Background: Synaptic activity induces translation of mRNAs in dendrites of neurons.
Results: Makorin 1 (MKRN1) interacts with poly(A)-binding protein, stimulates translation, accumulates in neuronal dendrites after plasticity-inducing stimuli, and is associated with dendritic mRNAs.
Conclusion: MKRN1 has the potential to locally control the translation of dendritic mRNAs at synapses.
Significance: MKRN1 is a novel positive regulator of translation.
Keywords: mRNA, Neurobiology, Ribonuclear Protein (RNP), RNA-binding Protein, Synaptic Plasticity, Translation Control
Abstract
The poly(A)-binding protein (PABP), a key component of different ribonucleoprotein complexes, plays a crucial role in the control of mRNA translation rates, stability, and subcellular targeting. In this study we identify RING zinc finger protein Makorin 1 (MKRN1), a bona fide RNA-binding protein, as a binding partner of PABP that interacts with PABP in an RNA-independent manner. In rat brain, a so far uncharacterized short MKRN1 isoform, MKRN1-short, predominates and is detected in forebrain nerve cells. In neuronal dendrites, MKRN1-short co-localizes with PABP in granule-like structures, which are morphological correlates of sites of mRNA metabolism. Moreover, in primary rat neurons MKRN1-short associates with dendritically localized mRNAs. When tethered to a reporter mRNA, MKRN1-short significantly enhances reporter protein synthesis. Furthermore, after induction of synaptic plasticity via electrical stimulation of the perforant path in vivo, MKRN1-short specifically accumulates in the activated dendritic lamina, the middle molecular layer of the hippocampal dentate gyrus. Collectively, these data indicate that in mammalian neurons MKRN1-short interacts with PABP to locally control the translation of dendritic mRNAs at synapses.
Introduction
Neurons are considered the most complex cell type of higher organisms. To ensure appropriate function within an intricate network, their molecular constituents have to be synthesized, modified, sorted, and degraded in due time. Synapses in particular are constantly modified in an activity-dependent manner (1), and synaptic function requires a well regulated interplay of mechanisms to maintain accurately balanced protein levels (2). In part, this is achieved by local translation of mRNAs that are transported to dendrites to await translational activation (3–6). RNA transport is specified by sequences called “zip codes,” and proteins that collectively build up messenger ribonucleoprotein complexes referred to as RNA granules (7). These complexes are most likely remodeled to allow for translational activation, translational silencing, or degradation of mRNA molecules. Different types of granules exist with distinct contents of proteins and RNAs (8–12), yet further work is needed to entirely reveal the diversity and functional significance of RNA granules in neurons.
The cytoplasmic poly(A)-binding protein (PABP)2 associates with the poly(A) tail of mRNAs and thus promotes translation (13). Yet the protein also interacts with various non-poly(A) sequences including the zip code of the dendritically localized vasopressin mRNA (14, 15). As a part of distinct multiprotein complexes, PABP is involved in translational stimulation (16–18) and silencing (19, 20), RNA localization (21), and mRNA turnover (22). In neurons, PABP localizes to RNA granules (8, 11, 23), indicating that it is involved in dendritic mRNA metabolism.
To better understand the molecular action of PABP in regulating mRNA fate in nerve cells, we have employed yeast two-hybrid techniques to identify binding partners required in support of its function. Here, we report the characterization of a novel PABP-interacting protein, RING zinc finger protein Makorin-1 (MKRN1) (24). The mkrn1 gene belongs to family members that encode putative RNA-binding proteins. MKRN1 is a modular protein with distinct arrays of C3H zinc finger (ZF) motifs, a ZF structure with unusual cysteine/histidine spacing, and a RING domain typically found in E3 ubiquitin ligases (25). Apparently, MKRN1 exhibits divergent functions both in the cell nucleus and the cytoplasm. As an E3 ubiquitin ligase it acts on itself and the catalytic subunit of human telomerase reverse transcriptase (26), p53 and p21 (27). Furthermore, MKRN1 modulates RNA polymerase II-mediated transcription (28) and may play a role in mRNA decay (29).
In our yeast two-hybrid screen with PABP bait, we have exclusively isolated a shorter isoform (called MKRN1-short) of hitherto unknown function encoded by exons 1–5 of the mkrn1 gene. We show that this protein is the major isoform in rat brain. MKRN1-short expression in forebrain neurons is more abundant than elsewhere in the brain, and the protein resides in both the nucleus as well as the cell body and dendrites. MKRN1-short contains a PAM2 (PCI/PINT associated module 2)-like motif that mediates its interaction with PABP in an RNA-independent manner. PAM2 motifs are found in several PABP-interacting proteins, for example the PABP-interacting protein 1 (PAIP1) and PAIP2 (30) that affect translation in a positive and negative manner, respectively (31, 32). MKRN1-short exerts a strong positive effect on translation when it is tethered to a reporter mRNA in primary neurons. In vivo, MKRN1-short accumulates in dendrites after induction of long term potentiation (LTP), a cellular process that requires de novo protein synthesis (33, 34). Taken together, these findings suggest that in mammalian brain neurons MKRN1-short functions as a modulator of local protein synthesis in dendrites.
EXPERIMENTAL PROCEDURES
Experimental Animals
Wistar- or Sprague-Dawley rats were used. Animals were bred and handled in accordance with national guidelines for animal welfare.
Electrophysiological Manipulation and Brain Tissue Preparation
Adult male Sprague-Dawley rats (250–500 g; Charles River) were deeply anesthetized with urethane (1.25 g/kg body weight, subcutaneously initially, and additional injections as needed). Surgery and stimulation procedures were performed as described (35). Briefly, stimulating electrodes were placed in the angular bundle of the medial perforant path. Recording microelectrodes were placed in the dorsal blade of the granule cell layer. High frequency stimulation was applied for 2 h to maximally evoke population spikes and induce robust LTP in granule cells as has been described (36). One train consisted of 8 pulses (500 μA, 0.1-ms pulse duration) of 400 Hz once per 10 s. Immediately after the end of the stimulation, rats were transcardially perfused with 4% paraformaldehyde.
Cloning Procedures
DNAs encoding PABP, MKRN1, DDX6, and Shank3 were either amplified by PCR techniques, or constructs were generated by subcloning procedures. Constructs generated by PCR were subjected to DNA sequencing. The clones employed in this study are summarized in supplemental Table 1. The following vectors were used: pGEX-6P-3 (GE Healthcare), pGBKT7 (Clontech), pcDNA6/myc-His (Invitrogen), pEGFP-C (Clontech). pN22-C1 and pN22-FLAG3-C1 are derivatives of pEGFP-C1 (Clontech), in which the EGFP cDNA has been replaced by regions encoding 22 amino acid residues from the N protein of the phage λ (N22; 37) and a fusion protein consisting of N22 and three consecutive FLAG epitopes, respectively. The eukaryotic expression vector pinFiRein-boxB16B is based on the previously described plasmid pFiRe-basic (38). It contains two recombinant genes, both of which are controlled by independent CMV immediate-early promoters, contain a chimeric intron from pFN21 (Promega) upstream of the coding region, and encode Photinus (PhoLuc) and Renilla luciferase (RenLuc), respectively. In their 3′-UTRs, PhoLuc transcripts include 16 consecutive copies of the 15-nucleotide RNA hairpin termed box B that specifically interacts with the N22 domain (37). The 3′-UTR was chosen for box B insertions because this part of mRNAs often regulates translation (39). pcDNA-T7 is a pcDNA3 derivative (Invitrogen) containing a T7 tag-encoding sequence (kindly provided by Dr. Hans-Jürgen Kreienkamp, University Medical Center Hamburg-Eppendorf, Hamburg, Germany).
Antibodies
Rabbit polyclonal antibodies were generated against full-size human MKRN1-short and rat PABP C terminus fused to GST. Antisera were produced by Pineda Antibody-Service (Berlin, Germany) and used at a dilution of 1:10 000 (anti-MKRN1) and 1:2000 (anti-PABP) unless stated otherwise. The following antibodies and antibody matrices were employed at the manufacturer recommended dilutions for the respective applications: GFP-Trap®_A (Chromotek), monoclonal mouse anti-T7 (Novagen), monoclonal mouse anti-T7-agarose (Novagen), monoclonal mouse anti-FLAG M2 (Stratagene), monoclonal mouse anti-FLAG M2-agarose (Sigma), monoclonal mouse anti-MAP2 (Chemicon), monoclonal mouse anti-myc (Sigma), rabbit anti-GFP (Abcam), goat anti-rabbit IgG-HRP (Dianova), goat anti-mouse IgG-HRP (Dianova), mouse anti-rabbit IgG (light chain specific)-HRP (Jackson ImmunoResearch Laboratories, Inc.), rabbit anti-mouse IgG (light chain specific)-HRP (Jackson ImmunoResearch Laboratories), Cy3-labeled goat anti-mouse IgG (Dianova), Cy3-labeled goat anti-rabbit IgG (Dianova), Alexa Fluor® 488-labeled goat anti-mouse IgG (Invitrogen), and Alexa Fluor® 488-labeled goat anti-rabbit IgG (Invitrogen).
Cell Culture and Transfection
HEK-293 and HeLa cells (DSMZ, Braunschweig, Germany) were grown in DMEM containing 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C, 5% CO2. For transfection experiments, antibiotics were omitted. Cells were plated at densities of 9–10 × 105 cells/6-cm dish and 2–3 × 106 cells/10 cm dish, respectively. Transfections were done with FuGENE 6 (Roche Applied Science) at a 3:1 ratio of FuGENE 6:DNA. In some cases cells were grown in the presence of 10 μm MG-132 (Calbiochem) for 1–4 h. Cells were lysed 24–36 h after transfections.
Rat hippocampal and cortical neurons were prepared from rat embryos (E19) according to published procedures (40) with the following modifications; neurons were grown in Neurobasal A medium containing B27 supplement (Invitrogen) without a glial feeder layer, and transfections were performed at day 7 in vitro (7 DIV) using calcium phosphate co-precipitation techniques (40). For luciferase assays and quantitative PCR analyses (see below), respective constructs were transfected in at least five independent series of transfections.
Yeast Two-hybrid Technique
Rat PABP cDNA was cloned into pGBKT7 (see “Cloning Procedures”). PABP-interacting proteins were identified by using the matchmaker® yeast-pretransformed human brain cDNA library (Clontech) as outlined in the manufacturer's instruction manual.
RNA Extraction and Purification
Total RNA from cultured cells and tissues was prepared with PeqGold RNAPure (PeqLab) according to the manufacturer's instructions followed by a cleanup using RNeasy (Qiagen). RNA purity and concentration was determined spectrophotometrically.
cDNA Synthesis
2–5 μg of total RNA were reverse-transcribed using Superscript II RT (Invitrogen) or RevertAid Premium Reverse Transcriptase (Fermentas) and 0.5 μg of oligo(dT) according to manufacturer instructions. Subsequently, RNA was digested with 2 units of RNase H (Invitrogen or Fermentas) for 20 min at 37 °C. For quantitative PCR analyses (see below) RNAs were subjected to two independent cDNA syntheses.
Qualitative PCR
2 μl of cDNAs were subjected to PCR in 50-μl reaction volumes using the ProofStart DNA polymerase kit (Qiagen) according to the manufacturer's instructions. The following primers were employed: MKRN1-short forward primer 5′-TGTTCAGCGCAGCAAGGAC-3′ (accession number NM_001004233, nucleotides 819–837) and reverse primer 5′-AGATTTAACGGATGGTGGATTTTT-3′ (accession number NM_001004233, nucleotides 1347–1372); MKRN1-long forward primer 5′-GTGGGATCTGCATGGAGGTGGTCT-3′ (accession number AAHX01058705, nucleotide residues 23043–23068) and reverse primer 5′-CCTTCGTTGGGCCTGGTATCTGCT-3′ (accession number AAHX01058705, nucleotide residues 22662–22685). PCR products were subjected to DNA sequencing.
Real-time PCR with TaqMan® Probes
A TaqMan® PCR reaction (20 μl) included 1.4 μl of cDNA, 1× TaqMan® Universal PCR Master Mix (Applied Biosystems), and either the TaqMan® Gene Expression Assays RenLuc (Applied Biosystems custom-made part no./assay ID 4332079/AIGJPCV) or PhoLuc (Applied Biosystems custom-made part no./assay ID 4332079/AIFAQ6N), respectively. TaqMan® PCR was carried out in a Sequence Detection System Applied Biosystems PRISM® 7900 HT thermocycler using the standard program (2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C and 1 min at 60 °C) and analyzed with Applied Biosystems PRISM® 7900 HT software. Data were evaluated using the comparative CT (threshold cycle) method for the relative quantification of the amount of PhoLuc cDNA normalized to RenLuc cDNA as a reference, given by 2−ΔΔCT. The CT parameter is defined as the fractional cycle number at which the fluorescence exceeds a fixed threshold value. Each cDNA was analyzed in triplicate with both TaqMan® Gene Expression Assays. Non-template and no reverse transcriptase controls were included in each run.
SYBR® Green Real-time RT-PCR
Semiquantitative determination of mRNA levels in immunoprecipitated messenger ribonucleoproteins (see below) was performed with the QuantiTectTM SYBR® Green RT-PCR kit (Qiagen) and QuantiTectTM Primer assays Arc (QT00373086, Qiagen), MAP2 (QT01084244), and β-tubulin 3 (QT00188818) according to the manufacturer's protocol on a Rotor-Gene 3000 cycler (Corbett/Qiagen). PCR reactions (20 μl each) were done in duplicate and included 1 μl of RNA solution. Non-template and no reverse transcriptase controls were included in each run. Data analysis was performed using REST (relative expression software tool) 2008 software for group-wise comparison and statistical analysis of relative expression results in real-time PCR (41).
Northern Blot Hybridization
Hybridization was performed with a multiple tissue Northern blot (Clontech). 25 ng of MKRN1-short cDNA was labeled with [32P]dCTP (3000 Ci/mmol, 10 mCi/ml, GE Healthcare) to high specific activity using the Prime-It II random primer labeling kit (Stratagene). Non-incorporated nucleotides were removed by PCR column purification (Qiagen). RNA blots were prehybridized for 2 h at 42 °C with 10 ml of ULTRAhyb solution (Ambion). Hybridization was performed in 10 ml of fresh ULTRAhyb solution with the addition of labeled probe (2 × 106 cpm/ml) at 42 °C overnight. Blots were washed in 2× saline, sodium citrate (SSC; 20× 3 m NaCl, 0.3 m sodium citrate, pH 7.0), 0.1% SDS (2 × 10 min at room temperature), 1 × SSC, 0.1% SDS (15 min, room temperature), 0.2 × SSC, 0.1% SDS (15 min, room temperature), 0.2 × SSC, 1% SDS (30 min, 50 °C), and exposed to x-ray film.
Fractionation of Polysomes
Separation of polysomes from freshly prepared rat hippocampal homogenates by sucrose gradient ultracentrifugation (15–45% sucrose) was done as described (42). Individual 1-ml gradient fractions were collected, immediately frozen in liquid nitrogen, and stored at −80 °C until use. For RNA precipitation, ⅕ of each fraction was incubated in 70% ethanol, 10 mm EDTA, and 0.044 mg/ml yeast tRNA for 2 h at 4 °C and subsequently centrifuged for 15 min, 12,000 × g, and 4 °C. RNA pellets were dissolved in RNase-free H2O and further purified as described above.
Protein Extraction
Protein Extracts from Cultured Cell lines
Cells were washed twice with ice-cold PBS. Cells were lysed in 0.8 ml/6-cm dish of radioimmunoprecipitation assay buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1% (v:v) Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 5 mm EDTA) containing protease inhibitors (Complete®, Roche Applied Science). Lysates were incubated on ice for 30 min and centrifuged for 15 min at 4 °C and 16000 × g.
Protein Extracts from Rat Hippocampi
Two rat hippocampi were homogenized in 1 ml of ice-cold buffer (50 mm HEPES, pH 7.5, 500 mm NaCl, 1.5 mm MgCl2, 1 mm EGTA, 10% (v:v) glycerol, 1% (w:v) Triton X-100, and Complete®) and centrifuged at 20,000 × g for 30 min and 4 °C.
Protein Extracts from Transfected Primary Neurons for Messenger Ribonucleoprotein Immunoprecipitations
Neurons were washed twice with ice-cold Hanks' balanced salt solution. Per 10-cm dish, cells were lysed in 0.425 ml of lysis buffer (120 mm NaCl, 50 mm HEPES, pH 7.5, 5 mm EDTA, 0.5% (w:v) Triton X-100) containing protease inhibitors (Complete®, Roche Applied Science and 200 units/ml RiboLock RNase inhibitor (Fermentas). Lysates were incubated on ice for 15 min and centrifuged for 15 min at 4 °C and 13,000 × g. Aliquots for RNA extraction (10%; input RNA) and Western blot analyses (2.5%, input protein) were withdrawn. The remainder was subjected to immunoprecipitation (see below).
Protein concentration was determined using the Protein Assay Reagent (Bio-Rad) with bovine serum albumin as standard. Proteins were snap-frozen in liquid nitrogen and stored at −80 °C until use.
Immunoprecipitation
Immunoprecipitations of proteins dissolved in radioimmunoprecipitation assay buffer (see below) were usually performed with 700–1000 μg of proteins at a concentration of ∼1 μg/μl.
For FLAG-agarose, T7-agarose, and GFP-Trap®_A (Chromotek), 20 μl of monoclonal mouse anti-FLAG M2- or anti-T7- agarose or 10 μl of GFP-Trap®_A/reaction were washed 5× with radioimmunoprecipitation assay buffer. Protein lysate was added and rotated overnight at 4 °C. Supernatant was removed, and the agarose was washed 3× with 1 ml of radioimmunoprecipitation assay buffer. Bound proteins were eluted with 1× concentrated Laemmli sample buffer at 95–99 °C for 5 min. In some cases immunoprecipitations were performed in the presence of RNase A as described (8).
Immunoprecipitations of recombinant proteins from transfected primary neurons were performed with 40 μl of GFP-Trap®_A/reaction. Lysates from two 10-cm dishes were combined, NaCl was added (final concentration, 200 mm), and lysates were incubated overnight at 4 °C with constant rotation. Beads were washed 6× with 500 μl each of lysis buffer containing 200 mm NaCl. Aliquots of beads (∼5%) were withdrawn for Western blot analysis. From the remaining beads RNA was eluted with 100 μl of elution buffer (200 mm sodium acetate, pH 5.5, 1 mm EDTA, 1% (w:v) SDS) for 10 min at 70 °C. RNAs were purified using PeqGold RNAPure as described above.
SDS-PAGE and Western Blotting
Proteins were separated by SDS-PAGE and semi-dry-blotted onto Protran BA 85 (Schleicher & Schuell) in 25 mm Tris base, 150 mm glycine, 0.05% SDS, 10% (v:v) methanol, pH 8.3, for 50–70 min at 150 mA, 30 V. Membranes were incubated in blocking solution (PBS-Tween 20: 150 mm NaCl, 2 mm NaH2PO4, 8 mm Na2HPO4, pH 7.4, 0.3% (w:v) Tween 20 containing 5% (w:v) nonfat dry milk) for 1 h at room temperature. Primary and secondary antibodies were diluted in blocking solution and incubated for 3 h at room temperature or overnight at 4 °C (primary antibodies) and for 1 h at room temperature (secondary antibodies), respectively. Blots were washed in PBS-Tween 20 (2 × 1 min, 1 × 15 min, 3 × 5 min). Antibody detection was done using Lumi-Light Western blotting substrate (Roche Applied Science) or Covalight chemiluminescent reagent kit (Covalab), and blots were exposed to x-ray films.
Luciferase Assays
The Dual-Luciferase Reporter Assay System (Promega) was used according to the manufacturer's recommendations with cortical neuron extracts prepared about 24 h after transfection.
Immunocytochemistry
Cells grown on coverslips and 15-μm rat brain tissue cryosections were fixed with 1 or 4% paraformaldehyde in PBS (150 mm NaCl, 10 mm sodium phosphate, pH 7.2) for 15 min at room temperature. After 2 washes in PBS (5 min each), cells and tissues were permeabilized with 0.3% Triton X-100 in PBS (5 min), washed twice as above, and blocked for 1 h in blocking solution (10% normal goat serum, 0.05% Triton X-100 in PBS). Primary and secondary antibodies diluted in blocking solution were incubated overnight at 4 °C and 2–3 h at room temperature, respectively. Cells and tissue sections were washed 3 times (5 min each) in PBS. Finally, they were mounted in Permafluor (PerkinElmer Life Sciences) or ProLong Gold antifade reagent containing DAPI (Invitrogen). Images were acquired using either a Zeiss Axiovert 135 microscope (10/0.3, 25/0.8, 40/1.3, 63/1.25 objectives) equipped with a Hamamatsu camera using Openlab software (Improvision) or a confocal microscope (Leica SP2, 40/1.25 objective) using Leica SP2 software. Images were exported in TIFF.
Free-floating sections (50 μm) derived from electrically stimulated animals (n = 10) were incubated for 10 min in 1% sodium borohydride in PBS followed by 2 h at room temperature in blocking buffer (0.1 m Tris-HCl, pH 7.4, 0.5% Triton X-100 (anti-MKRN1) or 0.05% Triton X-100 (anti-PABP), 10% normal goat serum) and incubated overnight at 4 °C with rabbit anti-MKRN1 (1:1000) or rabbit anti-PABP (1:500). Sections were washed 3× with TBS (0.1 m Tris-HCl, pH 7.4, 0.15 m NaCl) and subsequently incubated with biotin-conjugated goat anti-rabbit IgG (1:1000, Vector) for 2 h at room temperature. After washing, sections were incubated in Vector ABC (1:1000) for 1 h and reacted with diaminobenzedine and H2O2. Sections were mounted, dehydrated, and coverslipped. Micrographs were taken using a Kappa camera on a Zeiss Axiophot microscope. Images were exported in TIFF.
RESULTS
MKRN1 Is Novel PAPB-interacting Protein
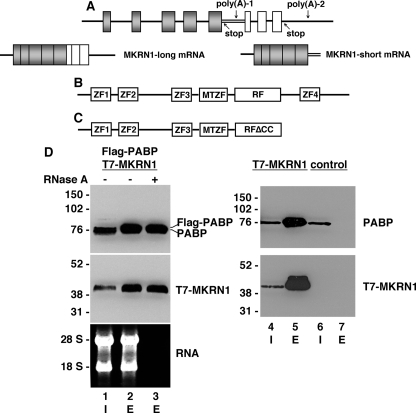
To better understand the molecular action of PABP in mammalian neurons, we employed a yeast two-hybrid screen utilizing full-length PABP bait and a human brain cDNA library. Among 69 positive clones, 20 independent cDNAs encoded MKRN1. Interestingly, all of the isolated clones encoded a short isoform, referred to as MKRN1-short, that is a C-terminal-truncated variant of full-size MKRN1 (MKRN1-long). As schematically shown in Fig. 1A, these isoforms are encoded by a single mkrn1 gene and arise by alternative splicing and differential polyadenylation (Ref. 24 and this study). MKRN1-long consists of four C3H-type ZFs, a MKRN-type ZF (MTZF), and a highly conserved C3HC4-type RING domain (Ref. 24, schematically outlined in Fig. 1B). C3H-type ZFs are RNA binding motifs (43, 44), whereas the RING finger domain is a protein/protein interaction module characteristic for RING-class E3 ubiquitin ligases (45). MKRN1-short lacks the C-terminal ZF and the last six amino acids of the RING finger domain (RFΔCC) essential for binding the second zinc ion (Fig. 1C). Human MKRN1-short consists of 329 amino acids and exhibits a calculated molecular mass of 35.2 kDa, whereas the 53.3-kDa MKRN1-long protein described by Gray et al. (24) is composed of 482 amino acids. To verify the MKRN1-short/PABP interaction, epitope-tagged proteins (T7-MKRN1-short and FLAG-PABP) were transiently expressed in HEK-293 cells and immunoprecipitated with appropriate antibodies, and precipitates were analyzed by Western blotting. As shown in Fig. 1D, immunoprecipitation of FLAG-PABP co-precipitated T7-MKRN1-short (Fig. 1D, lanes 1–3). This interaction persisted after digestion with RNase A (Fig. 1D, lane 3), indicating that it is not mediated by RNA. Essentially the same results were obtained by using RNase ONE (data not shown) that cleaves phosphodiester bonds between any two ribonucleotides. Consequently, a possible interaction bridged by the poly(A) tail of mRNAs can be ruled out. In HEK-293 cells neither of the MKRN1 isoforms is expressed at detectable levels (Fig. 1D, lane 6). Thus, an interaction between MKRN1-short and PABP could not be investigated for the endogenous components. However, immunoprecipitation of T7-MKRN1-short pulled down endogenous PABP (Fig. 1D, lanes 4 and 5). Taken together, these data show that in living cells MKRN1-short associates with PABP in an RNA-independent manner.
FIGURE 1.
PABP interacts with the RING zinc finger protein MKRN1 in an RNA-independent manner. A, the upper panel shows the genomic organization of the human MKRN1 gene, which consists of exons 1–8. The mRNAs encoding MKRN1-long and MKRN1-short, respectively, are generated by alternative splicing and differential polyadenylation (lower panel). Consequently, MKRN1-long and -short mRNAs possess different 3′-untranslated regions. Poly(A), polyadenylation signal; stop, stop codon. B, MKRN1-long is a modular protein consisting of four zinc fingers (ZF), a MKRN-type zinc finger (MTZF), and a RING finger domain (RF). C, MKRN1-short is C-terminally truncated and lacks ZF4 and the last six amino acids of the RING finger domain including two essential cysteine residues (RFΔCC). D, FLAG-PABP and/or T7-MKRN1-short were expressed in HEK-293 cells. The control shows non-transfected cells. Protein extracts were subjected to immunoprecipitation with either anti-FLAG-agarose (lanes 2, 3, and 7) or anti-T7-agarose (lane 5) in the absence (−) or presence (+) of RNase A. The inputs (I) are shown in lanes 1, 4, and 6. Western blots were probed with anti-PABP- and anti-MKRN1-short antibodies, respectively. The non-transfected control (lanes 6 and 7) shows no unspecific binding of PABP to anti-FLAG-agarose. RNAs extracted before (lane 1) and after immunoprecipitation (lanes 2 and 3) were resolved by agarose gel electrophoresis followed by ethidium bromide staining (lower panel, left). I, input; E, protein eluted from anti-FLAG- or anti-T7-agarose. The positions of molecular size marker proteins (in kDa) and the 18 S and 28 S ribosomal RNAs are indicated on the left.
Interaction of MKRN1-short with PABP Is Mediated by PAM2-like Motif
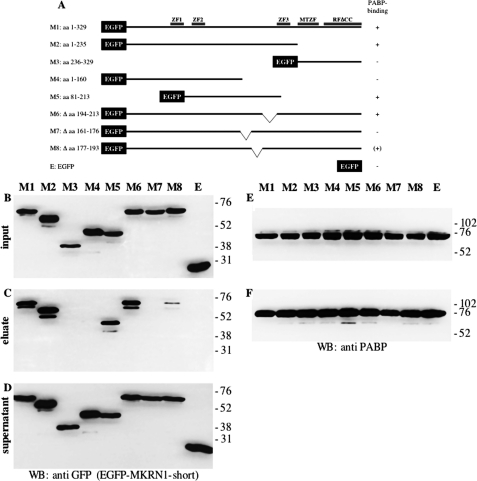
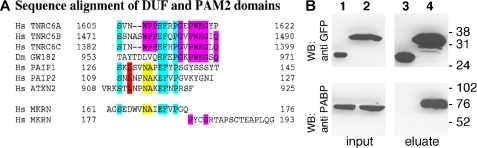
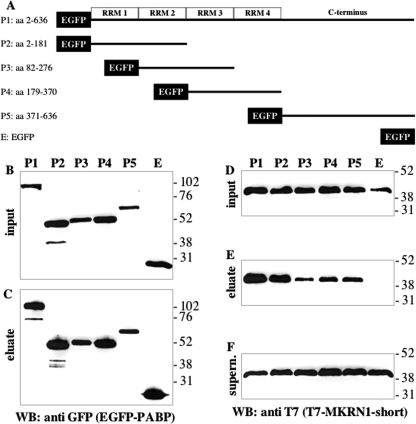
To determine which part of MKRN1-short mediates binding to PABP, full-length EGFP-MKRN1-short (M1, schematically depicted in Fig. 2A) as well as different EGFP-MKRN1-short deletion mutants (M2–M8, Fig. 2A) were expressed in HEK-293 cells along with FLAG-tagged PABP. Cell lysates were subjected to immunoprecipitation using FLAG-agarose beads. The Western blots shown in Fig. 2, B—F, demonstrate that the interaction of MKRN1-short with PABP depended on the N-terminal amino acids 1–235, whereas the C-terminal part of the molecule did not bind (Fig. 2, B–D, compare M2 and M3). Further analysis revealed that amino acids 161–176 were required for MKRN1-short/PABP interaction (Fig. 2, B–D, M7). However, they were not sufficient as the interaction was highly impaired upon deletion of adjacent amino acids 177–193 (Fig. 2, B–D, M8), indicating that these contribute substantially to PABP binding. A closer inspection (Fig. 3A) revealed that the sequence spanning amino acids 161–176 is very similar to PAM2 motifs within PAIP1 and -2 and Ataxin-2 (ATXN2), respectively (46). Yet in MKRN1-short a highly conserved leucine (Leu128 in PAIP1, Leu111 in PAIP2, and Leu913 in ATXN2) is replaced by aspartate (Asp165). The following region containing amino acid residues 177–193 exhibits some similarity to the C-terminal part of so-called domains of unknown function (DUF). In PABP-interacting proteins belonging to the GW182 family (such as human trinucleotide repeat containing gene protein TNRC6 and Drosophila GW182) DUF serve as PABP binding domains and are equivalents of PAM2 motifs (30). Obviously, the MKRN1-short PABP-interacting domain (henceforth referred to as PAM2-like domain) exhibits characteristics of both PAM2 motifs and DUF. To assess whether the PAM2-like domain binds to PABP by itself, MKRN1-short amino acids 161–193 were expressed as EGFP fusion protein (EGFP-PAM2MKRN) in HEK-293 cells. Immunoprecipitation using GFP-Trap® beads efficiently pulled down endogenous PABP (Fig. 3B). Other domains of MKRN1-short do not contribute to this interaction as deletion of various parts of the molecule, including ZF2 and -3, did not impair PABP binding (supplemental Fig. 1). With the exception of Drosophila GW182 DUF (47), all of the PAM2 motifs and DUF characterized so far bind to a stretch of some 70 amino acids within the C-terminal part of PABP called MLLE-domain (Met-Leu-Leu-Glu; Ref. 30). To elucidate the contact site(s) between PABP and MKRN1-short, plasmids encoding full-length EGFP-PABP or different deletion mutants (P1–P5, Fig. 4A) were transfected together with a T7-MKRN1-short expression vector. Immunoprecipitates obtained from cell lysates using GFP-Trap® beads were analyzed by Western blotting. Our results indicate that MKRN1-short contacts several sites within PABP but with different efficiencies (Fig. 4, B–F). The main interaction was mediated by RNA recognition motifs 1 and 2 (RRM1 and -2), being almost as strong as that seen with the full-size protein (Fig. 4E, compare P1 and P2). Yet other parts of the molecule such as RRMs 2 and 3, RRMs 3 and 4, and the C terminus bound to MKRN1-short as well (Fig. 4E, P3–P5). Hence, the PABP binding properties of the MKRN1-short PAM2-like domain and PAM2/DUF domains of the PABP-interacting proteins mentioned above are clearly distinct. This difference was further underscored by a direct comparison of the PABP/ATXN2 interaction in the same extracts. Distinct from MKRN1-short ATXN2 bound only to the MLLE-containing C terminus of PABP but not to any of the RRM-containing regions (supplemental Fig. 2). Moreover, the PAM2-like domain in MKRN1-short is by itself sufficient to confer binding to both the RRMs and C terminus of PABP in a manner largely identical to that seen with full-length MKRN1-short (supplemental Fig. 3). This raises the question of whether MKRN1-short may interact with further RRM-containing proteins involved in posttranscriptional regulation of gene expression. Insulin-like growth factor II mRNA-binding protein 1 (IMP1), for example, is a translational regulator that harbors two N-terminal RRMs (48). Co-immunoprecipitation experiments performed with EGFP-tagged full-length IMP1 and truncated protein revealed an RNA-independent binding of T7-MKRN1-short to the RRMs of IMP1 (supplemental Fig. 4).
FIGURE 2.
Identification of the PABP binding site of MKRN1-short. A, shown is a schematic representation of recombinant EGFP-MKRN1-short fusion proteins (encoded by M1-M8) that were transiently expressed in HEK-293 cells together with FLAG-PABP. B–F, protein extracts were subjected to immunoprecipitation with anti-FLAG-agarose followed by SDS-PAGE and Western blot (WB) analyses using anti-GFP antibodies for detection of EGFP-MKRN1 deletion mutants (B–D) or anti-PABP antibodies (E and F). B and E: inputs; C and F, eluted immunoprecipitates; D, supernatant after immunoprecipitation. The positions of molecular size marker proteins (in kDa) are indicated on the right. E, MTZF, Makorin-type zinc-finger; RFΔCC, truncated RING finger domain.
FIGURE 3.
MKRN1-short contains a PAM2-like motif that mediates its interaction with PABP. A, shown is sequence alignment of GW182 family DUF domains and PAM2 motifs of the PABP-binding proteins PAIP1, PAIP2, and ATXN2 with the PABP-interaction motif in MKRN1-short. Strictly conserved residues are highlighted in turquoise. Residues conserved within the DUF and PAM2 domains are shown in magenta and yellow, respectively. The second most critical residue, leucine, within the PAM2 domain, is highlighted in red. Numbers refer to the amino acid position within the respective proteins. B, recombinant EGFP (lanes 1 and 3) or the MKRN1-short PAM2-like domain (amino acids 161–193) fused to EGFP (EGFP-PAM2MKRN; lanes 2 and 4) were transiently expressed in HEK-293 cells. Protein extracts were subjected to immunoprecipitation with GFP-Trap®_A beads followed by SDS-PAGE and Western blot (WB) analyses using anti-GFP antibodies for detection of EGFP and EGFP-PAM2MKRN (upper panel) or anti-PABP antibodies for detection of endogenous PABP (lower panel), respectively. The positions of molecular size marker proteins (in kDa) are indicated on the right. Dm, Drosophila melanogaster; Hs, Homo sapiens; TNRC, trinucleotide repeat containing gene protein.
FIGURE 4.
MKRN1-short binds to several domains of PABP. A, shown is a schematic representation of recombinant EGFP-PABP fusion proteins (encoded by P1-P5) that were transiently expressed in HEK-293 cells together with T7-MKRN1-short. WB, Western blot. B–F, protein extracts were subjected to immunoprecipitation with GFP-Trap®_A followed by SDS-PAGE and Western blot analyses using anti-GFP antibodies for detection of EGFP-PABP (B and C) or anti-T7 antibodies (D–F) for detection of recombinant MKRN1-short. B and D, inputs; C and E, eluted immunoprecipitates; F, supernatant after immunoprecipitation. The positions of molecular size marker proteins (in kDa) are indicated on the right. aa, amino acids.
Characterization of mkrn1 Gene Expression in Rat Tissues
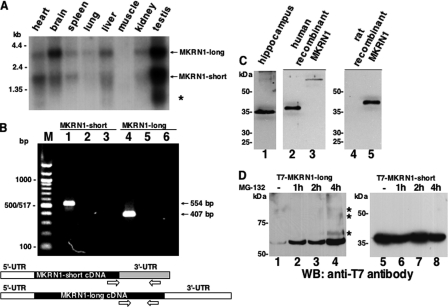
In human and mouse, MKRN1 mRNA is present in diverse non-neuronal and neuronal tissues (24). Our Northern blot analysis with RNA isolated from different rat tissues confirmed and extended these observations (Fig. 5A). Thus, highest transcript levels were detected in brain and testis. The 3.2- and 2.0-kb transcripts most likely encode rat MKRN1-long and -short, respectively. In testis, an additional smaller transcript of 0.75 kb was detected and presumably represents a further alternatively spliced MKRN1 mRNA encompassing exons 1–3 (49). PCR analysis with primers specific for MKRN1-long and -short cDNA, respectively, confirmed the presence of both variants in rat brain (Fig. 5B).
FIGURE 5.
Characterization of MKRN1 expression in rat brain at the mRNA and protein level. A, poly(A)-RNAs from various rat tissues were hybridized with a 32P-labeled MKRN1 probe that detects both MKRN1-long (3.2 kb) and MKRN1-short (2 kb) transcripts. The asterisk denotes a third transcript variant of 0.75 kb. The positions of marker RNAs (in kb) are indicated on the left. B, RT-PCR with rat hippocampal RNA was performed to confirm the expression of MKRN1-short (lane 1, 554 bp) and MKRN1-long (lane 4, 407 bp) in this brain region. The primers used for the amplification reactions (open arrows) are schematically shown in the lower part of this panel. As controls, PCRs were done in the absence of template (lanes 2 and 5) as well as with non-reverse transcribed RNA (lanes 3 and 6). PCR products were resolved by agarose gel electrophoresis along with a 100-bp ladder marker (left) followed by ethidium bromide staining. C, rat hippocampal proteins were probed with rabbit anti-MKRN1 antiserum (lane 1). Human T7-tagged MKRN1-short (lane 2) and MKRN1-long (lane 3) expressed in HEK-293 cells were run in parallel on the same gel. Antibodies raised against human MKRN1-short recognize recombinant myc-His-tagged rat MKRN1-short (lane 5). Non-transfected control extract is shown in lane 4. D, MKRN1-long is only efficiently expressed in HEK-293 cells if proteasomal degradation is inhibited by the addition of MG-132 (final concentration, 10 μm). The compound was added 1, 2, or 4 h (lanes 2–4) before protein extraction. Bands marked by asterisks most likely correspond to ubiquitinated forms of MKRN1-long. MKRN1-short lacks autoubiquitination properties, probably due to the C-terminal deletion of six amino acids of its RING finger domain. Destabilization of the short MKRN1 variant is not seen if cells are grown in the absence of MG-132 (lane 5). The addition of the compound for various periods of time does not alter the steady state concentration of MKRN1-short (lanes 6–8). The positions of molecular size marker proteins (in kDa) are indicated on the left. WB, Western blot.
To address the question of MKRN1 expression at the protein level in rat brain, Western blot analyses were performed with a polyclonal antiserum raised against recombinant GST-MKRN1-short. We have observed that the forebrain contained the highest level of MKRN1 (see below). Hence, Western blot analyses were performed with proteins extracted from the hippocampal formation. The antiserum detected a protein with an apparent molecular mass of ∼35 kDa (Fig. 5C, lane 1) that appears to represent MKRN1-short as it migrated just slightly faster than epitope-tagged recombinant human T7-MKRN1-short (Fig. 5C, lane 2). In addition, a very faint band of about 55 kDa may correspond to MKRN1-long as it co-migrates with human T7-MKRN1-long (Fig. 5C, compare lanes 1 and 3). Recombinant rat MKRN1-short was recognized by the antibody raised against the human protein (Fig. 5C, lane 5; lane 4 contains a protein extract from non-transfected cells). We have noted that in HEK-293 (and HeLa) cells recombinant MKRN1-long expression was very low unless cell cultures were treated with the proteasome inhibitor MG-132 (Fig. 5D, lanes 1–4). This is consistent with data reported by Kim et al. (26) showing that human MKRN1-long is subject to autoubiquitination, which leads to its very low abundance in cell lysates. Bands with lower electrophoretic mobility detected after 4 h of MG-132-treatment (Fig. 5D, lane 4, marked by asterisks) probably represent ubiquitinated forms of MKRN1-long. Thus, extremely low levels of MKRN1-long in rat brain (Fig. 5C, lane 1) despite the relative high abundance of its mRNA (Fig. 5A) may be explained by its autoubiquitination properties. In contrast, MKRN1-short does not appear to catalyze its own ubiquitination and proteasomal degradation (Fig. 5D, lanes 5–8).
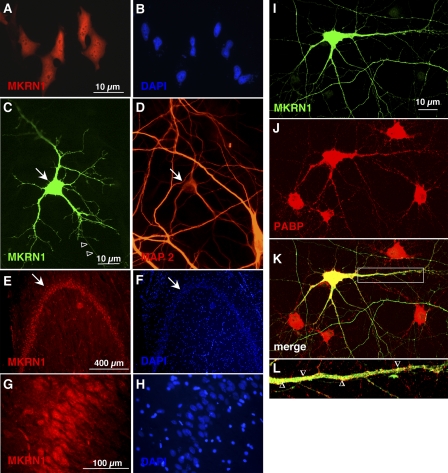
To determine the subcellular localization of both MKRN1 isoforms, we immunocytochemically analyzed HeLa cells and primary rat hippocampal neurons as well as rat brain sections. Despite the lack of any of the well characterized canonical nuclear localization signals, Myc-tagged MKRN1-short is present in both HeLa cell nuclei and cytoplasm (Fig. 6, A and B). In MG-132-treated HeLa cells, recombinant MKRN1-long essentially exhibited an identical distribution (data not shown). In primary neurons, Myc-tagged MKRN1-short was observed in both cell bodies and microtubule-associated protein 2 (MAP2)-positive dendrites as well as putative axons (MAP2-negative neurites; Fig. 6, C and D). In immunohistochemical investigations of rat brain sections, MKRN1 was detected in most major brain areas (data not shown). Highest immunoreactivity was observed in neurons of the hippocampal formation (Fig. 6, E–H) and in the neocortex (data not shown). In these neurons, MKRN1 is present in nuclei, cell bodies, and proximal dendrites (Fig. 6, G and H). Because MKRN1-long was hardly detectable in rat brain lysates by Western blotting (see Fig. 5C, lane 1) the staining pattern is likely to primarily reflect the brain distribution of MKRN1-short. Moreover, in transfected primary neurons recombinant MKRN1-short partially co-localized with endogenous PABP within dendrites (Fig. 6, I–L). However, many PABP-positive puncta did not include MKRN1-short, indicating that only part of the cellular PABP interacts with MKRN1-short or that the contact is rather transient in nature. In addition, MKRN1-short partially resided in cellular subareas devoid of PABP.
FIGURE 6.
MKRN1 is located in the cell nuclei and in the cytoplasm and localizes to neurites of nerve cells where it is partially co-localized with PABP. A and B, HeLa cells transfected with an Myc-tagged MKRN1-short encoding construct were immunostained with a monoclonal mouse anti-myc antibody. Recombinant protein is detected in the cell nuclei as well as in the cytoplasm. C and D, in vitro cultured rat hippocampal neurons transfected with Myc-tagged MKRN1-short encoding construct were immunostained with rabbit anti-MKRN1-short antiserum. Recombinant protein is detected in the cell body as well as in the dendritic tree. A neurite devoid of MAP2-staining (shown in D), a marker of the dendritic cytoskeleton, is also decorated by antibodies (C open arrowheads). E and F, sagittal rat hippocampal section (dentate gyrus) stained with rabbit anti-MKRN1-short antiserum is shown. Strongest immunoreactivity is seen in the granule cell body layer (arrow). G and H, higher power magnification of granule cells shown in E reveals MKRN1-staining in the nuclei and in the cytoplasm. Staining proceeds to the proximal parts of dendrites. I–K, in vitro cultured rat hippocampal neurons transfected with a T7-tagged MKRN1-short encoding construct were immunostained with mouse anti-T7 and rabbit anti-PABP antibodies. L, shown is higher power magnification of the dendritic segment encircled by the white rectangle in panel K. The open arrowheads denote examples of yellow-stained regions of MKRN1-short and PABP colocalization.
MKRN1-short Stimulates Translation in Nerve Cells and Is Associated with mRNAs Localized to Dendrites
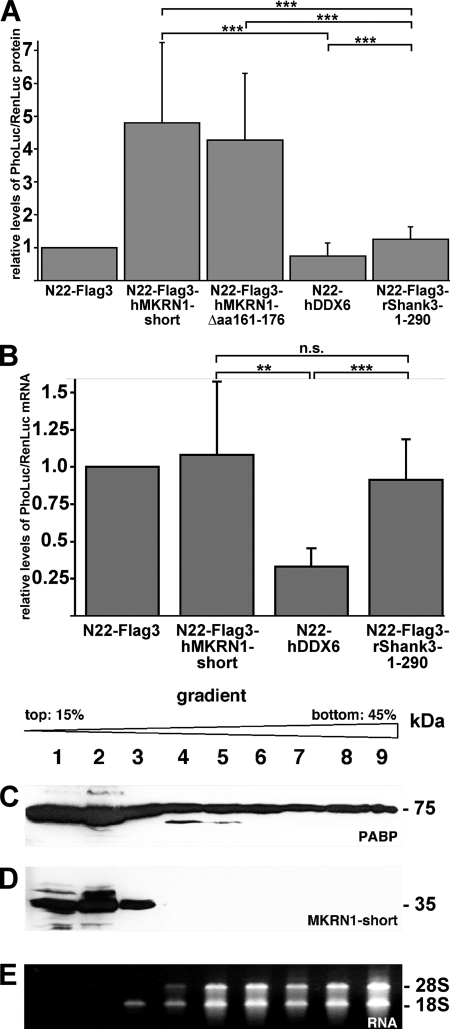
To investigate if MKRN1-short, as a PABP-interacting protein, regulates translation efficiency and/or mRNA stability, we performed a so-called N22-tethering experiment followed by either a dual luciferase reporter assay or a TaqMan® probe-based semiquantitative RT-PCR analysis (for a schematic representation of these assays, see supplemental Fig. 5). For this purpose we constructed a eukaryotic expression vector (pinFiRein-boxB16B) that contains two separate genes, each comprising its own CMV promoter, driving transcription from opposite DNA strands. Both genes possess an intron upstream of the region encoding either PhoLuc or RenLuc, respectively. In its 3′-UTR, the PhoLuc mRNA contains 16 copies of a stem-loop structure referred to as box B, which is specifically bound by a small protein domain (N22; Ref. 50). Notably, RenLuc transcripts lack these binding sites (internal tethering control). Thereby, any given N22-tagged protein is selectively tethered to PhoLuc but not RenLuc mRNAs. We co-transfected 7-day-old primary rat cortical neurons with both pinFiRein-boxB16B and an additional vector encoding one of several N22-tagged proteins. Two days later the neurons were harvested, and the activity of both luciferases in cell lysates was determined. To compensate for possible differences in the transfection rate, PhoLuc activity was normalized against RenLuc (henceforth referred to as “nPhoLuc activity”). The nPhoLuc activity observed in cells expressing a recombinant protein consisting of N22 and three tandem FLAG epitopes (N22-FLAG3) served as a reference and was arbitrarily set to 1. Selective tethering of N22-FLAG3 to PhoLuc mRNAs is thought to not significantly alter the metabolism of bound reporter transcripts. As compared with this control, tethering of both full-length MKRN1-short (N22-FLAG3-hMKRN1-short) and a mutant version lacking PABP binding activity (N22-FLAG3-hMKRN1-Δaa161–176; see Fig. 2, M7) lead to a more than 4-fold increased nPhoLuc activity (Fig. 7A). In contrast, N22-tagged human DDX6 (N22-hDDX6), a DEAD-box helicase promoting recruitment and degradation of mRNAs in P-bodies (processing bodies; Ref. 51) reduced nPhoLuc activity to about 50%. Finally, the fusion protein N22-rShank Shank3-1–290 containing an N-terminal part of the postsynaptic scaffold protein that is not known to play any role in RNA metabolism (52) did not significantly alter nPhoLuc activity as compared with the control value. To assess whether MKRN1-short tethered to reporter transcript control mRNA translation and/or stability, we performed real-time RT-PCR analysis with total RNA isolated from transfected neurons and TaqMan® probes hybridizing to sequences derived from two neighboring exons of RenLuc- and PhoLuc genes, respectively. Similar to the dual luciferase assay, PhoLuc mRNA levels were normalized to the concentration of RenLuc transcripts (nPhoLuc mRNA levels), and nPhoLuc transcript levels observed in N22-FLAG3-expressing cells again served as reference control that was arbitrarily set to 1. As shown in Fig. 7B, recombinant MKRN1-short (and, as expected, N22-FLAG3-Shank3-1–290) did not affect nPhoLuc mRNA levels as compared with the N22-FLAG3 control. However, consistent with its reported role in mRNA decay, N22-hDDX6 significantly reduced nPhoLuc mRNA abundance. As tethering of MKRN1-short significantly increased nPhoLuc activity without altering PhoLuc mRNA levels, this effect can only be accounted for by an increased translation rate and not by enhanced stability or nuclear export of reporter transcripts. Taken together, our findings show that MKRN1-short significantly enhances translation efficiency of bound reporter transcripts via a molecular mechanism that does not require its direct interaction with PABP. In hippocampal homogenates separated on a sucrose gradient MKRN1-short resides only in the light messenger ribonucleoprotein fraction (Fig. 7, C–E), suggesting that it might stimulate protein synthesis at the level of initiation.
FIGURE 7.
MKRN1-short stimulates translation in nerve cells. The eukaryotic expression vector pinFiRein-boxB16B was co-transfected with vectors encoding fusion proteins consisting of an N-terminal N22 peptide and either hMKRN1-short, hMKRN1-Δ487–535, hDDX6, or rShank3–1-299 into dispersed cortical neurons at 7 DIV. The empty vector N22-FLAG3 served as a control (for details see “Experimental Procedures”). A, dual luciferase assays were performed at 9 DIV. The relative levels of PhoLuc/RenLuc proteins are shown (in arbitrary units). B, RNAs from transfected neurons were prepared on 9 DIV, transcribed into cDNAs, and subjected to real-time PCR analyses using Pho/Luc- and RenLuc-specific TaqMan assays. The relative levels of PhoLuc/RenLuc mRNAs are shown (in arbitrary units). Bars represent S.E. Statistical analyses were done using Student's t test (**, p < 0.01; ***, p < 0.001, n.s., not significant). C and D, ribosomes/polysomes from adult rat hippocampi were fractionated by sucrose gradient ultracentrifugation. Individual fractions (lanes 1–9) were subjected to SDS-PAGE and Western blot analyses using anti-PABP (C) or anti-MKRN1 antibodies (D). The positions of molecular size marker proteins (in kDa) are indicated on the right. E, RNAs purified from individual gradient fractions were separated by agarose gel electrophoresis and stained with ethidium bromide. 28 S and 18 S, large and small ribosomal RNAs.
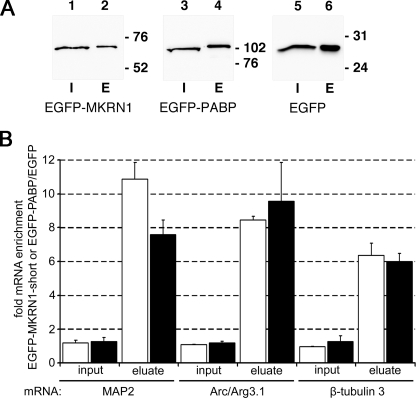
Dendrites harbor a number of mRNA species that are locally translated (3–6), among them transcripts encoding the MAP2 and activity-regulated cytoskeleton-associated protein (Arc/Arg3.1). To assess whether these mRNAs are targets of MKRN1-short we transfected cultured cortical neurons with vectors encoding EGFP-MKRN1-short, EGFP-PABP (positive control), and EGFP alone (empty vector control), respectively. Recombinant proteins were immunoprecipitated with GFP-Trap® (Fig. 8A), and individual associated mRNAs were detected by semiquantitative real-time RT-PCR analysis. As shown in Fig. 8B, immunoprecipitation of recombinant MKRN1-short led to an 11- and 8-fold enrichment of MAP2 and Arc/Arg3.1 mRNAs compared with the empty vector control. Enrichment of β-tubulin 3 transcripts was somewhat lower (6-fold). Similar data were obtained in experiments performed with recombinant PABP. Hence, in living neurons both proteins are associated with dendritic and somatically restricted mRNAs.
FIGURE 8.
MKRN1-short is associated with dendritically localized mRNAs. A, shown are protein lysates from rat primary cortical neurons expressing recombinant EGFP-MKRN1-short, EGFP-PABP, or EGFP alone that were subjected to immunoprecipitation with GFP Trap®_A beads followed by SDS-PAGE and Western blot analyses using anti-GFP antibodies for detection of EGFP-MKRN1 (lanes 1 and 2), EGFP-PABP (lanes 3 and 4), and EGFP (lanes 5 and 6), respectively. I, input proteins (lanes 1, 3, and 5); E, immunoprecipitated proteins eluted from the beads (lanes 2, 4, and 6). The positions of molecular size marker proteins (in kDa) are indicated on the right. B, RNAs extracted from whole cell lysates (input) and immunoprecipitated protein fractions (eluate) were subjected to semiquantitative real-time RT-PCR using primers for MAP2-, Arc/Arg3.1-, and neuron-specific β-tubulin 3 cDNAs. The graph depicts the enrichment of individual transcripts in inputs and in eluates obtained by immunoprecipitation of EGFP-MKRN1-short (open bars) and EGFP-PABP (closed bars), respectively, compared with the empty vector control (EGFP-IP). Data were obtained using REST (relative expression software tool) 2008 software for group-wise comparison and statistical analysis of relative expression results in real-time PCR (41). S.E., vertical lines, enrichment/eluates, p < 0.001; enrichment/inputs, not significant. Arc/Arg3.1, activity-regulated cytoskeleton-associated.
MKRN1-short Accumulates in Middle Molecular Layer of Dentate Gyrus after Plasticity-inducing Stimulation
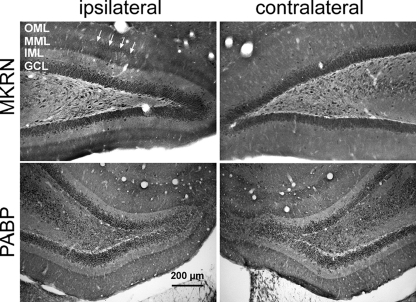
Specific forms of LTP, a model system for mechanisms underlying long term memory formation, require activity-induced local synthesis of proteins at synapses (33, 53). Because MKRN1-short is located in dendrites and involved in translation control (see above), we asked whether synaptic activity might alter MKRN1-short abundance at activated synapses in vivo. To answer this question we took advantage of the possibility to selectively stimulate the perforant path in the brain of living rats. These projections from the medial entorhinal cortex terminate in the middle molecular layer of the dentate gyrus (54). High frequency stimulation of this path leads to activity-dependent translation of dendritic mRNAs encoding the Ca2+/calmodulin-dependent kinase II α subunit (55), MAP2 (55), and Arc within the activated dendritic layer (36). Two hours after high frequency stimulation of the perforant path, MKRN1-short levels strongly increased in a sharply defined dendritic lamina of the dentate gyrus (Fig. 9, upper panel, left). In contrast, on the contralateral side this regional accumulation of the protein was not evident (Fig. 9, upper panel, right), indicating that the observed local increase in protein levels is triggered by the activity of nearby synapses. As in brain neurons MKRN1 mRNA is not detected in dendrites (supplemental Fig. 6) the accumulation of MKRN1-short in the stimulated dendritic lamina appears to reflect an activity-dependent redistribution of the protein rather than a local de novo protein synthesis. Distinct from MKRN1-short, the dendritic distribution of PABP did not change after stimulation of the perforant path (Fig. 9, lower panel). The experimental design used here regularly induced local accumulation of F-actin in the activated dendritic lamina as well as robust c-fos gene expression in dentate gyrus granule cells of the stimulated side (supplemental Fig. 7), one of the hallmarks of LTP (55–57). Taken together, the data underscore that the MKRN1-short accumulation in the middle molecular layer after LTP induction is specific and strongly suggest that the protein likely plays a role in synaptic plasticity.
FIGURE 9.
Induction of LTP in the perforant pathway leads to accumulation of MKRN1-short in the stimulated dendritic lamina of the dentate gyrus. Brain sections from rats subjected to plasticity-inducing unilateral stimulation of the perforant pathway were immunolabeled with anti-MKRN1-short antibodies (upper panel) and anti-PABP antibodies (lower panel), respectively. Arrowheads point to MKRN1-short accumulation in the middle molecular layer (MML) of the dentate gyrus. GCL, granule cell layer; IML, inner molecular layer; OML, outer molecular layer.
DISCUSSION
Our studies provide the first extensive characterization of MKRN1 expression at the protein level in the central nervous system. In rat brain a short isoform, MKRN1-short, predominates and is detectable in somata, dendrites, and nuclei of nerve cells. As this isoform lacks two of the cysteine residues essential for the formation of a functional C3HC4-type RING domain (58), it is unlikely to act as an E3 ubiquitin ligase such as MKRN1-long. The loss of autoubiquitination activity of the short MKRN1 isoform (see Fig. 5D) supports this assumption.
MKRN1-short interacts with PABP, a protein with numerous functions in mRNA metabolic pathways (15). Binding to PABP is mediated by a PAM2-like motif within MKRN1-short. A variety of PABP-interacting proteins possess such motifs, for example PAIP1, PAIP2, and ATXN2 (30). However, in contrast to PAM2 domains of those proteins, which exclusively associate with the PABP C-terminal MLLE domain (Refs. 59 and 60; see also supplemental Fig. 2), the MKRN1-short PAM2-like domain is peculiar as it preferentially interacts with RRMs 1 and 2 and to a lesser extent with RRMs 2 and 3, RRMs 3 and 4, and the C-terminal part of PABP. Whether MKRN1-short binding to these different parts of PABP occurs cooperatively or independently from each other remains to be seen. All in all, this binding behavior of MKRN1-short to PABP is somewhat reminiscent of that of PAIP1 and -2 (59, 60). The major difference is that a single motif in MKRN1-short is able to contact several sites within PABP, whereas binding of PAIP1 and -2 to the RRMs in PABP is mediated by a second motif called PAM1, which in fact represents the high affinity PABP interaction site (59, 60). It is this interaction with PABP that is responsible for translational stimulation by PAIP1 (31) and translational silencing by PAIP2 (32), whereas the functional consequence of the PAM2/PABP C-terminal MLLE domain interaction is less clear.
In this study we have demonstrated that MKRN1-short significantly stimulates protein synthesis when tethered to a reporter mRNA while not influencing mRNA stability, thus implying an interaction of MKRN1-short with the translation machinery. As a MKRN1-short mutant that lacks the PABP binding site still enhances reporter mRNA translation similar to the full-length protein, it seems likely that a molecule distinct from PABP mediates the functional link of MKRN1-short to the translation machinery. Yet, in vivo, PABP may provide a platform for MKRN1-short to tie it to an mRNA molecule, a function that is not required in the described N22-box B tethering assay. Such a role of PABP has been described for recruitment of the transducer of ErbB2 (TOB), a protein implicated in cytoplasmic mRNA deadenylation, to target mRNAs (61).
Plasticity producing high frequency stimulation of the perforant path in vivo leads to the accumulation MKRN1-short in the activated dendritic lamina of dentate gyrus granule cells. The same stimulus promotes translation of mRNAs encoding MAP2 and Ca2+/calmodulin-dependent kinase II α subunit in dendrites (55). Presumably, many more mRNA species are subject to activity-dependent translation activation given that dendrites harbor at least 150 different mRNA species (62) and that synaptic activation, which induces major changes in synapse composition, most likely requires local synthesis of many distinct protein species. Yet, there is no evidence for an increase in overall dendritic protein synthesis subsequent to neuronal activation (53). Furthermore, the number of polysomes at synapses is quite low (53). Hence, the available data suggest that dendritic mRNAs compete for a limited translational machinery and that synaptic activity triggers translation of selected mRNAs at the expense of other transcript species. The translation regulatory capacity of MKRN1-short, its association with dendritically localized mRNAs in living neurons, and its in vivo accumulation at activated synapses suggest that MKRN1-short controls activity-dependent reprogramming of the protein synthesis profile in dendrites. Due to the ability to bind to different parts of PABP, MKRN1-short could modulate recruitment, displacement, and/or binding properties of factors both at the N-terminal end harboring RRM1–4 and the C-terminal MLLE domain of PABP, each of which interacts with distinct sets of proteins (63). The ability of MKRN1-short to interact with IMP1 (see supplemental Fig. 4), a PABP-binding protein involved in translational regulation (64), is consistent with this view. Why does dendritic PABP distribution not change after synaptic stimulation? The most plausible interpretation is that PABP is already bound to all polyadenylated dendritic mRNAs. Unless there was gross redistribution of those transcripts as a result of the stimulus, an accumulation of PABP in the activated lamina is not to be expected.
Supplementary Material
Acknowledgments
We thank Dr. Hans-Jürgen Kreienkamp (Department of Human Genetics, University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for generous gifts of cloning vectors and Dr. H. Christian Peters (Animal Facility, University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for initial advice on real-time PCR.
This work was supported by Grants of the Deutsche Forschungsgemeinschaft DFG/GRK-336 (to E. M.), Ki488/2-6 (to S. K.), and KR1321/4-1 (to S. K)., the Fritz Thyssen Stiftung (Az. 10.05.2.185 (to S. K.), the National Ataxia Foundation (to S. K.), and the Werner Otto Stiftung (to C. S.). This work forms part of the thesis of H. Miroci, Faculty of Biology (University of Hamburg, Germany).

This article contains supplemental Table S1 and Figs. 1–7.
- PABP
- poly(A)-binding protein
- Arc/Arg3.1
- activity-regulated cytoskeleton-associated protein (also known as Arg3.1)
- ATXN
- Ataxin
- DIV
- days in vitro
- DUF
- domain of unknown function
- EGFP
- enhanced green fluorescent protein
- IMP
- insulin-like growth factor II mRNA-binding protein
- LTP
- long term potentiation
- MAP
- microtubule-associated protein
- MKRN
- makorin
- PAIP
- poly(A)-binding protein-interacting protein
- PAM
- PCI/PINT associated module
- P-bodies
- processing bodies
- PhoLuc
- Photinus luciferase
- RenLuc
- Renilla luciferase
- RRM
- RNA recognition motif
- ZF
- zinc-finger.
REFERENCES
- 1. Alvarez V. A., Sabatini B. L. (2007) Anatomical and physiological plasticity of dendritic spines. Annu. Rev. Neurosci. 30, 79–97 [DOI] [PubMed] [Google Scholar]
- 2. Cajigas I. J., Will T., Schuman E. M. (2010) Protein homeostasis and synaptic plasticity. EMBO J. 29, 2746–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hirokawa N. (2006) mRNA transport in dendrites. RNA granules, motors, and tracks. J. Neurosci. 26, 7139–7142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Martin K. C., Zukin R. S. (2006) RNA trafficking and local protein synthesis in dendrites. An overview. J. Neurosci. 26, 7131–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bramham C. R., Wells D. G. (2007) Dendritic mRNA. Transport, translation, and function. Nat. Rev. Neurosci. 8, 776–789 [DOI] [PubMed] [Google Scholar]
- 6. Lin A. C., Holt C. E. (2007) Local translation and directional steering in axons. EMBO J. 26, 3729–3736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Donnelly C. J., Fainzilber M., Twiss J. L. (2010) Subcellular communication through RNA transport and localized protein synthesis. Traffic 11, 1498–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brendel C., Rehbein M., Kreienkamp H. J., Buck F., Richter D., Kindler S. (2004) Characterization of Staufen 1 ribonucleoprotein complexes. Biochem. J. 384, 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanai Y., Dohmae N., Hirokawa N. (2004) Kinesin transports RNA. Isolation and characterization of an RNA-transporting granule. Neuron 43, 513–525 [DOI] [PubMed] [Google Scholar]
- 10. Anderson P., Kedersha N. (2006) RNA granules. J. Cell Biol. 172, 803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barbee S. A., Estes P. S., Cziko A. M., Hillebrand J., Luedeman R. A., Coller J. M., Johnson N., Howlett I. C., Geng C., Ueda R., Brand A. H., Newbury S. F., Wilhelm J. E., Levine R. B., Nakamura A., Parker R., Ramaswami M. (2006) Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 52, 997–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sossin W. S., DesGroseillers L. (2006) Intracellular trafficking of RNA in neurons. Traffic 7, 1581–1589 [DOI] [PubMed] [Google Scholar]
- 13. Kahvejian A., Roy G., Sonenberg N. (2001) The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb. Symp. Quant. Biol. 66, 293–300 [DOI] [PubMed] [Google Scholar]
- 14. Mohr E., Prakash N., Vieluf K., Fuhrmann C., Buck F., Richter D. (2001) Vasopressin mRNA localization in nerve cells. Characterization of cis-acting elements and trans-acting factors. Proc. Natl. Acad. Sci. U.S.A. 98, 7072–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohr E., Richter D. (2004) Subcellular vasopressin mRNA trafficking and local translation in dendrites. J. Neuroendocrinol. 16, 333–339 [DOI] [PubMed] [Google Scholar]
- 16. Skabkina O. V., Skabkin M. A., Popova N. V., Lyabin D. N., Penalva L. O., Ovchinnikov L. P. (2003) Poly(A)-binding protein positively affects YB-1 mRNA translation through specific interaction with YB-1 mRNA. J. Biol. Chem. 278, 18191–18198 [DOI] [PubMed] [Google Scholar]
- 17. Nelson M. R., Luo H., Vari H. K., Cox B. J., Simmonds A. J., Krause H. M., Lipshitz H. D., Smibert C. A. (2007) A multiprotein complex that mediates translational enhancement in Drosophila. J. Biol. Chem. 282, 34031–34038 [DOI] [PubMed] [Google Scholar]
- 18. Kulkarni S. D., Muralidharan B., Panda A. C., Bakthavachalu B., Vindu A., Seshadri V. (2011) Glucose-stimulated translation regulation of insulin by the 5′-UTR-binding proteins. J. Biol. Chem. 286, 14146–14156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Melo Neto O. P., Standart N., Martins de Sa C. (1995) Autoregulation of poly(A)-binding protein synthesis in vitro. Nucleic Acids Res. 23, 2198–2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bag J., Wu J. (1996) Translational control of poly(A)-binding protein expression. Eur. J. Biochem. 237, 143–152 [DOI] [PubMed] [Google Scholar]
- 21. Arn E. A., Cha B. J., Theurkauf W. E., Macdonald P. M. (2003) Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell 4, 41–51 [DOI] [PubMed] [Google Scholar]
- 22. Grosset C., Chen C. Y., Xu N., Sonenberg N., Jacquemin-Sablon H., Shyu A. B. (2000) A mechanism for translationally coupled mRNA turnover. Interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103, 29–40 [DOI] [PubMed] [Google Scholar]
- 23. Krichevsky A. M., Kosik K. S. (2001) Neuronal RNA granules. A link between RNA localization and stimulation-dependent translation. Neuron 32, 683–696 [DOI] [PubMed] [Google Scholar]
- 24. Gray T. A., Hernandez L., Carey A. H., Schaldach M. A., Smithwick M. J., Rus K., Marshall Graves J. A., Stewart C. L., Nicholls R. D. (2000) The ancient source of a distinct gene family encoding proteins featuring RING and C(3)H zinc finger motifs with abundant expression in developing brain and nervous system. Genomics 66, 76–86 [DOI] [PubMed] [Google Scholar]
- 25. Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 96, 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J. H., Park S. M., Kang M. R., Oh S. Y., Lee T. H., Muller M. T., Chung I. K. (2005) Ubiquitin ligase MKRN1 modulates telomere length homeostasis through a proteolysis of hTERT. Genes Dev. 19, 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee E. W., Lee M. S., Camus S., Ghim J., Yang M. R., Oh W., Ha N. C., Lane D. P., Song J. (2009) Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 28, 2100–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Omwancha J., Zhou X. F., Chen S. Y., Baslan T., Fisher C. J., Zheng Z., Cai C., Shemshedini L. (2006) Makorin RING finger protein 1 (MKRN1) has negative and positive effects on RNA polymerase II-dependent transcription. Endocrine 29, 363–373 [DOI] [PubMed] [Google Scholar]
- 29. Lehner B., Sanderson C. M. (2004) A protein interaction framework for human mRNA degradation. Genome Res. 14, 1315–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kozlov G., Safaee N., Rosenauer A., Gehring K. (2010) Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J. Biol. Chem. 285, 13599–13606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Craig A. W., Haghighat A., Yu A. T., Sonenberg N. (1998) Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392, 520–523 [DOI] [PubMed] [Google Scholar]
- 32. Karim M. M., Svitkin Y. V., Kahvejian A., De Crescenzo G., Costa-Mattioli M., Sonenberg N. (2006) A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A)-binding protein (PABP) binding. Proc. Natl. Acad. Sci. U.S.A. 103, 9494–9499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sutton M. A., Schuman E. M. (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58 [DOI] [PubMed] [Google Scholar]
- 34. Richter J. D. (2010) Translational control of synaptic plasticity. Biochem. Soc. Trans. 38, 1527–1530 [DOI] [PubMed] [Google Scholar]
- 35. Schwarzacher S. W., Vuksic M., Haas C. A., Burbach G. J., Sloviter R. S., Deller T. (2006) Neuronal hyperactivity induces astrocytic expression of neurocan in the adult rat hippocampus. Glia 53, 704–714 [DOI] [PubMed] [Google Scholar]
- 36. Steward O., Wallace C. S., Lyford G. L., Worley P. F. (1998) Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 21, 741–751 [DOI] [PubMed] [Google Scholar]
- 37. Baron-Benhamou J., Gehring N. H., Kulozik A. E., Hentze M. W. (2004) Using the λN peptide to tether proteins to RNAs. Methods Mol. Biol. 257, 135–154 [DOI] [PubMed] [Google Scholar]
- 38. Schütt J., Falley K., Richter D., Kreienkamp H. J., Kindler S. (2009) Fragile X mental retardation protein regulates the levels of scaffold proteins and glutamate receptors in postsynaptic densities. J. Biol. Chem. 284, 25479–25487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jackson R. J., Hellen C. U., Pestova T. V. (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blichenberg A., Schwanke B., Rehbein M., Garner C. C., Richter D., Kindler S. (1999) Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J. Neurosci. 19, 8818–8829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pfaffl M. W., Horgan G. W., Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mohr E., Morris J. F., Richter D. (1995) Differential subcellular mRNA targeting. Deletion of a single nucleotide prevents the transport to axons but not to dendrites of rat hypothalamic magnocellular neurons. Proc. Natl. Acad. Sci. U.S.A. 92, 4377–4381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lunde B. M., Moore C., Varani G. (2007) RNA-binding proteins. Modular design for efficient function. Nat. Rev. Mol. Cell Biol. 8, 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liang J., Song W., Tromp G., Kolattukudy P. E., Fu M. (2008) Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE 3, e2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hunter T. (2007) The age of cross-talk. Phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 46. Kozlov G., De Crescenzo G., Lim N. S., Siddiqui N., Fantus D., Kahvejian A., Trempe J. F., Elias D., Ekiel I., Sonenberg N., O'Connor-McCourt M., Gehring K. (2004) Structural basis of ligand recognition by PABC, a highly specific peptide binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 23, 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zekri L., Huntzinger E., Heimstädt S., Izaurralde E. (2009) The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 29, 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nielsen J., Christiansen J., Lykke-Andersen J., Johnsen A. H., Wewer U. M., Nielsen F. C. (1999) A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol. Cell. Biol. 19, 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gray T. A., Wilson A., Fortin P. J., Nicholls R. D. (2006) The putatively functional Mkrn1-p1 pseudogene is neither expressed nor imprinted nor does it regulate its source gene in trans. Proc. Natl. Acad. Sci. U.S.A. 103, 12039–12044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Franklin N. C. (1985) Conservation of genome form but not sequence in the transcription antitermination determinants of bacteriophages λ, φ21 and P22. J. Mol. Biol. 181, 75–84 [DOI] [PubMed] [Google Scholar]
- 51. Naarmann I. S., Harnisch C., Müller-Newen G., Urlaub H., Ostareck-Lederer A., Ostareck D. H. (2010) DDX6 recruits translational silenced human reticulocyte 15-lipoxygenase mRNA to RNP granules. RNA 16, 2189–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Naisbitt S., Kim E., Tu J. C., Xiao B., Sala C., Valtschanoff J., Weinberg R. J., Worley P. F., Sheng M. (1999) Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron 23, 569–582 [DOI] [PubMed] [Google Scholar]
- 53. Schuman E. M., Dynes J. L., Steward O. (2006) Synaptic regulation of translation of dendritic mRNAs. J. Neurosci. 26, 7143–7146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Steward O. (1976) Reinnervation of dentate gyrus by homologous afferents following entorhinal cortical lesions in adult rats. Science 194, 426–428 [DOI] [PubMed] [Google Scholar]
- 55. Steward O., Halpain S. (1999) Lamina-specific synaptic activation causes domain-specific alterations in dendritic immunostaining for MAP2 and CAM kinase II. J. Neurosci. 19, 7834–7845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dragunow M., Abraham W. C., Goulding M., Mason S. E., Robertson H. A., Faull R. L. (1989) Long term potentiation and the induction of c-fos mRNA and proteins in the dentate gyrus of unanesthetized rats. Neurosci. Lett. 101, 274–280 [DOI] [PubMed] [Google Scholar]
- 57. Fukazawa Y., Saitoh Y., Ozawa F., Ohta Y., Mizuno K., Inokuchi K. (2003) Hippocampal LTP is accompanied by enhanced F-actin content within the dendritic spine that is essential for late LTP maintenance in vivo. Neuron 38, 447–460 [DOI] [PubMed] [Google Scholar]
- 58. Laity J. H., Lee B. M., Wright P. E. (2001) Zinc finger proteins. New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 [DOI] [PubMed] [Google Scholar]
- 59. Roy G., De Crescenzo G., Khaleghpour K., Kahvejian A., O'Connor-McCourt M., Sonenberg N. (2002) Paip1 interacts with poly(A)-binding protein through two independent binding motifs. Mol. Cell. Biol. 22, 3769–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Khaleghpour K., Kahvejian A., De Crescenzo G., Roy G., Svitkin Y. V., Imataka H., O'Connor-McCourt M., Sonenberg N. (2001) Dual interactions of the translational repressor Paip2 with poly(A)-binding protein. Mol. Cell. Biol. 21, 5200–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ezzeddine N., Chang T. C., Zhu W., Yamashita A., Chen C. Y., Zhong Z., Yamashita Y., Zheng D., Shyu A. B. (2007) Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 27, 7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhong J., Zhang T., Bloch L. M. (2006) Dendritic mRNAs encode diversified functionalities in hippocampal pyramidal neurons. BMC Neuroscience 7, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kozlov G., Trempe J. F., Khaleghpour K., Kahvejian A., Ekiel I., Gehring K. (2001) Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. U.S.A. 98, 4409–4413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Patel G. P., Bag J. (2006) IMP1 interacts with poly(A)-binding protein (PABP) and the autoregulatory translational control element of PABP-mRNA through the KH III-IV domain. FEBS J. 273, 5678–5690 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.