Background: Dual-specific antibodies can bind to several antigens via different mechanisms.
Results: Mutagenesis of CXCL9/CXCL10 from different species identifies serine 13 as a key residue targeted by anti-human CXCL9/CXCL10 dual-specific scFvs.
Conclusion: Structural mimicry between human CXCL9 and CXCL10 governs dual-specific scFv binding.
Significance: Dissecting the binding mechanism of dual-specific antibodies is important for the development of this class of antibodies.
Keywords: Antibodies, Chemokines, Epitope Mapping, Glycosaminoglycan, Site-directed Mutagenesis, Dual Specificity
Abstract
Dual-specific antibodies are characterized by an antigen-combining site mediating specific interactions with two different antigens. We have generated five dual-specific single chain variable fragments (scFv) that neutralize the activity of the two chemokines, CXCL9 and CXCL10, to bind to their receptor CXCR3. To better understand how these dual-specific scFvs bind these two chemokines that only share a 37% sequence identity, we mapped their epitopes on human CXCL9 and CXCL10 and identified serine 13 (Ser13) as a critical residue. It is conserved between the two chemokines but not in the third ligand for CXCR3, CXCL11. Furthermore, Ser13 is exposed in the tetrameric structure of CXCL10, which is consistent with our finding that the scFvs are able to bind to CXCL9 and CXCL10 immobilized on glycosaminoglycans. Overall, the data indicate that these dual-specific scFvs bind to a conserved surface involved in CXCR3 receptor interaction for CXCL10 and CXCL9. Thus, structural mimicry between the two targets is likely to be responsible for the observed dual specificity of these antibody fragments.
Introduction
Monoclonal antibodies (mAbs) are characterized by high affinity binding to their cognate antigen and thus demonstrate exquisite target specificity. This feature is useful for research purposes as well as attractive for diagnostic or therapeutic applications. Indeed, this lowered off-target binding or neutralization risk leads to enhanced safety profiles and has contributed to making mAbs the fastest growing class of therapeutic molecules (1). However, it becomes increasingly clear that multiple mediators contribute to a clinical situation. Thus, the neutralization of a single target might not be sufficient to achieve therapeutic efficacy and thus limit the scope of mAbs. It is therefore not surprising that in recent years, the generation of numerous novel antibody formats capable of engaging multiple targets has been the focus of intense efforts (2). Antibody formats featuring two or more independent binding sites, collectively called bispecific antibodies, clearly represent the major class of multitargeting molecules under development (3). However, other avenues, such as the use of recombinant polyclonal antibodies or dual-specific antibodies, have also been explored (4–11). Dual-specific antibodies are mAbs having an antigen-combining site that can mediate specific binding to two targets or more. As their format does not differ from a standard IgG, their manufacturing and development path is simplified compared with many bispecific antibody formats or recombinant polyclonal antibodies. Such antibodies have been raised against proteins sharing a high degree of identity, structural similarity, or even completely unrelated molecules and therefore challenging the “one antibody, one antigen” dogma (4–8, 12, 13).
We previously isolated and described a panel of dual-specific antibody fragments and demonstrated that it is possible to tune the specificity of single-chain antibody fragments (scFv)2 to neutralize two human chemokines, CXCL10 (interferon inducible protein 10, IP-10) and CXCL9 (monokine induced by interferon γ, MIG), that only share a 37% sequence identity (5). These chemokines belong to the same CXC family, share a conserved three-dimensional structure, and are ligands of the CXCR3 receptor to which they bind in a similar fashion (14). In contrast, the third ligand of CXCR3, CXCL11, has been shown to engage the receptor at a different site and with the highest affinity (14). Although the presence of three ligands for the same receptor is commonly seen as a sign of redundancy of this system, studies have shown that the absence of one of the ligands cannot always be compensated in vivo (15). In addition, expression of the three CXCR3 ligands is differentially induced by pro-inflammatory cytokines and regulated by distinct promoter elements that lead to differential timing and expression patterns (16). The precise biological function of the different CXCR3 ligands remains to be fully understood.
In this study, we aimed at elucidating how dual-specific scFvs can specifically engage CXCL9 and CXCL10 but not the third CXCR3 ligand. For this, the epitopes of the scFvs were characterized. The dual specificity of the scFvs toward CXCL9 and CXCL10 was determined using the sequences of different species to identify important regions and residues. Site-directed mutagenesis was used to generate multiple mutants of CXCL10, CXCL9, and CXCL11 from these species allowing the identification of residues that restored binding and thus played a key role in the antibody-antigen interaction. The results indicate that the scFvs bind to the same region on CXCL9 and CXCL10, in a site that overlaps with receptor interaction. Furthermore, a critical residue for binding was identified that is conserved between human CXCL9 and CXCL10 but that is not sufficient to mediate binding when introduced into the third CXCR3 ligand, CXCL11. Structural analysis indicates that the main chain conformation differs between CXCL10 and CXCL11 in the epitope region, providing an explanation for the lack of binding of the scFv to CXCL11 despite the high local degree of amino acid identity. This study highlights a structural difference of a functionally important epitope within the different CXCR3 ligands.
EXPERIMENTAL PROCEDURES
Molecular Cloning
The genes encoding the mature protein human CXCL10 (accession number NM001565), mouse CXCL10 (accession number NM_021274), rat CXCL10 (accession number BC058444), rabbit CXCL10 (accession number EC618601), human CXCL9 (accession number NM002416), mouse CXCL9 (accession number NM_008599), and human CXCL11 (accession number AF030514) were cloned in an expression plasmid pET43.1a (Novagen Madison, WI) by PCR amplification. For cynomolgus chemokines, the rhesus monkey genes (accession numbers AY044446 and AY044445, for CXCL10 and CXCL9, respectively) were used for designing primers to amplify and clone the corresponding cynomolgus genes into the pET43.1a vector. Chemokines were produced as recombinant proteins fused to the Escherichia coli NusA protein, for solubilization purposes, as described previously (17). Thus, the sequence for the factor Xa protease cleavage site was introduced at the C terminus of NusA. The sequence for the AviTagTM (Avidity) biotinylation site was introduced at the C terminus of the chemokine coding sequence. The pET-derived plasmids were transformed into TunerTM (DE3) competent bacteria (Novagen).
Site-directed Mutagenesis
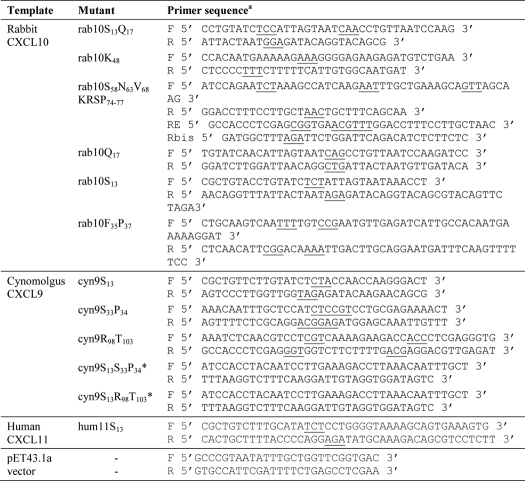
Five rabbit CXCL10 mutants, rab10S13, rab10K48, rab10S58N63V68KRSP74–77, rab10Q17, and rab10S13, and three cynomolgus CXCL9 mutants, cyn9S13, cyn9S33P34, and cyn9R98T103, were generated by site-directed mutagenesis. Residues were numbered according to the target sequence. The recombinant pET43.1a plasmids containing mature rabbit CXCL10 (rab10) or cynomolgus CXCL9 (cyn9) were used for overlapping PCR mutagenesis using specific primer pairs (Table 1). All PCR assembly products were digested with SacII and XhoI and ligated into pET43 expression vector. The recombinant plasmids were then transformed into competent E. coli strain XL1 cells, and the expected mutations were further confirmed by DNA sequencing. Plasmids were then transformed into TunerTM (DE3) competent bacteria (Novagen) for recombinant protein production.
TABLE 1.
PCR primers used to generate the rabbit CXCL10 and cynomolgus CXCL9 site-directed mutants
a The mutant codons are underlined. F is forward; R is reverse; E is elongation.
* These mutants were constructed by assembly of cyn9S13 and cyn9S33P34 or cyn9R98T103 as templates.
Expression and Purification of Recombinant His-tagged NusA Chemokine Fusion
Expression of wild type and mutated recombinant chemokines was performed as described previously (17). An overnight culture of bacteria harboring the chemokine construct was diluted into Terrific Broth (Invitrogen) containing 50 μg/ml ampicillin. The culture was incubated at 37 °C with shaking until the A600 = 0.6 was reached. Isopropyl 1-thio-β-d-galactopyranoside was then added to a final concentration of 1 mm, and the culture was incubated overnight at 28 °C with shaking (260 rpm). Bacterial pellets were resuspended in Bugbuster (Novagen) containing Benzonase® nuclease and protease inhibitor Complete EDTA-free (Roche Applied Science) and incubated for 1 h at 4 °C. The soluble and insoluble fractions were separated by centrifugation (10,000 × g, 15 min, 4 °C), and the soluble fraction was then purified by affinity chromatography, using nickel-nitrilotriacetic acid-agarose (Qiagen) and polyprep columns (Bio-Rad). 50% (v/v) of beads was added to soluble fraction and incubated for 30 min at room temperature with gentle shaking. After washing, the chemokines were eluted with imidazole-containing buffer (Tris-HCl 50 mm, pH 8.0, NaCl 200 mm, CaCl2 5 mm, and imidazole 400 mm). Elution fractions containing the protein were pooled and desalted in PBS (Sigma) using PD-10 columns (Amersham Biosciences).
Expression and Purification of Cross-reactive scFvs
The scFv E7, J9, P8, F13, C1, and J5 were produced as described previously (5). Briefly, a single colony was grown in 400 ml of 2× TYAG overnight at 30 °C. The next day scFv expression was induced by adding isopropyl 1-thio-β-d-galactopyranoside. The cells were collected by centrifugation and subjected to osmotic shock by incubating for 30 min on ice with ice-cold Tris/EDTA/sodium chloride (TES) buffer. Cells were centrifuged, and the supernatant was transferred to a fresh tube. scFvs were then purified by gravity flow by nickel-nitrilotriacetic acid affinity chromatography and desalted, as described above for chemokines.
Binding Enzyme-linked Immunosorbent Assay (Binding ELISA)
The capacity of cross-reactive scFvs E7, J9, F13, P8, C1, and J5 to bind to CXCL10, CXCL9, and CXCL11 from different species was investigated by ELISA. 96-Well Maxisorp plates (Nunc) were coated at 10 μg/ml of NusA fusion chemokine and incubated at 4 °C overnight. After washing with PBS, 0.05% Tween 20 (Sigma), plates were blocked with 3% milk/PBS. Then 50 μl of 1 μg/ml scFv in 1% milk/PBS buffer was added and incubated for 1 h at room temperature. Plates were washed, and 50 μl of monoclonal mouse anti-c-Myc IgG (1:2500, in house) in 1% milk/PBS buffer was added and incubated for 1 h at room temperature to detect bound scFv. Plates were washed and incubated for 1 h at room temperature with 50 μl of anti-mouse IgG Fcγ conjugated to HRP (1:5000, Jackson ImmunoResearch). After washing, binding was revealed with 50 μl of TMB substrate (Sigma) and stopped by adding 50 μl of H2SO4 (2 n). The absorbance at 450 nm was recorded using a precision microplate reader (Molecular Devices). Coating was controlled using specific anti-NusA mAb (Novagen), and NusA protein was also added to the assay as a negative control (data not shown).
GAG Enzyme-linked Immunosorbent Assay (GAG ELISA)
A 96-well Maxisorp plate was coated with 10 μg/ml of anti-c-Myc IgG (1:2500, in house) and incubated at 4 °C overnight. After washing with PBS, 0.05% Tween 20 (Sigma), the plate was blocked with 3% BSA/PBS buffer (Sigma). Then 5 μg/ml scFv E7, J9, F13, P8, C1, or J5 in 1% BSA/PBS buffer was added and incubated for 1 h at room temperature. In addition, two previously isolated scFvs, CF1 and A2, were used as positive and negative controls, respectively. CF1 was shown to bind to human CXCL10 in a GAG context, whereas A2 did not (data not shown). In parallel, 50 nm biotinylated heparin was preincubated with 125 nm of either hCXCL10 (PeproTech) or hCXCL9 (PeproTech) for 1 h, and the mixture was added to the washed plate for 1 h. The plate was washed, and streptavidin conjugated to HRP (Jackson ImmunoResearch, 1:5000) was added to the plate for 1 h. After washing, immobilized scFv capable of binding to chemokines in complex to biotinylated heparin was revealed with 50 μl of TMB substrate (Sigma) and stopped by adding 50 μl of H2SO4 (2 n). The absorbance at 450 nm was recorded using a precision microplate reader (Molecular Devices).
Sequence Alignments
The “UniProt/Swiss-Prot” data base was searched with a mature chemokine amino acid sequence, and species alignment was made by using ClustalW2 (18). Scores table shows the alignment score for each pair of chemokines. A pairwise score is calculated as the number of identities in the best alignment divided by the number of residues compared for every pair of sequences that are aligned. hCXCL10, hCXCL9, and hCXCL11 were retrieved from the Uniprot database and then aligned using T-Coffee (19). The final alignment was then represented using GeneDoc, and conserved residues were highlighted in black according to BLOSUM 62 substitution matrix.
Structural Representations
All the structure representations and in silico mutagenesis were prepared using PyMOL. Monomeric hCXCL10 crystal structure was retrieved from Protein Data Bank code 1LV9, and tetrameric hCXCL10 M-form crystal structure from Protein Data Bank code 1O7Y.
RESULTS
Cross-reactivity of Dual-specific scFv against CXCL10/CXCL9 from Different Species
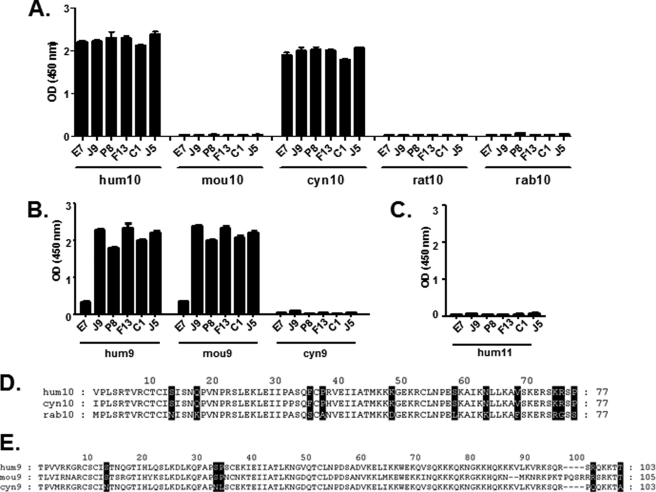
A panel of five dual-specific scFvs, J9, P8, F13, C1, and J5, have been previously derived from E7, an scFv binding and neutralizing human CXCL10 and weakly cross-reactive against human CXCL9 (5). These scFvs were obtained using a phage display selection of E7 variants in which diversity had been introduced into the complementary determining region 3 (CDR3) of the light chain (supplemental Table 1). They neutralized both human CXCL10 (hum10) and human CXCL9 (hum9), whereas no binding was observed on human CXCL11 (hum11) (Fig. 1, A–C). Their specificity was further characterized by ELISA against CXCL10 and CXCL9 from different species. The data demonstrated that, in addition to human CXCL10 (hum10), the scFvs were also able to bind to cynomolgus (cyn) CXCL10 but not to mouse (mou), rat, and rabbit (rab) CXCL10, with the exception of some weak binding of J9 to rabbit CXCL10 (Fig. 1A). In contrast, no binding was observed on cynomolgus CXCL9, whereas all five scFvs were able to bind to mouse CXCL9 (Fig. 1B).
FIGURE 1.
Determination of potential residues involved in chemokine interaction with dual-specific scFvs using binding specificity against CXCL10 and CXCL9 proteins from different species. A–C, specific binding of E7, J9, P8, F13, C1, and J5 dual-specific scFvs to a panel of immobilized CXCL10 (A), CXCL9 (B), and CXCL11 (C) chemokines from different species was assessed in an ELISA. The indicated NusA-fusion chemokines were captured and then incubated with scFvs. Coating was controlled using a specific anti-NusA mAb, and NusA protein was also added to the assay as a negative control (data not shown). Results are expressed as mean ± S.D. of duplicates. D and E, sequence alignments of mature CXCL10 (D) and CXCL9 (E) chemokine amino acid sequences from different species. Residues that are identical for human and cynomolgus but different from rabbit CXCL10 (D) or residues that are identical for human and mouse but different from cynomolgus CXCL9 (E) are shaded black. These residues were targeted for mutagenesis and substituted with the corresponding human residue. Numbering was done according to the template sequence, rabbit CXCL10 or cynomolgus CXCL9. hum10, human CXCL10; mou10, mouse CXCL10; cyn10, cynomolgus CXCL10; rat10, rat CXCL10; rab10, rabbit CXCL10; hum9, human CXCL9; mou9, mouse CXCL9; cyn9, cynomolgus CXCL9.
Identification of Epitope Candidate Residues on Human CXCL10 and CXCL9
Interestingly, despite 83% sequence identity between human and rabbit CXCL10 (Table 2), E7 and its derivatives were unable to bind to the latter (Fig. 1A). We therefore aligned the sequences of the human, cynomolgus, and rabbit CXCL10 to identify residues that are conserved in the two first proteins and not in the latter. Eleven residues were identified as potentially important for mediating scFv binding (Fig. 1D). The same method was applied to CXCL9. The fact that the E7 scFv derivatives were able to bind to mouse CXCL9 that has only 65% identity to human CXCL9 and not to the more closely related cynomolgus CXCL9 (91% identity; Table 2) allowed the identification of five residues as potentially key amino acids for the epitope on CXCL9 (Fig. 1E).
TABLE 2.
Sequence identities between mature proteins from different species of CXCL10 and CXCL9
| Protein 1a,b | Protein 2a,b | Pairwise scorec |
|---|---|---|
| % | ||
| hum10 (77 AA) | mou10 (77 AA) | 70 |
| hum10 (77 AA) | cyn10 (77 AA) | 96 |
| hum10 (77 AA) | rat10 (77 AA) | 72 |
| hum10 (77 AA) | rab10 (77 AA) | 83 |
| mou10 (77 AA) | cyn10 (77 AA) | 68 |
| mou10 (77 AA) | rat10 (77 AA) | 77 |
| mou10 (77 AA) | rab10 (77 AA) | 63 |
| cyn10 (77 AA) | rat10 (77 AA) | 74 |
| cyn10 (77 AA) | rab10 (77 AA) | 81 |
| rat10 (77 AA) | rab10 (77 AA) | 64 |
| hum9 (103 AA) | mou9 (105 AA) | 65 |
| hum9 (103 AA) | cyn9 (103 AA) | 91 |
| mou9 (105 AA) | cyn9 (103 AA) | 62 |
a The following abbreviations are used: hum10, human CXCL10; mou10, mouse CXCL10; cyn10, cynomolgus CXCL10; rat10, rat CXCL10; rab10, rabbit CXCL10; hum9, human CXCL9; mou9, mouse CXCL9; cyn9, cynomolgus CXCL9.
b The number of amino acid (AA) residues were compared.
c A pairwise score was calculated as the number of identical amino acids of the best alignment divided by the number of residues compared for each pair.
Site-directed Mutagenesis of Rabbit CXCL10 and Cynomolgus CXCL9
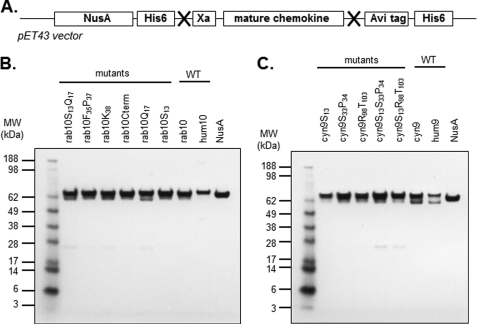
Instead of using a standard alanine-scanning mutagenesis approach to probe potentially important amino acids in the human chemokines, we attempted to restore binding to a chemokine to which the scFvs did not bind. Thus, we converted the previously identified amino acids of these nonbinding chemokines into the corresponding amino acids of the human proteins. As such, a first set of rabbit CXCL10 site-directed mutants was generated as follows: rab10S13Q17, rab10F35P37, rab10K48, and rab10S58N63V68KRSP74–77 also termed rab10Cterm. Similarly, three cynomolgus CXCL9 site-directed mutants were designed as follows: cyn9S13, cyn9S33P34, and cyn9R98T103. Mutated proteins were expressed as soluble NusA fusion proteins, using a pET43-derived expression vector, and purified by affinity chromatography via their histidine tag, as described elsewhere (Fig. 2A) (17). CXCL10 and CXCL9 mutants fused to NusA were purified, and an SDS-PAGE analysis was carried out to evaluate their degree of purity (Fig. 2, B and C). The size of the NusA-CXCL10 and NusA-CXCL9 fusion proteins corresponded to their expected molecular mass (65.8 and 68.9 kDa, respectively), and only occasional minor degradation products could be observed for some constructs.
FIGURE 2.
Generation by site-directed mutagenesis and soluble expression of CXCL10 and CXCL9 mutants in E. coli. A, schematic of chemokine construct. Recombinant chemokines were generated by site-directed mutagenesis, PCR assembly, enzymatic digestion, and cloning into pET43 vector for E. coli expression. All resulting chemokines were expressed as NusA fusions proteins and contained both N- and C-terminal histidine tags for purification purposes. His6, 6-histidine tag; Xa, cleavage factor; Avi, AviTagTM (avidity). X indicates an enzymatic restriction site. B and C, SDS-PAGE analysis of affinity-purified CXCL10 and CXCL9 mutants, respectively. Affinity-purified CXCL10 (B) and CXCL9 (C) mutants and NusA protein were denatured under reducing conditions and stained with Coomassie Blue. M, Seeblue Plus molecular weight marker (Invitrogen); hum10, human CXCL10; rab10, rabbit CXCL10; hum9, human CXCL9; cyn9, cynomolgus CXCL9.
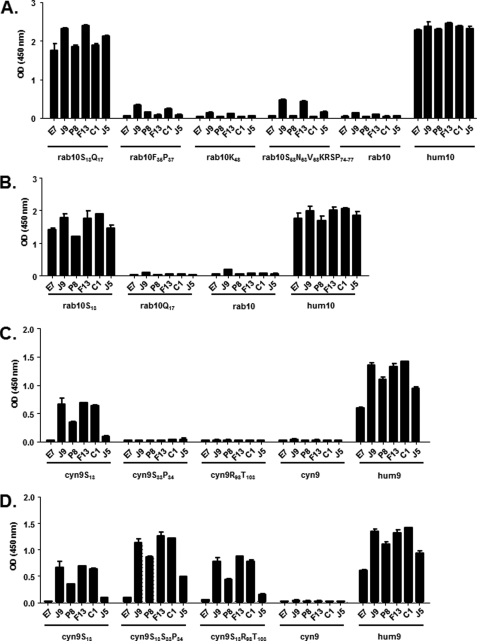
The CXCL10 mutants were probed by ELISA using E7 and the dual-specific scFvs (Fig. 3A). The data indicate that rab10S13Q17 was the only mutant to which the binding of all the scFvs was restored, and thus Ser13 and/or Gln17 are key epitope residues. A slight binding activity of scFv J9, F13, and J5 was also detected with rab10S58N63V68KRSP74–77. However, as the signals remained low, these amino acids were not considered to play a significant role in chemokine interaction. To further dissect the contribution of Ser13 and Gln17, two additional mutants, rab10S13 and rab10Q17, were generated and tested for purity by SDS-PAGE (Fig. 2B). The ELISA data demonstrated that all the scFvs were capable of binding to rab10S13 and not to rab10Q17 (Fig. 3B). Thus, Ser13 was identified as the key residue in the CXCL10 dual-specific scFv interaction.
FIGURE 3.
Restoration of binding capacity and epitope identification on human CXCL10 and CXCL9. Specific binding of E7, J9, P8, F13, C1, and J5 dual-specific scFvs to a panel of CXCL10 (A and B) and CXCL9 (C and D) mutants was assessed in an ELISA. The indicated NusA-fusion chemokines were captured and then incubated with scFvs. Coating was controlled using specific anti NusA mAb, and NusA protein was also added to the assay as negative control (data not shown). Results are expressed as mean ± S.D. of duplicates. hum10, human CXCL10; rab10, rabbit CXCL10; hum9, human CXCL9; cyn9, CXCL9.
The three CXCL9 mutants, cyn9S13, cyn9S33P34, and cyn9R98T103, were also tested, and the data showed that cyn9S13 was the only mutant able to partially restore the binding capacity to J9, F13, and C1, but not E7, J5, and to a lesser extent P8 (Fig. 3C). To determine whether a combination of mutations could further restore binding activity of the scFvs against cynomolgus CXCL9, two additional mutants, cyn9S13S33P34 and cyn9S13R98T103, were generated. The binding experiments demonstrated that the cyn9S13S33P34 combination mutant better restored the binding capacity (Fig. 3D). In contrast, cyn9S13R98T103 did not provide major improvement compared with the cyn9S13 single mutant (Fig. 3D).
Site-directed Mutagenesis of Human CXCL10 and CXCL9
To confirm the importance of Ser13, a new series of mutants, human CXCL9 and human CXCL10, was generated as follows: hum10N13, hum10H13, and hum10D13, in which Ser13 was replaced by the residues found at that position in rabbit, mouse, and rat CXCL10, respectively (supplemental “Experimental Procedures” and supplemental Table 2). Similarly, the mutant hum9N13 was generated by introducing the corresponding residue found in the cynomolgus CXCL9 sequence. These proteins were produced as NusA fusions and purified and probed by ELISA using the panel of scFvs. The results showed that replacement of Ser13 completely abolished binding of the scFv to both chemokines except for the hum10N13 mutant where the binding was significantly diminished (supplemental Fig. 1). The loss of binding observed with this panel of mutants complemented and confirmed the data obtained via the restoration of binding to the non-human chemokine approach, i.e. the importance of Ser13 for scFv binding to both human CXCL10 and CXCL9.
Epitope Accessibility in Context of Glycosaminoglycan
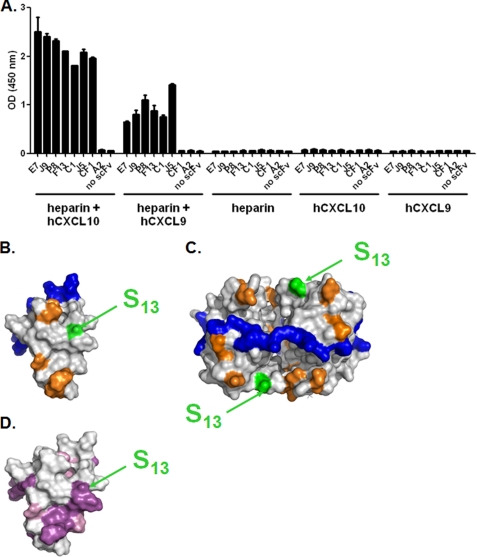
Chemokines can oligomerize and bind to GAGs and thus are retained on the surface of endothelial cells, forming a solid phase of accumulated substrate (20). The amino acids involved in GAG-chemokine interaction have been mapped (21, 22). Mutagenesis studies have shown that this interaction is crucial for the biological activity in vivo (23–25). To further define the epitope of the scFvs, their capacity to bind to CXCL10 and CXCL9, immobilized on the prototypic GAG, heparin, was tested (Fig. 4A). The results show that the epitopes of all the scFvs are accessible in the context of GAG. Ser13 is located in close vicinity to residues involved in the CXCR3 interaction (Fig. 4B) (26), which is consistent with the finding that E7 and its dual-specific variants neutralize the chemokine activity in chemotaxis assays (5). Similarly, Ser13 appears accessible in the tetrameric structure of CXCL10 and distant from a series of aligned lysine residues that were proposed to be involved in the GAG interaction (Fig. 4C) (22). This is in agreement with the ability of the scFvs to bind to their targets in the context of GAG (Fig. 4A). Finally, the amino acids that are conserved between CXCL9 and CXCL10 are highlighted on the structure of CXCL10 and in a sequence alignment (Figs. 4D and 5A). In the three-dimensional representation, a continuous patch of conserved residues between CXCL9 and CXCL10 that include Ser13 is apparent (Fig. 4D). Collectively, our data demonstrate that Ser13 is a critical epitope residue on both CXCL9 and CXCL10, suggesting that a conserved area is targeted by the scFvs on both chemokines. Therefore, dual specificity is conferred through local structural homology between the antigens.
FIGURE 4.
Epitope accessibility in the context of glycosaminoglycans. A, E7, J9, P8, F13, C1, and J5 dual-specific scFvs were immobilized in an ELISA-based assay and tested for their capacity to bind to either hCXCL10 or hCXCL9 in complex to biotinylated heparin. scFv CF1 was added as a positive control for an anti-hCXCL10, and scFv A2 was added as an irrelevant negative control. Unspecific coating of either chemokine-heparin complex, chemokine, or heparin, in the absence of scFv, was also evaluated. Results are expressed as the mean ± S.D. of duplicates. B and C, Connolly surface representations of monomeric (B) hCXCL10 (Protein Data Bank 1LV9) and tetrameric (C) hCXCL10. Color representations are as follows: blue, GAG-interacting residues; orange, hCXCR3-interacting residues; green and arrows, Ser13. CXCR3-binding residues were determined according to Booth et al. (26). Potential GAG binding residues were suggested by Swaminathan et al. (22). D, Connolly surface representation of monomeric hCXCL10. Conserved residues between hCXCL9 and hCXCL10 were determined according to the alignment in Fig. 5F. Color representation is as follows: purple, conserved residues; pink, amino acid residues with similar physico-chemical properties according to BLOSUM 62 substitution matrix; arrows indicate Ser13.
FIGURE 5.
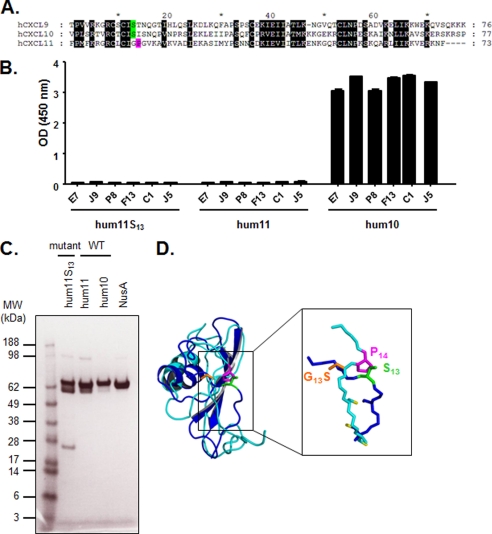
Introduction of Ser13 into CXCL11. A, alignment of mature hCXCL10, hCXCL9, and hCXCL11 chemokine amino acid sequences. Black shaded letters indicate conserved residues according to BLOSUM 62 substitution matrix. Ser13 is represented in green, and Pro14 is indicated in pink. B, specific binding of E7, J9, P8, F13, C1, and J5 dual-specific scFvs to human CXCL11, its mutant, and human CXCL10 was assessed in an ELISA. The indicated NusA-fusion chemokines were captured and then incubated with scFvs. Coating was controlled using specific anti-NusA mAb, and NusA protein was also added to the assay as negative control (data not shown). Results are expressed as mean ± S.D. of duplicates. hum11, human CXCL11; hum10, human CXCL10. C, SDS-PAGE analysis of affinity-purified CXCL11 mutant and wild type, CXCL10, and NusA proteins. The proteins were denatured under reducing conditions and stained with Coomassie Blue. M, Seeblue Plus SDS molecular weight marker (Invitrogen); hum11, human CXCL11; hum10, human CXCL10. D, superimposition of the ribbon representations of monomeric hCXCL10 (dark blue) and hCXCL11 (light blue). The critical amino acid side chains at positions 13 and 14 (Ser13, G13S, and Pro14), and cysteine side chains are represented as sticks in green, orange, pink, and yellow, respectively.
N-terminal Proline Kink Differentiates CXCL11 from Two Other CXCR3 Ligands
Interestingly, the amino acid sequence in this region is also conserved in CXCL11 with the exception of Ser13, which is replaced by a glycine (Fig. 5A). To further define the importance of the residue at position 13, the hum11S13 mutant was generated by introducing a serine at position 13 into human CXCL11 by site-directed mutagenesis (Table 1). After confirmation of hum11S13 purity by SDS-PAGE analysis, binding of the scFv to hum11S13 was probed by ELISA (Fig. 5, B and C). The introduction of a serine at position 13 of human CXCL11 did not allow binding of any of the scFvs (Fig. 5B). The superimposition of the structures of CXCL10 and CXCL11 shows that, in the epitope region, the main chain conformations differ between the two proteins and that the side chain orientation of Gly13 in CXCL11 is different from Ser13 in CXCL10. Indeed, the replacement of glycine by serine at position 13 (G13S) using in silico mutagenesis of CXCL11 shows an opposite side chain orientation (Fig. 5D). We observed that Pro14 of CXCL11 induces a proline kink in its N-terminal loop, which changes the orientation of the residue 13, as compared with the same position in the structure of CXCL10.
DISCUSSION
It is often considered that specificity is crucial for biochemical interactions and that a lack of specificity is overall detrimental. However, promiscuity of the interaction and function of proteins is a widespread mechanism in living organisms (27). Beyond the fundamental understanding of molecular recognition and protein evolution mechanisms, exploring protein promiscuity also has implications for the engineering of proteins with multiple functions. In particular, the opportunity of creating antibodies that are capable of engaging multiple targets can significantly expand the efficacy of antibodies and the range of their therapeutic applications. It is therefore not surprising that approaches to generate such multispecific antibodies are beginning to emerge (4–8, 13, 28).
In this study, we provide new insights into how multispecific recognition by antibodies can be achieved. We have characterized the epitope of a panel of dual-specific antibody fragments that were engineered to neutralize two inflammatory chemokines, human CXCL9 and CXCL10, that share only 37% sequence identity (Table 3). We first determined the cross-reactivity of the antibody fragments against the CXCL9 and CXCL10 homologues from different species and took advantage of their differential binding properties to identify residues that are potentially critical for the epitope. We took a “gain of function” approach to restore binding to CXCL9 and CXCL10 from species that are not recognized by the scFvs. We reasoned that this approach is preferable in particular for small proteins such as chemokines in which a mutation can more easily affect binding to a site distant from the actual epitope. We have identified Ser13 as a critical residue that is conserved between human CXCL9 and CXCL10 and is sufficient to restore binding of the scFvs to rabbit CXCL10 and cynomolgus CXCL9. The importance of this Ser13 was further confirmed by the loss of binding observed against human CXCL9 and CXCL10 in which this residue was mutated. Ser13 is located next to a patch of residues that are conserved between all three ligands of CXCR3. Despite this conservation, introduction of Ser13 into CXCL11 does not restore the epitope recognized by the dual-specific scFvs. Further analysis of the CXCL11 structure revealed that, due to Pro14, the orientation of the residue at position 13 differs between CXCL11 and CXCL10. Therefore, we hypothesize that mutation of the Gly13 residue into Ser13 (G13S) may not be able to restore binding to scFvs because of unfavorable side chain orientation at the scFv-CXCL11-binding interface.
TABLE 3.
Sequence identities for each pair of CXC mature chemokines
| Protein 1a,b | Protein 2a,b | Pairwise scorec |
|---|---|---|
| % | ||
| hum10 (77 AA) | hum9 (103 AA) | 37 |
| hum10 (77 AA) | hum11 (73 AA) | 32 |
| hum9 (103 AA) | hum11 (73 AA) | 35 |
a The following abbreviations are used: hum10, human CXCL10; hum9, human CXCL9; hum11, human CXCL11.
b The number of amino acid (AA) residues were compared.
c A pairwise score was calculated as the number of identical amino acids of the best alignment divided by the number of residues compared for each pair.
In addition, although CXCL11 signals through the same receptor as CXCL9 and CXCL10, it clearly interacts differently with CXCR3 as CXCL11 does not compete for binding with the two other ligands (29). Thus, collectively our results indicate that E7 and its derivatives target a conserved surface on CXCL9 and CXCL10 involved in the CXCR3 interaction. Furthermore, beyond the absence of Ser13, this surface differs in CXCL11, which is consistent with its alternative binding mode.
Chemokines can oligomerize and interact with GAGs expressed at the surface of cells. The capacity of a neutralizing antibody to interact with its target in a physiological context is an important parameter. The location of Ser13 in the tetrameric structure of CXCL10 supports our findings that the dual-specific scFvs are able to bind to CXCL10 and CXCL9 in the context of GAGs.
Various mechanisms have been proposed to mediate the promiscuity of protein-protein interactions (27). In the case of multiple antigen binding by an antibody, one mechanism conferring antibody multispecificity relies on the structural flexibility of the complementary determining region loops that adopt multiple conformations, some of which are stabilized upon antigen engagement (12). Another mechanism consists of two antigens interacting with the antibody-combining site in an asymmetric manner, one interacting mainly with the heavy chain and the other with the light chain. This mode is exemplified by a recently isolated variant of Herceptin® (6). The crystal structure revealed that an engineered antibody was able to interact with human epidermal growth factor receptor 2 (HER2) as well as vascular endothelial growth factor with different but partially overlapping regions of the paratope. Conformational flexibility of the light chain was also required to achieve target cross-reactivity. Finally, structural mimicry, in which regions of the targets are structurally similar, is also a means to attain antibody multispecificity. Our data suggest that antigen structural mimicry accounts for the dual specificity of the variants described in this study. Structural studies using scFv-chemokine co-crystals, if possible to generate, would help to further define even more precisely the interactions between the paratopes of the different E7 derivatives with the two chemokines. However, for the moment, our study already provides new insights for the in vitro evolution of multispecific antibodies directed against structurally related antigens such as chemokines and highlights a structural difference between CXCR3 ligands within a site that confers importance for biological activity.
Supplementary Material
The authors are employees of NovImmune SA.

This article contains supplemental Tables 1 and 2, Fig. 1, and “Experimental Procedures.”
- scFv
- single-chain variable fragment
- GAG
- glycosaminoglycan.
REFERENCES
- 1. Reichert J. M. (2011) MAbs 3, 76–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan A. C., Carter P. J. (2010) Nat. Rev. Immunol. 10, 301–316 [DOI] [PubMed] [Google Scholar]
- 3. Fischer N., Léger O. (2007) Pathobiology 74, 3–14 [DOI] [PubMed] [Google Scholar]
- 4. Zhang M. Y., Xiao X., Sidorov I. A., Choudhry V., Cham F., Zhang P. F., Bouma P., Zwick M., Choudhary A., Montefiori D. C., Broder C. C., Burton D. R., Quinnan G. V., Jr., Dimitrov D. S. (2004) J. Virol. 78, 9233–9242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fagète S., Ravn U., Gueneau F., Magistrelli G., Kosco-Vilbois M. H., Fischer N. (2009) MAbs. 1, 288–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bostrom J., Yu S. F., Kan D., Appleton B. A., Lee C. V., Billeci K., Man W., Peale F., Ross S., Wiesmann C., Fuh G. (2009) Science 323, 1610–1614 [DOI] [PubMed] [Google Scholar]
- 7. Garcia-Rodriguez C., Levy R., Arndt J. W., Forsyth C. M., Razai A., Lou J., Geren I., Stevens R. C., Marks J. D. (2007) Nat. Biotechnol. 25, 107–116 [DOI] [PubMed] [Google Scholar]
- 8. Reid C., Rushe M., Jarpe M., van Vlijmen H., Dolinski B., Qian F., Cachero T. G., Cuervo H., Yanachkova M., Nwankwo C., Wang X., Etienne N., Garber E., Bailly V., de Fougerolles A., Boriack-Sjodin P. A. (2006) Protein Eng. Des. Sel. 19, 317–324 [DOI] [PubMed] [Google Scholar]
- 9. Ishida I., Tomizuka K., Yoshida H., Tahara T., Takahashi N., Ohguma A., Tanaka S., Umehashi M., Maeda H., Nozaki C., Halk E., Lonberg N. (2002) Cloning Stem Cells 4, 91–102 [DOI] [PubMed] [Google Scholar]
- 10. Wiberg F. C., Rasmussen S. K., Frandsen T. P., Rasmussen L. K., Tengbjerg K., Coljee V. W., Sharon J., Yang C. Y., Bregenholt S., Nielsen L. S., Haurum J. S., Tolstrup A. B. (2006) Biotechnol. Bioeng. 94, 396–405 [DOI] [PubMed] [Google Scholar]
- 11. Bregenholt S., Jensen A., Lantto J., Hyldig S., Haurum J. S. (2006) Curr. Pharm. Des. 12, 2007–2015 [DOI] [PubMed] [Google Scholar]
- 12. James L. C., Roversi P., Tawfik D. S. (2003) Science 299, 1362–1367 [DOI] [PubMed] [Google Scholar]
- 13. Garcia-Rodriguez C., Geren I. N., Lou J., Conrad F., Forsyth C., Wen W., Chakraborti S., Zao H., Manzanarez G., Smith T. J., Brown J., Tepp W. H., Liu N., Wijesuriya S., Tomic M. T., Johnson E. A., Smith L. A., Marks J. D. (2011) Protein Eng. Des. Sel. 24, 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clark-Lewis I., Mattioli I., Gong J. H., Loetscher P. (2003) J. Biol. Chem. 278, 289–295 [DOI] [PubMed] [Google Scholar]
- 15. Hsieh M. F., Lai S. L., Chen J. P., Sung J. M., Lin Y. L., Wu-Hsieh B. A., Gerard C., Luster A., Liao F. (2006) J. Immunol. 177, 1855–1863 [DOI] [PubMed] [Google Scholar]
- 16. Groom J. R., Luster A. D. (2011) Immunol. Cell Biol. 89, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Magistrelli G., Gueneau F., Muslmani M., Ravn U., Kosco-Vilbois M., Fischer N. (2005) Biochem. Biophys. Res. Commun. 334, 370–375 [DOI] [PubMed] [Google Scholar]
- 18. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Notredame C., Higgins D. G., Heringa J. (2000) J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 20. Hoogewerf A. J., Kuschert G. S., Proudfoot A. E., Borlat F., Clark-Lewis I., Power C. A., Wells T. N. (1997) Biochemistry 36, 13570–13578 [DOI] [PubMed] [Google Scholar]
- 21. Johnson Z., Proudfoot A. E., Handel T. M. (2005) Cytokine Growth Factor Rev. 16, 625–636 [DOI] [PubMed] [Google Scholar]
- 22. Swaminathan G. J., Holloway D. E., Colvin R. A., Campanella G. K., Papageorgiou A. C., Luster A. D., Acharya K. R. (2003) Structure 11, 521–532 [DOI] [PubMed] [Google Scholar]
- 23. Proudfoot A. E., Handel T. M., Johnson Z., Lau E. K., LiWang P., Clark-Lewis I., Borlat F., Wells T. N., Kosco-Vilbois M. H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Proudfoot A. E. (2006) Biochem. Soc. Trans. 34, 422–426 [DOI] [PubMed] [Google Scholar]
- 25. Johnson Z., Kosco-Vilbois M. H., Herren S., Cirillo R., Muzio V., Zaratin P., Carbonatto M., Mack M., Smailbegovic A., Rose M., Lever R., Page C., Wells T. N., Proudfoot A. E. (2004) J. Immunol. 173, 5776–5785 [DOI] [PubMed] [Google Scholar]
- 26. Booth V., Keizer D. W., Kamphuis M. B., Clark-Lewis I., Sykes B. D. (2002) Biochemistry 41, 10418–10425 [DOI] [PubMed] [Google Scholar]
- 27. Nobeli I., Favia A. D., Thornton J. M. (2009) Nat. Biotechnol. 27, 157–167 [DOI] [PubMed] [Google Scholar]
- 28. Bostrom J., Haber L., Koenig P., Kelley R. F., Fuh G. (2011) PLoS One 6, e17887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cox M. A., Jenh C. H., Gonsiorek W., Fine J., Narula S. K., Zavodny P. J., Hipkin R. W. (2001) Mol. Pharmacol. 59, 707–715 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.