase liquid TY cultures). The plants were maintained at 19 °C with 14 h of light and 15 °C with 10 h of dark.
RESULTS
Discovery of the rgtD and rgtE Genes
The predicted peptide sequence from the previously described LPS core GalATs, RgtA, -B, and -C (4), were aligned with the R. leguminosarum bv. viciae 3841 genome using the basic local alignment search tool (22). Results revealed a chromosomally localized single cistronic open reading frame RL0684 (accession number NC_0008380.1) with appreciable similarities to RgtA, -B, and -C (supplemental Fig. S1). In addition, RL0684 is predicted to be an ArnT-like protein, a class of proteins responsible for the transfer of monosaccharides from bactoprenyl-phosphate monosaccharide lipid donors to the lipid A back bone (23, 24). Furthermore, the rgtD gene is found in the genomes of a number of R. etli and R. leguminosarum strains including R. etli CE3/CFN42, R. etli CIAT652, R. leguminosarum bv. viciae 3841, and R. leguminosarum bv. trifolii WSM1325. The LPS lipid A and core oligosaccharide structures are conserved in these Rhizobium species, and thus, it is expected that genes responsible for the synthesis of LPS lipid A and core oligosaccharide are also conserved.
The rgtE gene, previously referred to as orf3, was predicted to encode the GalAT that synthesizes Dod-P-GalA by Kanjilal-Kolar et al. (3, 4); however, the gene function was not determined. The predicted amino acid sequence of RgtE is similar to a class of glycosyl transferases that has been demonstrated experimentally to synthesize polyprenyl-phosphate glycosyl lipid donors necessary for the glycosylation of lipid A (supplemental Fig. S2) (24, 26, 27). The proteins are predicted bacterial dolichol-like phosphate mannosyl (dpm1) type 2 glycosyl transferases and are of similar sizes containing two predicted C-terminal transmembrane domains that likely tether the proteins to the inner membrane, whereas the catalytic portion resides on the inner membrane/cytoplasm interface.
Analysis of Total Lipids from rgtE− Mutant by LC-ESI-MS
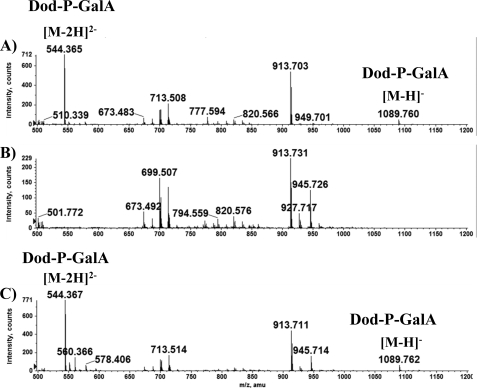
Total lipids were collected from cell pellets of the parent R. leguminosarum 3841, mutant EL202, and the complemented mutant strain using the Bligh and Dyer extraction method as described under “Experimental Procedures.” Kanjilal-Kolar et al. (3) previously reported the lipid donor Dod-P-GalA to be involved in the transfer of GalA to R. leguminosarum LPS by the Rhizobium GalATs: RgtA, B, and C. Therefore, if RgtE is required for the synthesis of Dod-P-GalA, then mutant EL202 lipids would not contain Dod-P-GalA. Extracted lipids were subjected to LC-ESI-MS analysis for the presence of Dod-P-GalA. As expected, R. leguminosarum 3841 lipids contained the appropriate ions observed previously for Dod-P-GalA (Fig. 2, panel A). At 20 min into the LC run a peak was observed that contained Dod-P-GalA single- and double-charged signature ions of m/z ratios of 1089 and 544, respectively. Ion m/z 913 agrees with the molecular weight of Dod-P. The signature ions for Dod-P-GalA were not detected in extracted lipids from mutant EL202 (Fig. 2, panel B). However, strain EL202 lipids did contain ion m/z 913 due to Dod-P. Complementing the mutant with the rgtE gene recovered the appearance of Dod-P-GalA in the lipid composition (Fig. 2, panel C).
FIGURE 2.
LC-ESI-MS of membrane lipid components. Total lipids were examined for the appearance of Dod-P-GalA by LC-ESI-MS (data were acquired in the negative mode). An ion scan for 1089 was performed on data acquired from the total lipid extracts of 3841 (A), EL202 (B), and complemented strain (C). The ion scan revealed the retention time of Dod-P-GalA to be 20 min where a sharp peak was observed on the LC chromatogram (not shown). Dod-P-GalA ions were not detectable in the total lipids from EL202. MS/MS data confirmed the identification of the labeled ions (not shown).
The Inability of the rgtE− Mutant Lipid Extracts to Act as GalA Donor in Enzyme Activity Assay with R. leguminosarum bv. viciae 3841 GalAT, RgtA
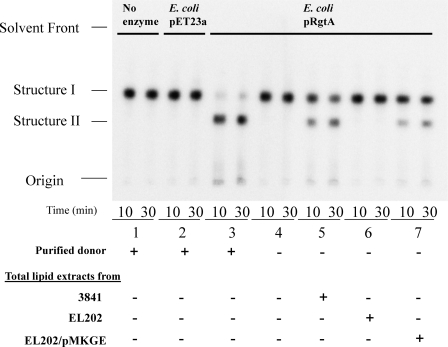
As previously described (3), extracted lipids from R. leguminosarum bv. viciae 3841, due to the presence of Dod-P-GalA, are able to act as a donor in the conversion of the radio-labeled substrate Kdo2-1-dephospho-[4′-32P]lipid IVa to the GalA-Kdo2-1-dephospho-[4′-32P]lipid IVa product in the presence of membranes from E. coli overexpressing the R. leguminosarum bv. viciae 3841 RgtA. Total lipids extracted from strains 3841, EL202, and complemented EL202 mutant were assayed for their ability to serve as a donor for this reaction. Standard reaction mixtures were prepared with equal amounts of lipid extracts, E. coli membranes containing RgtA, and radiolabeled Kdo2-1-dephospho-[4′-32P]lipid IVa substrate. Reactions were performed for 10 or 30 min and stopped by spotting 4 μl of the reaction mixture on a Silica Gel 60 plate. The silica gel plate containing each reaction was subsequently developed using the solvent system described (see “Experimental Procedures.”), and the products were observed on a PhosphorImager (Amersham Biosciences) (Fig. 3). Reaction mixtures containing parent strain 3841 lipid extract (lane 5) were able to convert the substrate into the product. In contrast, no product was observed from reactions containing mutant EL202 lipid extract (lane 6). Lipid extract from mutant EL202 complemented with the rgtE gene was able to carry out conversion of substrate to product (lane 7). These results are consistent with the requirement of RgtE for the production of Dod-P-GalA, which is necessary for the RgtA catalyzed transfer of GalA to Kdo2-1-dephospho-[4′-32P]lipid IVa.
FIGURE 3.
RgtA activity assay with total lipid extracts. Total lipids were tested for their ability to serve as a GalA donor in an activity assay with RgtA overexpressed in E. coli membranes and the radiolabeled acceptor substrate 1-dephospho-[4′-32P]lipid IVa (Structure I). Standard reaction mixtures are described under “Experimental Procedures.” Products were separated by thin layer chromatography and viewed by a radio imager. Lanes 1 and 2 lack enzyme. Lanes 3–7 contain, as an enzyme source, membranes from E. coli overexpressing RgtA. Lipid substrate was added where indicated by (+). The product in lane 3 was identified by Kanjilal-Kolar et al. (3, 4) to be GalA-modified 1-dephospho-[4′-32P]lipid IVa (Structure II). Conversion of substrate was not observed from reactions containing EL202 lipids (lane 6), indicating the lack of the necessary lipid donor (Dod-P-GalA) required for activity. Lipids from EL202 complemented with the rgtE gene (EL202/pMKGE) were able to recover RgtA activity (lane 7).
Composition and Gel Electrophoresis of Lipopolysaccharides from the rgt Mutants
The presence of LPS-derived GalA in each strain was determined by GC/MS of TMS-derived methyl glycosides and fatty acid methyl esters. As expected, strain EL202 (rgtE−) did not contain detectable amounts of GalA. Because there is a single galactosyl residue on each LPS molecule, the Gal:GalA ratio was used to determine differences in the amounts of GalA. Strains EL203 (rgtA−) (Gal:GalA = 1.0:2.0), EL205 (rgtC−) (Gal:GalA = 1.0:2.9), and EL206 (rgtD−) (Gal:GalA = 1.0:2.9) LPS had less GalA than that of the parent strain 3841 (Gal:GalA = 1.0:3.9). There was not an observable decrease in the amount of GalA in the LPS of strain EL204 (rgtB−) (Gal:GalA = 1.0:4.0). However, as discussed further below, linkage analysis showed that strain EL204 (rgtB−) was disrupted in its ability to add one of the two GalA residues to the branching Kdo residue; i.e. Kdo2 in Fig. 1. Together, these results suggest that all of the rgt mutants were disrupted to varying degrees in GalA addition to the LPS.
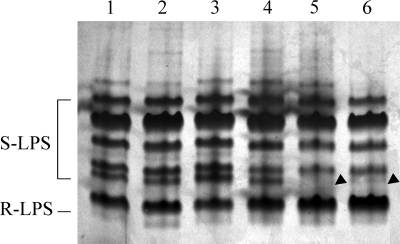
Gel electrophoresis (DOC-PAGE) of the rgt mutant LPSs was performed and compared with parent strain (Fig. 4). Surprisingly, there was no observable difference in the migration of the rgt mutants LPSs when compared with parent strain 3841. This was unexpected, particularly in the case of strain EL202 (rgtE−) as this strain lacks four GalA residues when compared with strain 3841, which calculates as a mass difference of ∼704 mass units. It may be that any change in migration due to the reduction in size is offset by the loss of charge due to the absence of GalA residues. The banding pattern of the rgt mutants suggests that there was no major disruption to OPS biosynthesis. In agreement, composition analysis of the LPS from each rgt mutant showed that they all contain the normal OPS glycosyl residues present in the parent strain LPS; e.g. N-acetyl quinovosamine, fucose (Fuc), and 3-O-methyl-6-deoxytalose. The LPSs from strains EL202 (rgtE−) and EL206 (rgtD−) displayed a slightly different banding pattern than those of the parent strain and the other rgt mutants (Fig. 4, lanes 5 and 6) in that the intensity of one band was greatly reduced. The molecular basis for this difference is not known.
FIGURE 4.
DOC-PAGE LPS profiles. Extracted LPS (1 μg) was loaded into each well and stained as described under “Experimental Procedures.” Lane 1, parent strain 3841; lane 2, strain EL203 (rgtA−); lane 3, strain EL204 (rgtB−); lane 4, strain EL205 (rgtC−). lane 5, strain EL206 (rgtD−); lane 6, strain EL202 (rgtE−). Arrowheads indicate the reduction of an LPS band from strains EL202 (rgtE−) and EL206 (rgtD−) that is present in the other LPS DOC-PAGE profiles. S-LPS, smooth LPS containing OPS; R-LPS, rough LPS lacking OPS.
MALDI-TOF/MS of the Lipid A from the rgt Mutants
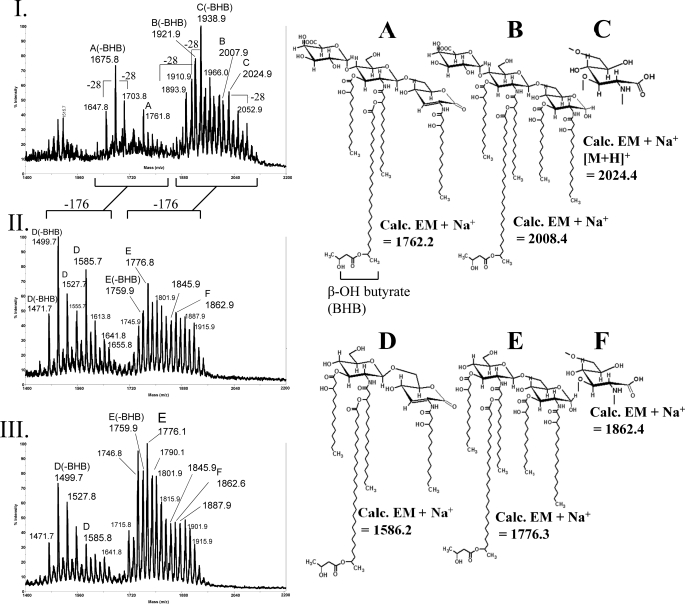
The rgt mutant lipid A preparations were studied in detail by MALDI-TOF/MS. The lipid A observed in Rhizobium species is microheterogeneous in nature. The microheterogeneity is due to several factors including the presence or absence of β-hydroxybutyryl linked at the C27 position of the secondary long chain fatty acid 27-hydroxyoctacosanoic acid (mass difference of 86), the non-stoichiometric oxidation of the proximal GlcN to GlcNonate (a mass difference of 16), and variation in acyl chain length (mass differences in series of 28 or 14 mass units). In addition, chemically modified lipid A species occur due to the mild acid hydrolysis procedure whereby acid leads conversion of the proximal GlcNonate to GlcNonolactone and the resulting acid-catalyzed β-elimination of β-hydroxymyristate from the 3-position, which results in the formation of 2,3-dideoxy-2-amino-2-ene-gluconolactone as the proximal lipid A glycosyl residue (Fig. 5, structures A and D).
FIGURE 5.
MALDI-TOF MS analysis of the rgtD− and rgtE− lipid A. Spectra were acquired in the positive reflectron mode. I, parent strain 3841 is shown. II, mutant strain EL202 (rgtE−) is shown. III, mutant strain EL206 (rgtD−) is shown. Structures A–F indicates ions observed in the spectra, and the calculated exact masses (Calc. EM) are labeled. Variations occur to structures A–F by the presence or absence of β-hydroxybutyryl (BHB; −86) and variation in the chain length of the hydroxy fatty acids (±28 and/or 14). Panels II and III contain ions with masses consistent with the absence of GalA (−176 mass units).
MALDI-TOF MS analysis of the rgtA−, B−, and C− mutant lipid A preparations (data not shown) produced ions identical to those observed for the parental lipid A (Fig. 5, structures A, B, and C), showing that each of these mutants produce a normal lipid A structure with GalA at the 4′-position. The EL202 (rgtE−) and EL206 (rgtD−) mutant lipid A lacked GalA (Fig. 5, panel III). The composition of the EL206 (rgtD−) lipid A was compared with that of the parent lipid A (Fig. 6) and confirmed that the EL206 (rgtD−) mutant produces lipid A that lacks any detectable GalA. This was also the case for the EL202 (rgtE−) mutant (data not shown).
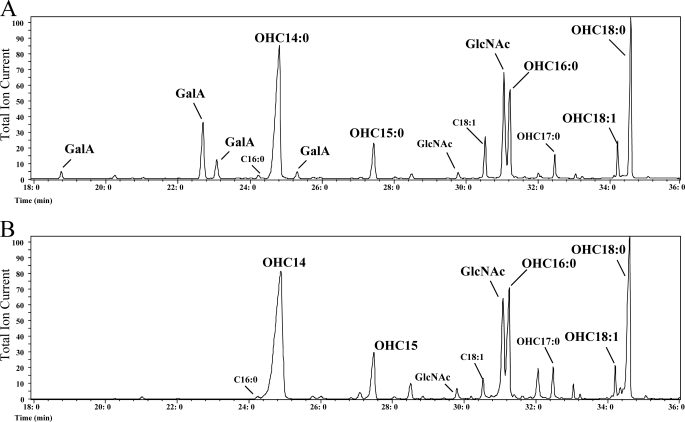
FIGURE 6.
GC/MS of the EL206 (rgtD−) mutant lipid A. A, parent strain 3841. B, mutant strain EL206 (rgtD−) is shown. Trimethylsilyl methylglycosides and fatty acid methyl esters were derived from partitioned lipid A and analyzed by GC/MS. The lipid A from the rgtA−, B−, and C− mutants gave profiles similar to that of the parent strain, whereas the lipid A from the rgtE− mutant gave a GC/MS profile similar to that of the rgtD− mutant; i.e. it was devoid of GalA residues. GlcNAc is derived from the lipid A GlcN residues.
Linkage Analysis of rgt Mutants
To determine the structures of the rgtA−, B−, C−, and D− mutant LPS core oligosaccharides, glycosyl linkage analysis was performed on their LPSs as well as the LPS from the parent 3841 strain, which contains all LPS GalA residues, and the LPS from EL202 (rgtE−) mutant, which lacks all the LPS GalA residues. The results of this analysis are shown in Table 3.
TABLE 3.
Linkage analysis of the rgt mutant LPSs
PMAA were prepared from isolated LPS and analyzed by GC/MS to determine carbohydrate linkages. The relative abundance of Kdo residues having different linkages was estimated by selective ion monitoring for the diagnostic m/z 89 primary fragment ion derived from carbons 7 and 8 of each Kdo derivative. The relative abundance of Gal/GalA and Man residues was determined from the intensity of the shared m/z 118 primary ion. Numbers before each residue (i.e. 4-Kdo) represent the linked position. Derivatives and primary ion fragments: 6-linked Man, 1,5,6-tri-O-acetyl-2,3,4-tri-O-methyl-mannitol (m/z 118, 162, 189, 233); 4,6-linked Man, 1,4,5,6-tetra-O-acetyl-2,3-di-O-methyl-mannitol (m/z 118, 261); 6-linked Gal, 1,5,6-tri-O-acetyl-2,3,4-tri-O-methyl-galactitol (m/z 118, 162, 189, 233); T-GalA, 1,5,6-tri-O-acetyl-2,3,4-tri-O-methyl-6,6-2H-galactitol (m/z 118, 162, 191, 235); T-Kdo, 1,2,6-tri-O-acetyl-3-deoxy-4,5,7,8-tetra-O-methyl-1,1-2H-octitol (m/z 89, 146, 205, 206, 250, 366); 4,5-linked Kdo, 1,2,4,5,6-penta-O-acetyl-3-deoxy-7,8-di-O-methyl-1,1-2H-octitol (m/z 89, 186, 228, 348, 422); 4-linked Kdo, 1,2,4,6-tetra-O-acetyl-3-deoxy-5,7,8-tri-O-methyl-1,1-2H-octitol (m/z 89, 205, 278, 320, 394).
| Strains | 89 m/z ion extraction |
118 m/z ion extraction |
|||||
|---|---|---|---|---|---|---|---|
| T-Kdo | 4-Kdo | 5-Kdo | 4,5-Kdo | 6-Gal (T-GalA)a | 6-Man | 4,6-Man | |
| 3841 (Parent Strain) | 9.8 | 28 | 0.0 | 62 | 77 | 0.0 | 23 |
| EL202 (rgtE−) | 49 | 15 | 0.0 | 36 | 51 | 49 | 0.0 |
| EL203 (rgtA−) | 45 | 24 | 0.0 | 31 | 66 | 14 | 20 |
| EL204 (rgtB−) | 6.2 | 63 | 0.0 | 31 | 66 | 2.9 | 31 |
| EL205 (rgtC−) | 16 | 22 | 0.0 | 62 | 77 | 18 | 4.4 |
| EL206 (rgtD−) | 20 | 43 | 0.0 | 36 | 67 | 18 | 15 |
a 6-linked Gal and terminal GalA can be distinguished by the relative abundance of ions m/z 189/191 and m/z 233/235. T, terminally linked residue.
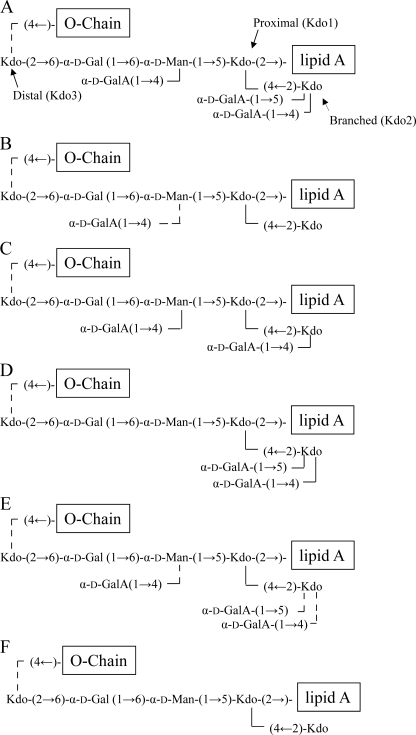
The core oligosaccharide structure of the parent R. leguminosarum bv. viciae 3841 LPS has been determined (1, 2) and shown to contain three Kdo residues as shown in Figs. 1. The proximal Kdo residue (Kdo1) is ketosidically linked the to the distal GlcN residue of the lipid A backbone, the branching Kdo residue (Kdo2) is linked to the 4-position of the proximal Kdo1 residue, and the distal Kdo residue (Kdo3) is linked to the 6-position of the core oligosaccharide Gal residue. Two of the three core GalA residues are linked to the 4- and 5-positions of the branching Kdo2 residue; i.e. Kdo2 is 4,5-linked. The core Man residue is linked to the 5-position of the proximal Kdo1 residue; i.e. Kdo1 is also 4,5-linked. The OPS, which is present in non-stoichiometric amounts, is linked to the 4-position of the distal Kdo3 residue; i.e. Kdo3 is 4-linked when OPS is present and terminally linked on LPS molecules without OPS. Thus, if one considers the parent LPS structure (Fig. 1), the ratio of 4,5-linked Kdo (Kdo1+Kdo2):Kdo3 (4-linked Kdo3 + terminally linked Kdo3) would be 2:1, and the ratio of terminally linked Kdo:4-linked Kdo, i.e. Kdo3 without and with OPS attached, would indicate the ratio of LPS structures that lack OPS. Table 3 shows that the ratio of 4,5-linked Kdo:4-linked + terminally linked Kdo is 1.7, which is consistent with the expected 2:1 ratio of (Kdo1 + Kdo2):Kdo3. The ratio of terminally linked to 4-linked Kdo3 from Table 3 is 0.35, which reflects the ratio of LPS molecules without OPS to those with OPS. In addition to the linkage of the Kdo residues, the expected ratio of 6-linked Gal + terminally linked GalA (which would also appear as 6-linked residue in the method used) to 4,6-linked Man would be 4:1, and Table 3 shows that it is 3.3, which is reasonably consistent with the expected value as the recovery of terminally linked GalA is always low with the method used. Thus, the linkage analysis results of the core Kdo, Gal, GalA, and Man residues are consistent with the known core oligosaccharide structure (see Fig. 1 and structure A in Fig. 7) for this LPS.
FIGURE 7.
The proposed core structures of the LPS from rgt mutants based on glycosyl linkage analysis, as described under “Results.” Dashed lines represent non-stoichiometric amounts of the attached residue.
The rgtE− mutant produces an LPS that is devoid of GalA. Thus, if one considers the parental LPS core structure shown in Fig. 1 or 7A, the absence of all GalA residues would result in one 4,5-linked Kdo (the proximal Kdo1 residue), one terminally linked Kdo (the branching Kdo2 residue), and one 4-linked Kdo (the distal Kdo3 residue); i.e. a 1:1:1 ratio of 4,5-: 4-:terminally linked Kdo residues. Because the LPS preparation consists of LPS with and without OPS, a certain amount of terminally linked Kdo3 would be present due to structures in which there is no OPS attached to the 4-position of the distal Kdo3 residue. The actual ratio of 4,5-:4-:terminally linked Kdo is 1.0:0.41:1.3. Again, the lower than expected value of 4-linked Kdo and the higher than expected value of terminally linked Kdo are due to the fact that there is, as with the parent LPS, a portion of LPS that lacks OPS. The reduction in the relative amount of 4-linked Kdo3 compared with the parent LPS suggests that the rgtE− mutant may be somewhat reduced in the proportion of LPS molecules containing OPS in comparison to parental LPS; however, this possible reduction is apparently not sufficient to detect with regard to the LPS composition and DOC-PAGE results described above. The observed linkages of the Gal, GalA, and Man residues are also consistent with the absence of terminal GalA. The mass spectrum of the PMAA derived from the 6-linked Gal residue shows that it does not contain deuterium at C6, confirming that terminal GalA is not present (the PMAA derivatives and their diagnostic primary ion fragments are listed in Table 3). The fact that there is no detectable 4,6-linked Man and only 6-linked Man confirms that there is no GalA attached to the 4-position of the Man residue, and the 1:1 ratio of 6-linked Gal:6-Man is completely consistent with a core oligosaccharide that is devoid of GalA residues. Thus, the rgtE− mutant has the core structure F shown in Fig. 7.
It is known that RgtA is the GalAT that adds one of the two GalA residues to either the 4- or 5-position of the branching Kdo2 residue in an in vitro assay using Man-Kdo2-lipid-IVa as the substrate (4). It was not known which of these two GalA residues is added by RgtA or how the lack of either one of these GalA residues might affect the addition of the remaining GalA residues to the LPS in R. leguminosarum bv. viciae 3841. The results for the Kdo linkages (see Table 3) show that the 4,5-:4-:terminal ratio of Kdo is 1:0.77:1.5, which is quite similar to the results for the Kdo linkages of the rgtE− mutant, which lacks both of the GalA residues attached to the branching Kdo2 residue. These results support the conclusion that the rgtA− is unable to add either GalA residue to the branching Kdo2 residue. Analysis, GC/MS, of the PMAAs for the Gal, GalA, and Man residues shows that a portion of the PMAA of the 6-linked Gal is due to terminal GalA and that the presence of 4,6-linked Man indicates that some GalA is added to the 4-position of the Man residue. The percentage of Man residues containing this GalA residue is 59%, whereas 41% lack the GalA residue. Thus, inactivation of rgtA prevents GalA addition to both the 4- and 5-positions of the branching Kdo2 residue, i.e. the absence of RgtA activity also prevents RgtB activity, and also reduces the level of GalA added to the Man residue, i.e. negatively affects the activity of RgtC. Thus, the rgtA− mutant contains a mixture of the core structures B and F shown in Fig. 7.
As with RgtA, it is known that RgtB adds one of the two GalA residues to the 4- or 5-position of the branching Kdo2 residue in an in vitro assay using Man-Kdo2-lipid-IVa as the substrate, but the identity of the position was unknown (4). Table 3 shows that the ratio of 4,5-:4-:terminal Kdo in the rgtB− mutant is 1.0:2.0:0.20. This result supports that RgtA activity in the absence of RgtB adds GalA to the 4-position of the branching Kdo2 residue. Therefore, the functions of RgtB and RgtA are to add GalA to the 5- and 4-positions, respectively, of the branching Kdo2 residue. Furthermore, given the fact that inactivation of RgtA entirely prevents the addition of either of these GalA residues, these results strongly support the conclusion that RgtA addition of GalA to the 4-position of the branching Kdo2 residue is required for RgtB activity. The linkages of the Gal, GalA, and Man residues of the rgtB− mutant, Table 3, show a 4,6-Man:6-Man:6-Gal + terminal-GalA ratio of 1.0:0.09:2.1. This result supports a core structure in which the Man residue is almost completely substituted with GalA at the 4-position. Thus, in the rgtB− mutant, RgtA activity is able to add GalA to the 4-position of the branching Kdo2 residue, and in contrast to the rgtA− mutant, almost complete GalA substitution of the Man residue at O-4 occurs. Therefore, the rgtB− mutant largely contains structure C shown in Fig. 7.
Previous work showed that RgtC added GalA to the 4-position of the Man residue in an in vitro assay using Man-Kdo2-lipid-IVa as the substrate (4). However, it was not known if the absence of this activity would affect the addition of any of the other core GalA residues during LPS synthesis in R. leguminosarum bv. viciae 3841. Table 3 shows that the ratio of 4,5-:4-:terminally linked Kdo residues is similar to that observed for the 3841 parent strain with the exception that the relative level of 4-linked Kdo is somewhat reduced (22 rather that 28%), whereas the level of terminal Kdo is somewhat increased (16 rather than 9.8%). These results support the conclusion that the rgtC−-branching Kdo2 residue is fully substituted by GalA, whereas the level of OPS substitution at O-4 of the distal Kdo3 may be slightly reduced compared with the parent LPS, resulting in the slight decrease in 4-linked Kdo and slight increase of terminally linked Kdo. The linkages of Gal, GalA, and Man residues (Table 3) clearly indicate a large decrease in 4,6-linked Man and an increase in 6-linked Man; i.e. the ratio of these residues is 0.24:1.0, indicating that the rgtC− mutant is essentially unable to add GalA to the 6-position of the core Man residue. This result is consistent with those previously published regarding RgtC activity in an in vitro assay using Man-Kdo2-lipid-IVa as the substrate (4). The small amounts of 4,6-linked Man indicates that the rgtC− mutant may be slightly leaky, and therefore, there is a low level of RgtC activity. In summary, these results support the conclusion that the rgtC− mutant mainly produces structure D shown in Fig. 7 with a small amount of the parent structure A.
We describe above that rgtD encodes the GalAT, which adds GalA to the 4′-position of the lipid A. To determine whether the rgtD− mutant was altered in ability to add GalA to the core oligosaccharide, glycosyl linkages were determined, and the results are shown in Table 3. The level of 4,5-linked Kdo is decreased in comparison to the parent LPS to levels similar to those observed in the rgtA−, rgtB−, and rgtE− mutants. This result indicates that the branching Kdo2 residue may not be fully substituted with GalA at either the 4- or both the 4- and 5-positions. However, the increased level of 4-linked Kdo relative to that observed in the rgtA− and the rgtE− mutants (43% compared with 15 and 24%, respectively) indicates that the rgtD− mutant is able to add some GalA to the 4-position of the branching Kdo2 residue. The level of terminally linked Kdo in the rgtD− mutant is also increased compared to that observed in the parent and rgtB− mutant but less than that observed for either the rgtE− or rgtA− mutants. Because the high levels of terminally linked Kdo in the latter mutants are due to a complete inability to add GalA to either the 4- or 5-positions of the branching Kdo2, the relatively reduced level of terminally linked Kdo in the rgtD− mutant is likely due to its ability to partially substitute the branching Kdo2 residue at the 4- or both the 4- and 5-positions. The linkage positions of Gal, GalA, and Man are also given in Table 3. Gal is 6-linked, and GalA is terminally linked as would be expected. The relative level of 6-linked Man (18%) is very similar to the level of 4,6-linked Man (15%), indicating that a little less than 50% of the Man residues is substituted with GalA at the 4-position. Thus, these data indicate that inactivating rgtD, in addition causing the loss of GalA on the 4′-position of the lipid A, results in a product that is affected in the addition of GalA to all positions in the core oligosaccharide, possibly by acting as a less than an optimal substrate for RgtA, -B, and -C. The linkage data indicate that the rgtD− mutant is able to produce a mixture of core structures A–F shown in Fig. 7.
Susceptibility of rgt Mutants to Deoxycholic Acid and PmxB
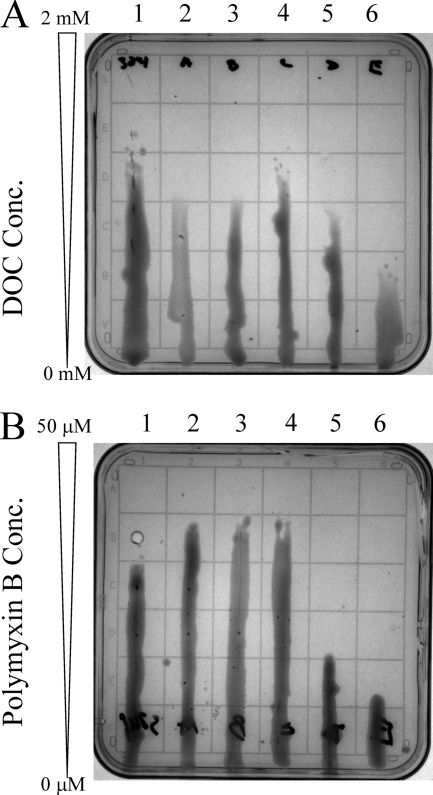
Because the rgt mutants were disrupted in their ability to add GalA to the LPS and it has been previously suggested that negatively charged moieties provide stability to the outer-membrane of Gram-negative bacteria (28, 29), we hypothesized that the membranes may be less stable due to the loss of charge provided by GalA residues. Therefore, we assayed their relative susceptibility to DOC using agar plates prepared with a DOC gradient (Fig. 8). With the exception of strain EL205 (rgtC−), the rgt mutants were somewhat more susceptible to DOC when compared with the parent strain (Fig. 8, panel A). Of the susceptible rgt mutants, strain EL202 (rgtE−), which lacks all GalA residues, showed the highest degree of susceptibility, whereas EL203 (rgtA−), EL204 (rgtB−), and EL206 (rgtD−) showed comparable susceptibility to each other and slightly more than the parent. These results suggest that, of the four GalA residues, those on the branched Kdo2 residue and lipid A contribute to the greatest extent to the stability of the outer-membrane in response to DOC.
FIGURE 8.
Susceptibility of the rgt mutants to DOC and PmxB. Panel A shows growth of bacteria on DOC gradient plates. 1, parent strain 3841; 2, mutant strain EL203 (rgtA−); 3, mutant strain EL204 (rgtB−); 4, mutant strain EL205 (rgtC−); 5, mutant strain EL206 (rgtD−); 6, mutant strain EL202. Panel B shows growth of bacteria on PmxB gradient plates. 1, parent strain 3841; 2, mutant strain EL203 (rgtA−); 3, mutant strain EL204 (rgtB−); 4, mutant strain EL205 (rgtC−); 5, mutant strain EL206 (rgtD−); 6, mutant strain EL202 (rgtE−).
In addition, we tested the relative susceptibilities of the rgt mutants to the antimicrobial compound PmxB. PmxB is a polycationic antimicrobial peptide that reacts with bacterial LPS due to interactions with the negative charged groups, e.g. phosphates, on the LPS as well as hydrophobic interactions. It is known that reduction of the negative charges on the LPS (e.g. from Salmonella) via introduction of positively charged groups such as 4-aminoarabinose or phosphoethanol amine increase resistance toward PmxB (28, 30). Strains EL203 (rgtA−), EL204 (rgtB−), and EL205 (rgtC−) were more resistant to PmxB when compared with parent strain (Fig. 8, panel B). As described above, these strains produce LPS with less GalA on the core region as compared with parent strain (Table 3) while maintaining normal amounts of GalA on the lipid A. Strains EL202 (rgtE−) and EL206 (rgtD−), both of which produce an LPS that lacks GalA on the 4′-position of the lipid A, were more susceptible to PmxB than the parent and the other rgt mutants (with EL202 (rgtE−), being slightly more susceptible than EL206 (rgtD−). Because removal of GalA reduced the negative charge of the LPS, it was expected that these mutants would have increased resistance to PmxB, and this indeed appears to be the case for mutants EL203 (rgtA−), EL204 (rgtB−), and EL205 (rgtC−). However, unexpectedly, mutants EL202 (rgtE−) and EL206 (rgtD−) that, respectively, produce LPSs that lack the GalA attached to the 4′-position of the lipid A as well as all or a portion of the core GalA residues were significantly more sensitive to PmxB than the parent and other rgt mutants. Thus, the GalA residue attached to the 4′-position of the lipid A appears to be important in conferring resistance rather than susceptibility to PmxB.
Symbiotic Phenotype between rgt Mutants and Host Plant (Pea)
Pea plants were inoculated with the rgtA−, -B−, -C−, and -D− mutants to determine whether disruption of these genes resulted in any obvious symbiotic defects. At 7 days post-inoculation (dpi), nodules were formed in plants inoculated with either the parent strain or the mutant strains. At 21 dpi the parent and mutant strains contained pink nodules, indicative of infected and nitrogen-fixing nodules. Thus, it appears that nitrogen-fixing symbiosis occurred in each of these rgt mutants.
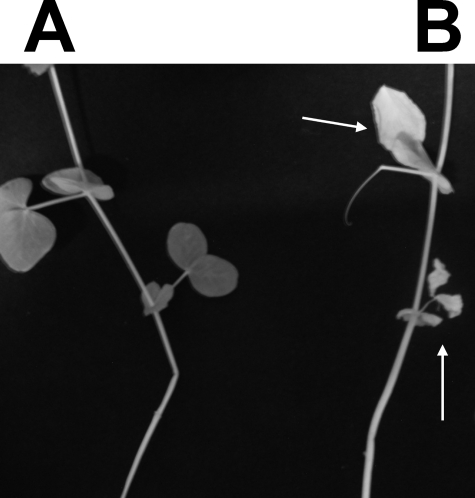
Interestingly, in the case with the rgtE− mutant, even though at 28 dpi plants inoculated with parent strain R. leguminosarum bv. viciae 3841 or rgtE− mutant both displayed pink nodules, developed comparable numbers of nodules, and had equal levels of acetylene reduction, indicating that nitrogenase activity in the rgtE− mutant was not impaired, a phenotype was observed. All rgtE− mutant-inoculated plants showed discoloration and wilting of the early leaves between 10 and 14 dpi, and by 28 dpi all mutant-inoculated plants (40 of 40) showed this wilting response (Fig. 9). None of the parent strain-inoculated plants had wilted leaves. Detailed microscopy studies on the development of nodules and bacteroids for each of these mutants is required to fully understand any possible symbiotic function of these GalA residues. These more detailed studies are needed particularly in view of the already published data showing that disruption of acpXL, the gene that encodes the acyl carrier protein required for the synthesis of the very long chain fatty acid that is present on the lipid A, also results in nitrogen-fixing nodules; however, microscopic examination showed that bacteroid formation was severely affected (31–33). This work is currently in progress for each of the rgt mutants and will be described in an additional manuscript.
FIGURE 9.
The symbiotic phenotype of plants inoculated with the rgtE− mutant inoculated. A, shown is a representative plant of 40 plants at 21 dpi inoculated with the parent strain 3841. B, shown is a representative plant of 40 plants at 21 d.p.i. inoculated with mutant EL202. The early leaves of all 40 mutant inoculated plants displayed necrosis between 7 and 14 dpi and were completely wilted by 28 dpi. All of the parent strain 3841 inoculated plants maintained green non-wilted leaves even at 28 dpi.
DISCUSSION
A number of Gram-negative bacteria contain GalA as part of their LPS core and/or lipid A. To study the functions of these GalA residues with regard to LPS synthesis, symbiosis, pathogenesis, and bacterial physiology, it was necessary to identify and disrupt each of the genes that encode for the various GalATs that add these GalA residues to the LPS. In this study we describe the identification and function of the rgtD and rgtE genes from the nitrogen-fixing endosymbiont R. leguminosarum bv. viciae 3841. We demonstrate that the rgtD gene product is responsible for the addition of GalA to the 4′-position of the lipid A distal GlcN and that the rgtE product is responsible for the synthesis of the Dod-P-GalA lipid donor. We also additionally prepared single gene mutations in each of the rgtA, -B, and -C genes. Our results together with the known structure of the R leguminosarum bv viciae 3841 LPS allowed us to determine the structures for each of the rgt mutant LPSs, demonstrate the order of GalA substitution during LPS synthesis, and determine the effects of each mutation on membrane stability and resistance to the antimicrobial peptide, PmxB.
Analysis of the lipid A from mutant EL206 (rgtD−) demonstrated the absence of GalA from the 4′-position and, therefore, showed that rgtD encodes the GalAT that transfers GalA to this position of the lipid A. Bacteria that contain 4′-galacturonosylated lipid A include R. leguminosarum and R. etli strains, which are members of the Rhizobiales (1), the stalk forming Caulobacter crescentus (34), and the hyperthermophile Aquifex aeolicus (35). Indeed, strains C. crescentus CB15 and A. aeolicus VF5 contain RgtD homologues (supplemental Fig. S3), and these proteins are likely involved in the synthesis of 4′-galacturonosylated lipid A in these bacteria as well. The information provided in this study concerning the rgtD will aid in future studies regarding 4′ galacturonylated lipid A in a variety of organisms.
Total lipid extracts from rgtE− mutant did not contain detectable amounts of the lipid donor Dod-P-GalA (Fig. 2) showing that the rgtE gene is necessary for the synthesis of Dod-P-GalA. In addition, a membrane preparation from the rgtE− mutant could not serve as a substrate to the support RgtA-catalyzed addition of GalA to Kdo2lipid-IVa, and it produces an LPS devoid of GalA. These results along with amino acid comparisons with other proteins that synthesize lipid-linked sugars (supplemental Fig. S2) support the conclusion that RgtE is the GalAT responsible for the synthesis of the Dod-P-GalA lipid donor required for the addition of GalA residues to the core region by RgtA, -B, and -C and to the lipid A by RgtD.
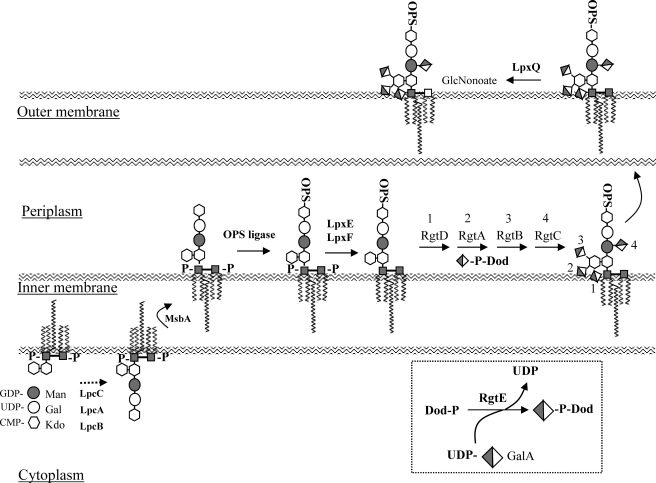
The determination of each of the rgt mutant core structures allowed us to predict the order of the transfer of the various GalA residues during LPS biosynthesis in R. leguminosarum bv. viciae 3841, shown in Fig. 10. Disruption of RgtD, which adds the GalA to the 4′-position of the lipid A, perturbed the addition of all of the remaining GalA residues, resulting in a mixture of core oligosaccharides containing various structures with and without GalA addition to the branching Kdo2 residue as well as to the Man residue. Because the parent LPS contains stoichiometric levels of all four GalA residues, it is likely that the RgtD product is required as the optimal substrate for the subsequent GalAT and, therefore, that RgtD adds the first of the LPS GalA residues. In addition, for all of the remaining rgtABC− mutants, the 4′-lipid A GalA residue is present, showing that RgtD activity is not affected in any of these mutants. Disruption of RgtA results in the complete absence of GalA on both the 4- and 5- positions of the branching Kdo2 residue and incomplete addition of GalA to the Man residue, whereas disruption of RgtB results in addition of GalA to the 4-position of Kdo2 and nearly complete addition of GalA to the Man residue. This implies that the RgtA activity is needed before RgtB and, further, that RgtA adds GalA to the 4-position of Kdo2, and therefore, RgtB adds GalA to the 5-position of Kdo2. In addition, because a small percentage of the Man residue in the rgtB mutant LPS lacks GalA, the likely order is RgtA followed by RgtB and then RgtC. This is further supported by the observation that whereas disruption of RgtC eliminated the addition of GalA to the Man residue, it did not affect the addition of GalA to the 4- and 5-positions of Kdo2. Thus, in R. leguminosarum bv. viciae 3841, the predicted order for the transfer of the GalA residues, as shown in Fig. 10, is transfer to the 4′-position of the lipid A by RgtD followed by transfer to the 4-position of the branching Kdo2 by RgtA, then to the 5-position of the Kdo2 residue by RgtB, and last, to the 4-position of the Man residue by RgtC.
FIGURE 10.
Schematic diagram showing the biosynthetic steps for R. leguminosarum bv. viciae LPS and indicating the order by which the GalA residues are added to the lipid A and core oligosaccharide. The LPS core backbone is built off of the lipid A on the cytosolic side of the innermembrane and is flipped to the periplasmic side of the innermembrane where the transmembrane Rgt proteins transfer GalA from the lipid donor Dod-P-GalA to the LPS (1). The LPS is transported to the outer membrane where the oxidase LpxQ oxidizes the proximal GlcN to GlcNonate. The Rgt proteins are labeled 1–4 to signify the predicted order of GalA addition. LpcA, -B, and -C = glycosyl transferases, rgtA, -B, -C, -D, and -E = GalA transferases. The boxed region shows the synthesis of Dod-P-GalA from UDP-GalA by RgtE.
With regard to membrane stability, all of the rgt mutants were more sensitive to DOC than the parent strain (Fig. 8, panel B) except for EL205 (rgtC−). Strain EL202 (rgtE−) was the most sensitive, whereas EL203 (rgtA−), EL204 (rgtB−), and EL206 (rgtD−) were comparable with each other in their DOC susceptibility but less sensitive than EL202 (rgtE−). The complemented strains recovered parent strain levels of DOC resistance. These results suggest that the GalA residues present on the branching Kdo2 residue and on the 4′-position of the lipid A distal GlcN confer membrane stability toward disruption by DOC. It is possible that the presence of GalA contributes to the cross-linking of LPS molecules in the outer membrane through ionic bridging between the negatively charged carboxyl moiety and divalent cations such as Ca2+ and Mg2+. Typically, as is the case with enteric bacteria, lipid A is bis-phosphorylated at the 1- and 4′-positions and in some cases on inner core residues. These negatively charged phosphates contribute to membrane stability through LPS/divalent cation bridging (29, 36). R. leguminosarum and R. etli lipid A does not contain the 1- and 4′-phosphates but, rather, contains 4′-GalA and, to some degree, proximal 2-aminogluconic acid (Fig. 1), both negatively charged moieties. The GalA residues likely add negative charge in lieu of phosphate, and results presented in this study suggest that the LPS GalA residues function similarly to phosphate groups in stabilizing the outer membrane.
In addition to affecting sensitivity to DOC, the rgt mutants varied in their susceptibility to PmxB. PmxB is an acylated polycationic cyclic peptide that interacts with the negatively charged moieties and hydrophobic acyl portion of LPS. Mutant strains EL203 (rgtA−), EL204 (rgtB−), and EL205 (rgtC−) were somewhat more resistant to PmxB compared with parent strain. In the case of these mutants, reduction of the anionic GalA residues would be expected to reduce interaction with the cationic PmxB, and therefore, it is not surprising that increased PmxB resistance was observed. However, strains EL202 (rgtE−) and EL206 (rgtD−) were significantly less resistant to PmxB compared with parent strain. These strains differ from the other rgt mutant strains in that the LPSs from both completely lack the lipid A 4′-GalA residue, whereas the other rgt mutants and parent strain maintain this residue. It is possible that the lipid A 4′-GalA residue is a major contributor for conferring membrane stability against disruption by PmxB, and therefore, even though the absence of GalA may reduce PmxB binding, the greater reduction in membrane stability results in increased disruption of the outer membrane and, therefore, increased sensitivity to PmxB.
It is unclear why R. leguminosarum and R. etli strains modify their LPS with GalA. Perhaps bacteria that attach GalA instead of phosphate have a competitive advantage in phosphate-limiting environments. In addition, GalA is a common component of the plant cell wall (e.g. such as galacturonan polysaccharides) and the presence of GalA on the LPS may provide host mimicry, allowing it to escape or manipulate the host defense response. Because bacteroids maintain a tight association with the plant-derived peribacteroid membrane, it is tempting to speculate that LPS GalA residues may interact in an ionic manner with the peribacteroid membrane during endosymbiosis to help maintain this tight association. Furthermore, the GalA residues present on the LPS may aid in the recruitment of plant defensin-like molecules. The causative agent for terminal bacteroid differentiation of Sinorhizobum meliloti was recently reported (37) to be host-derived defensin-like peptides, nodule-specific cysteine-rich peptides (NCRs). Nodule-specific cysteine-rich peptides act at the cell surface and cause elongation, branching, and endoreduplication of free-living S. meliloti. A similar process likely occurs during symbiosis between R. leguminosarum bv. viciae 3841 and its host, Pisum sativum (pea), and it is possible that LPS GalA residues contribute to the interaction between pea NCRs and the bacterial cell surface. Further work is in progress to determine the role of the LPS GalA residues with regard to its interaction with pea defensins.
Supplementary Material
Acknowledgments
We acknowledge Hak Suk Chung, Ziqiang Guan, and Chris Raetz for help in preparing and performing LC-ESI-MS analysis of the total membrane extracts and in performing the RgtA assays.
This work was supported, in whole or in part, by National Institutes of Health Grant GM39583 (to R. W. C.). This work was also supported by Department of Energy Grant DE-FG-02-93ER20097 (to the Complex Carbohydrate Research Center). This work is in partial fulfillment for the Ph.D. degree of D. B. B.

This article contains supplemental Figs. 1–3.
- GalA
- galacturonic acid
- Kdo
- 3-deoxy-d-manno-2 octulosonic acid
- GlcNonate
- 2-aminogluconate
- Dod-P-GalA
- dodecaprenyl phosphate galacturonic acid
- ESI
- electrospray ionization
- DOC
- deoxycholic acid
- PmxB
- polymyxin B
- PMAA
- partially methylated alditol acetate
- dpi
- days post-inoculation
- GalAT
- galacturonic acid transferase
- TY
- Tryptone-yeast
- OPS
- O-chain polysaccharide.
REFERENCES
- 1. Carlson R. W., Forsberg L. S., Kannenberg E. L. (2010) Lipopolysaccharides in Rhizobium legume symbioses in Subcellular Biochemistry: Endotoxins: Structure, Function, and Recognition (Quinn P. J., Wang X., eds) pp. 339–386, Springer-Verlag New York Inc; [DOI] [PubMed] [Google Scholar]
- 2. Forsberg L. S., Carlson R. W. (1998) The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359. The complete structure of the core region of R. etli lipopolysaccharides. J. Biol. Chem. 273, 2747–2757 [DOI] [PubMed] [Google Scholar]
- 3. Kanjilal-Kolar S., Raetz C. R. (2006) Dodecaprenyl phosphate-galacturonic acid as a donor substrate for lipopolysaccharide core glycosylation in Rhizobium leguminosarum. J. Biol. Chem. 281, 12879–12887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kanjilal-Kolar S., Basu S. S., Kanipes M. I., Guan Z., Garrett T. A., Raetz C. R. (2006) Expression cloning of three Rhizobium leguminosarum lipopolysaccharide core galacturonosyltransferases. J. Biol. Chem. 281, 12865–12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Karbarz M. J., Six D. A., Raetz C. R. (2009) Purification and characterization of the lipid A 1-phosphatase LpxE of Rhizobium leguminosarum. J. Biol. Chem. 284, 414–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Price N. P., Jeyaretnam B., Carlson R. W., Kadrmas J. L., Raetz C. R., Brozek K. A. (1995) Lipid A biosynthesis in Rhizobium leguminosarum, Role of a 2-keto-3-deoxyoctulosonate-activated 4′ phosphatase. Proc. Natl. Acad. Sci. U.S.A. 92, 7352–7356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Que-Gewirth N. L., Karbarz M. J., Kalb S. R., Cotter R. J., Raetz C. R. (2003) Origin of the 2-amino-2-deoxy-gluconate unit in Rhizobium leguminosarum lipid A. Expression cloning of the outer membrane oxidase LpxQ. J. Biol. Chem. 278, 12120–12129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beringer J. E. (1974) R factor transfer in Rhizobium leguminosarum. Microbiology 84, 188–198 [DOI] [PubMed] [Google Scholar]
- 9. Becker A., Schmidt M., Jäger W., Pühler A. (1995) New gentamicin-resistance and lacZ promoter-probe cassettes suitable for insertion mutagenesis and generation of transcriptional fusions. Gene 162, 37–39 [DOI] [PubMed] [Google Scholar]
- 10. Hoang T. T., Karkhoff-Schweizer R. R., Kutchma A. J., Schweizer H. P. (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally located DNA sequences. Application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212, 77–86 [DOI] [PubMed] [Google Scholar]
- 11. Westphal O., Jann K. (1965) Endotoxins: Extraction with phenol-water and further applications of the procedure. Methods Carbohydr. Chem. 5, 83–91 [Google Scholar]
- 12. Fraysse N., Jabbouri S., Treilhou M., Couderc F., Poinsot V. (2002) Symbiotic conditions induce structural modifications of Sinorhizobium sp. NGR234 surface polysaccharides. Glycobiology 12, 741–748 [DOI] [PubMed] [Google Scholar]
- 13. Reuhs B. L., Carlson R. W., Kim J. S. (1993) Rhizobium fredii and Rhizobium meliloti produce 3-deoxy-d-manno-2-octulosonic acid-containing polysaccharides that are structurally analogous to group II K antigens (capsular polysaccharides) found in Escherichia coli. J. Bacteriol. 175, 3570–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kittelberger R., Hilbink F. (1993) Sensitive silver-staining detection of bacterial lipopolysaccharides in polyacrylamide gels. J. Biochem. Biophys. Methods 26, 81–86 [DOI] [PubMed] [Google Scholar]
- 15. York W. S., Darvill A. G., McNeil M., Stevenson T. T., Albersheim P. (1986) Isolation and characterization of plant-cell walls and cell-wall components. Methods Enzymol. 118, 3–40 [Google Scholar]
- 16. Bhat U. R., Forsberg L. S., Carlson R. W. (1994) Structure of lipid A component of Rhizobium leguminosarum bv. phaseoli lipopolysaccharide. Unique nonphosphorylated lipid A containing 2-amino-2-deoxygluconate, galacturonate, and glucosamine. J. Biol. Chem. 269, 14402–14410 [PubMed] [Google Scholar]
- 17. Ryan J. M., Conrad H. E. (1974) Structural heterogeneity in the lipopolysaccharide of Salmonella newington. Arch. Biochem. Biophys. 162, 530–535 [DOI] [PubMed] [Google Scholar]
- 18. Forsberg L. S., Noel K. D., Box J., Carlson R. W. (2003) Genetic locus and structural characterization of the biochemical defect in the O-antigenic polysaccharide of the symbiotically deficient Rhizobium etli mutant, CE166. Replacement of N-acetylquinovosamine with its hexosyl-4-ulose precursor. J. Biol. Chem. 278, 51347–51359 [DOI] [PubMed] [Google Scholar]
- 19. Ciucanu I., Kerek F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res. 131, 209–217 [Google Scholar]
- 20. Cointe D., Leroy Y., Chirat F. (1998) Determination of the sialylation level and of the ratio α-(2→3)/α-(2→6) sialyl linkages of N-glycans by methylation and GC/MS analysis. Carbohydr. Res. 311, 51–59 [DOI] [PubMed] [Google Scholar]
- 21. Anumula K. R., Taylor P. B. (1992) A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal. Biochem. 203, 101–108 [DOI] [PubMed] [Google Scholar]
- 22. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST, a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., Lu F., Marchler G. H., Mullokandov M., Omelchenko M. V., Robertson C. L., Song J. S., Thanki N., Yamashita R. A., Zhang D., Zhang N., Zheng C., Bryant S. H. (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 39 (Database issue): D225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trent M. S., Ribeiro A. A., Lin S., Cotter R. J., Raetz C. R. (2001) An inner membrane enzyme in Salmonella and Escherichia coli that transfers 4-amino-4-deoxy-l-arabinose to lipid A. Induction on polymyxin-resistant mutants and role of a novel lipid-linked donor. J. Biol. Chem. 276, 43122–43131 [DOI] [PubMed] [Google Scholar]
- 25. Phadnis S. H., Das H. K. (1987) Use of the plasmid pRK 2013 as a vehicle for transposition in Azotobacter vinelandii. J. Biosci. 12, 131–135 [Google Scholar]
- 26. Song F., Guan Z., Raetz C. R. (2009) Biosynthesis of undecaprenyl phosphate-galactosamine and undecaprenyl phosphate-glucose in Francisella novicida. Biochemistry 48, 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang X., Ribeiro A. A., Guan Z., Raetz C. R. (2009) Identification of undecaprenyl phosphate-β-d-galactosamine in Francisella novicida and its function in lipid A modification. Biochemistry 48, 1162–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Snyder S., Kim D., McIntosh T. J. (1999) Lipopolysaccharide bilayer structure. Effect of chemotype, core mutations, divalent cations, and temperature. Biochemistry 38, 10758–10767 [DOI] [PubMed] [Google Scholar]
- 30. Trent M. S., Stead C. M., Tran A. X., Hankins J. V. (2006) Diversity of endotoxin and its impact on pathogenesis. J. Endotoxin Res. 12, 205–223 [DOI] [PubMed] [Google Scholar]
- 31. Vedam V., Haynes J. G., Kannenberg E. L., Carlson R. W., Sherrier D. J. (2004) A Rhizobium leguminosarum lipopolysaccharide lipid-A mutant induces nitrogen-fixing nodules with delayed and defective bacteroid formation. Mol. Plant-Microbe Interact. 17, 283–291 [DOI] [PubMed] [Google Scholar]
- 32. Brown D. B., Huang Y. C., Kannenberg E. L., Sherrier D. J., Carlson R. W. (2011) An acpXL mutant of Rhizobium leguminosarum bv. phaseoli lacks 27-hydroxyoctacosanoic acid in its lipid A and is developmentally delayed during symbiotic infection of the determinate nodulating host plant Phaseolus vulgaris. J. Bacteriol. 193, 4766–4778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Haag A. F., Wehmeier S., Beck S., Marlow V. L., Fletcher V., James E. K., Ferguson G. P. (2009) The Sinorhizobium meliloti LpxXL and AcpXL proteins play important roles in bacteroid development within alfalfa. J. Bacteriol. 191, 4681–4686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smit J., Kaltashov I. A., Kaltoshov I. A., Cotter R. J., Vinogradov E., Perry M. B., Haider H., Qureshi N. (2008) Structure of a novel lipid A obtained from the lipopolysaccharide of Caulobacter crescentus. Innate Immun. 14, 25–37 [DOI] [PubMed] [Google Scholar]
- 35. Plötz B. M., Lindner B., Stetter K. O., Holst O. (2000) Characterization of a novel lipid A containing d-galacturonic acid that replaces phosphate residues. The structure of the lipid A of the lipopolysaccharide from the hyperthermophilic bacterium Aquifex pyrophilus. J. Biol. Chem. 275, 11222–11228 [DOI] [PubMed] [Google Scholar]
- 36. Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Van de Velde W., Zehirov G., Szatmari A., Debreczeny M., Ishihara H., Kevei Z., Farkas A., Mikulass K., Nagy A., Tiricz H., Satiat-Jeunemaître B., Alunni B., Bourge M., Kucho K., Abe M., Kereszt A., Maroti G., Uchiumi T., Kondorosi E., Mergaert P. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126 [DOI] [PubMed] [Google Scholar]
- 38. Wang T. L., Wood E. A., Brewin N. J. (1982) Growth regulators, Rhizobium, and nodulation of peas. Planta 155, 345–349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.