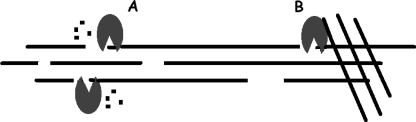Abstract
The kinetics of cellulose hydrolysis have longbeen described by an initial fast hydrolysis rate, tapering rapidly off, leading to a process that takes days rather than hours to complete. This behavior has been mainly attributed to the action of cellobiohydrolases and often linked to the processive mechanism of this exo-acting group of enzymes. The initial kinetics of endo-glucanases (EGs) is far less investigated, partly due to a limited availability of quantitative assay technologies. We have used isothermal calorimetry to monitor the early time course of the hydrolysis of insoluble cellulose by the three main EGs from Trichoderma reesei (Tr): TrCel7B (formerly EG I), TrCel5A (EG II), and TrCel12A (EG III). These endo-glucanases show a distinctive initial burst with a maximal rate that is about 5-fold higher than the rate after 5 min of hydrolysis. The burst is particularly conspicuous for TrCel7B, which reaches a maximal turnover of about 20 s−1 at 30 °C and conducts about 1200 catalytic cycles per enzyme molecule in the initial fast phase. For TrCel5A and TrCel12A the extent of the burst is 2–300 cycles per enzyme molecule. The availability of continuous data on EG activity allows an analysis of the mechanisms underlying the initial kinetics, and it is suggested that the slowdown is linked to transient inactivation of enzyme on the cellulose surface. We propose, therefore, that the frequency of structures on the substrate surface that cause transient inactivation determine the extent of the burst phase.
Keywords: Cellulase, Enzyme Inactivation, Enzyme Kinetics, Enzyme Mechanisms, Isothermal Titration Calorimetry, Burst Kinetics, Endo-Glucanase, T. reesei
Introduction
Biomass provides a carbon neutral alternative to fossil fuels, and the optimization of the enzymatic breakdown of lignocellulose has intensified greatly in recent times (1, 2). The hydrolysis of cellulose is a complex process involving the synergistic action of a combination of glycoside hydrolases (1) collectively called cellulases (2, 3). endo-1,4-β-d-Glucan-4-glucanohydrolases (EG)2 (EC 3.2.1.4) randomly cleave the cellulose polymer backbone in the amorphous areas (2, 4). In addition to this, there are normally two exoenzymes, exo-1,4-β-d-glucancellobiohydrolase I and II (EC 3.2.1.91), which cleave cellobiose from the reducing and non-reducing ends (5) of the cellulose chain, and degrade the crystalline areas of cellulose (3, 6). Finally, β-d-glucoside glucohydrolase (cellobiase) (EC 3.2.1.21) hydrolyzes the cellobiose into glucose monomers (6).
The activity of many cellulolytic enzymes shows an unusual time course including an initial burst and a subsequent slowdown. The latter can sometimes be detected already within seconds or minutes of the reaction onset, and the gradual reduction in the hydrolytic rate often persists for days (7, 8). This behavior is of particular interest both in attempts to elucidate the molecular mechanisms and regulation of cellulolytic enzymes and in the application of cellulases for the industrial breakdown of biomass. Thus, a typical biomass saccharification starts with a conversion of up to 50% in the first 24 h but takes 2–5 days to achieve appreciable yields (>80%) of glucose (9). For the breakdown of complex biomass the slowdown is undoubtedly controlled by a variety of factors, but the observation of a distinct burst and slowdown for pure cellulose/cellulase systems at very low degrees of conversion has directed the focus to the intrinsic molecular properties of the enzymes and substrate (10–14). The general conclusion is that the early slowdown in pure systems depends on the processive mode of action for the cellobiohydrolases (i.e. their tendency to sequentially hydrolyze stretches of one cellulose strand without dissociation). Clearly, this explanation is not immediately applicable to EGs, which are thought to hydrolyze glycosidic bonds randomly (that is non-processively). Here we study the initial kinetics of the three main EGs produced by Hypocrea jecorina (anamorph: Trichoderma reesei). TrCel7B, TrCel5A, and TrCel12A (4) belong to glycoside hydrolase families 7 (15), 5 (16), and 12 (17), respectively. TrCel7B has a C-terminal carbohydrate binding module I and shares 40–45% overall homology to the exo-cellulase TrCel7A (18, 19). TrCel5A has the least homology to the other T. reesei cellulases and has an N-terminal carbohydrate binding module I (16). TrCel12A is smaller (25 kDa) and has no carbohydrate binding module (20), All three are retaining glycoside hydrolases (21, 22).
Methods to determine the activity of EGs have been based on the quantitative spectrophotometric measurement of reducing ends produced on soluble and insoluble substrates (23–29). These assays often do not correlate well with each other, are often subject to exoenzyme interferences (30), and are often influenced by different stoichiometric reactivities based on the cello-oligomer length (31, 32). Being spectrophotometric, they are also subject to optical limitations in that complex biomass samples cannot be assayed in this way, and being end-point assays, time resolution must be estimated by differentiating a series of stopped aliquots or separate parallel reactions. EG activity is also often determined through the use of viscosimetry, based on the depolymerization of carboxymethylcellulose (CMC) or hydroxyethylcellulose substrates (30, 33–35). CMC provides a cellulose that cannot be significantly depolymerized by processive (e.g. cellobiohydrolase) enzymes due to the steric hindrance of the carboxymethyl groups (36, 37), and EGs have even been referred to as CMCases (30, 38). However, it is a model substrate, so the activity of an enzyme on it does not reflect the activity on a more realistic insoluble substrate (39, 40). CMC, being an ionic substituted substrate, is also subject to changes in viscosity dependent on ionic strength, pH, and polyvalent cations (41).
To follow the initial kinetics of EG action on an unmodified cellulose substrate, we have used isothermal titration calorimetry (ITC). The heat-flow measured may be directly converted to a rate of hydrolysis (42–44). In the current study this new method is introduced and validated against both the widely used p-hydroxy benzoic acid hydrazide assay (24, 25) and bicinchoninic acid assay (BCA) (29) reducing sugar assays. Based on the time course of the calorimetric results and data from chromatography and continuous amperometric detection of glucose and cellobiose production, we analyze the molecular origins of the initial burst and rapid decline in the activity of endo-glucanases on insoluble cellulose.
MATERIALS AND METHODS
TrCel7B
TrCel7B was cloned and expressed in Aspergillus oryzae as described in WO 2005/067531 (87). The broth was filtered then buffer-exchanged using a 10-kDa polyethersulfone membrane with 20 mm Tris-HCl, pH 8.5. The sample was loaded onto a Q-Sepharose® high performance column (GE Healthcare), and bound proteins were eluted with a linear gradient from 0 to 600 mm NaCl. The fractions were desalted into 20 mm Tris, pH 8.0, 150 mm NaCl.
TrCel5A
TrCel5A was cloned and expressed in A. oryzae. Filtered broth was desalted and buffer-exchanged into 20 mm Tris-HCl, pH 8.0. Desalted TrCel5A was loaded onto a Mono Q™ HR 16/10 ion exchange column (GE Healthcare) and eluted with a 0–300 mm NaCl gradient in 20 mm Tris-HCl, pH 8.0.
TrCel12A
TrCel12A was cloned and expressed in A. oryzae. The purification procedure was adapted from Karlsson et al. (45). The protein was eluted from the phenyl-Sepharose FF column (GE Healthcare) with 20 mm sodium phosphate, pH 6.00, followed by 50:50% v/v 20 mm sodium phosphate, pH 6.0, 95% ethanol. Fractions were pooled based on the activity measured by AZCL-HE-Cellulose from Megazyme (Bray, Co., Wicklow, Ireland) and buffer-exchanged using a Sephadex G25 column (GE Healthcare) into 12 mm sodium acetate, pH 4.8.
Before ITC analysis, all proteins were buffer-changed to 50 mm sodium acetate, 2 mm calcium chloride, pH 5.0. The concentration of all enzymes was determined from A280 using the molar absorption coefficients of 72,770 m−1cm−1 (TrCel7B), 81,360 m−1cm−1 (TrCel5A), and 73,340 m−1cm−1 (TrCel12A). These are theoretical values calculated from the known amino acid sequences. The online program Protparam was used.
Megazyme EG Assay
The assay was carried out as per the manufacturer's instructions using 0.1% (w/v) azurine-cross-linked hydroxyethylcellulose substrate for 15 min at 50 °C, 1400 rpm in 1.5-ml Eppendorf tubes. The solutions were centrifuged at 13,500 rpm for 5 min, and A595 was read.
Substrates
Regenerated amorphous cellulose (RAC) was prepared from Sigmacell 20 as described previously (46). Total glucose was quantified (46–48) using a glucose (Sigma) standard curve at A490. The number-averaged degree of polymerization (DPN) was calculated from the ratio of reducing ends measured by BCA (see below) and total sugars. The crystallinity index of RAC was determined using solid state 13C cross-polarization/magic angle spinning nuclear magnetic resonance (NMR) with settings adapted from Matulova et al. (49) and described in Murphy et al. (46). Cello-oligosaccharides (COS) were prepared according to Zhang and Lynd (50), and purity was determined using high performance anion exchange chromatography with pulsed amperometric detection (46).
Reducing Ends Assays
The p-hydroxy benzoic acid hydrazide assay was adapted from Lever (24, 25), and samples were quantified using a glucose and cellotetraose standard curve at A410. The 2,2′-BCA assay was performed as outlined in Zhang and Lynd (48) at 80 °C in a thermomixer at 1400 rpm and a 30-min incubation time. Samples were quantified using a glucose and cellotetraose standard curve at A560.
Reducing Ends Assays on Hydrolysis Reactions
Reactions were performed in the ITC cell (with and without 400 rpm mixing), and after data collection, the samples were immediately quenched. This was achieved by taking a 1-ml sample and pipetting 3 × 50-μl aliquots to 450 μl of buffer with 500 μl BCA reagent, and the remaining 850 μl was quenched using 50 μl of 2 m Na2CO3. BCA and p-hydroxy benzoic acid hydrazide reducing sugar determination were carried out on the quenched aliquots. The effect of 2 m Na2CO3 on the substrate was checked by allowing contact over several hours before assaying compared with a sample of RAC with freshly added 2 m Na2CO3. Protein controls were run to correct for BCA-protein interference.
Reducing Ends Assays to Determine Soluble and Insoluble Fractions
Both enzymes and substrates were preincubated in a thermomixer at 30 °C and allowed to reach the set temperature before initiation of hydrolysis by adding the enzyme to the 8 g/liter RAC substrate. When initially added, the enzyme:substrate mixture was vortexed for 30 s and then incubated in the thermomixer with no further stirring for a period of 15 min. After this time reactions were quenched immediately as outlined above. The total reducing ends were immediately determined from the collected samples. Samples were then centrifuged, and the supernatant was filtered through a 0.22-μm filter to remove all insoluble matter before determination of soluble reducing ends products.
ITC Hydrolysis Monitoring
ITC was carried out using power-compensated equipment (Nano ITC2G from TA Instruments, New Castle, DE). The Nano ITC2G has a gold cell of volume 0.957 ml. It has previously been reported that the rapid decrease in polymerization as a result of EG activity has a profound effect on the base line in ITC experiments that may be corrected for manually (44). To overcome this limitation a new method has been developed. A stable base line with no stirring is established, and then the enzyme is injected under maximum stirring and mixed for a total of 60 s. The stirring is then turned off, and the hydrolysis is monitored continuously. All data have been corrected for heat of dilution and calibration factors (46, 51, 52). Details of the new calorimetric method are given in the supplemental material Fig. S1.
ITC Depletion Experiments
The ITC cell was filled with degassed 800 μm COS-3, -4, or -5 in 50 mm sodium acetate, 2 mm calcium chloride, pH 5.0. After thermal equilibration at 30 °C with 400 rpm stirring, a 100 nm concentration of enzyme was added. Data were collected every 2 s until the substrate was depleted. The depletion of a soluble substrate monitored by ITC has often been used to determine kinetic constants (43, 52–55), and the derivation of the equations used in this analysis (Fig. 5) is presented elsewhere (43).
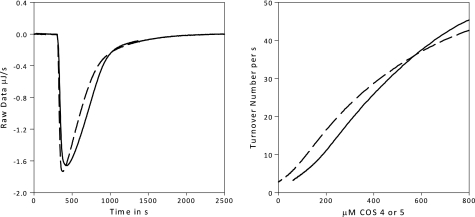
FIGURE 5.
ITC data (exothermic down). Left, 100 nm TrCel7B is titrated to 800 μm COS 5 (solid line) or COS 4 (dashed line) at 30 °C at 400 rpm. The return to the base line indicates the substrate has been completely hydrolyzed. Right, the raw data have been converted to rates and substrate concentrations using the ΔappH measured from Fig. 1. Note there can be no meaningful kinetic constants derived, as we appear to be under the Km value for both COS 5 and 4, given there is no Vmax reached. Previously reported values have varied from μm (45) to mm (16), so this is not unreasonable.
Biosensor Measurements of Cellobiose and Glucose
Amperometric biosensors for glucose (based on glucose oxidase) and cellobiose (based on cellobiose dehydrogenase) were made and used as described in detail elsewhere (56–59). Briefly, the sensors were based on a mediator-modified carbon paste electrode with surface-immobilized and cross-linked glucose oxidase or cellobiose dehydrogenase. In the kinetic measurements an aliquot of endo-glucanase was added to the 30 °C temperature-controlled, 5-ml reaction vessel stirred at 550 rpm after a stable base line with 8 g/liter RAC had been established. The separate currents from the two sensors generated by the enzymatic oxidation of the analytes were simultaneously recorded as a function of time. These currents were transformed into sugar concentrations using calibrations made immediately before and after the kinetic measurement. All data analysis has been performed using Origin Pro Version 8.0 and Sigmaplot Version 11.
RESULTS AND DISCUSSION
RAC
The RAC substrate was determined to have a number-averaged degree of polymerization (DPN) of 181 ± 9. There was no evidence of a C-4 peak on the cross-polarization/magic angle spinning NMR at 92 to 86 ppm (60), indicating a completely amorphous substrate (<0.05).
Reducing Ends Assays
The results from the BCA assay for measuring hydrolysis products are shown in Fig. 1, and Table 1 lists the soluble and insoluble reducing sugar equivalents determined. More information on calibration of the assays is outlined in the supplemental material Fig. S2.
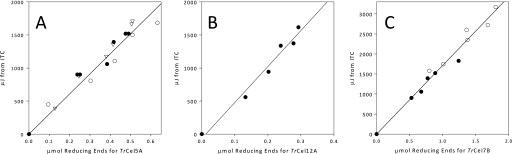
FIGURE 1.
Calibration data. A–C, shown are the calibration plots to determine ΔappH for each enzyme. Data on the ordinate are integrated values from the ITC runs shown in Fig. 2, and the data on the abscissa have been measured using the BCA assay on the same samples. The integration was carried out from time t = 0 to t = 970 s. All analyses are performed on 8 g/liter RAC, 30 °C, pH 5.0. Panel A shows varied doses (200–1000 nm) of TrCel5A added to RAC. Panel B shows varied doses of (100–500 nm) TrCel12A added to RAC. Panel C shows varied doses of TrCel7B (50–250 nm) added to 8g/liter RAC. In both A and C the different symbols represent runs on different batches of substrate. μmol values in A, B, and C were measured in triplicate, the coefficient of variation was under 5% in all cases (error bars are not shown). From the slopes: ΔappH of TrCel7B = −1.7 kJ/mol, TrCel5A = −3.0 kJ/mol, and TrCel12A = −5.4 kJ/mol.
TABLE 1.
Total soluble and insoluble reducing ends of TrEGs determined using BCA
The product profile was determined using high performance anion exchange chromatography with pulsed amperometric detection. Hydrolysis was carried out for 15 min in a thermomixer at 30 °C, pH 5.0, with no stirring, and stopped aliquots were measured for soluble (centrifuged and filtered) and total (before separation of solid and liquid phases) reducing sugar equivalents. ND, not detected.
| Enzyme | Soluble reducing ends/Total reducing ends | Soluble cello-oligosaccharide profile (fraction) |
|||||
|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | G5 | G6 | ||
| 100 nmTrCel7B | 0.97 ± 0.06 | 0.29 | 0.69 | 0.02 | ND | ND | ND |
| 200 nmTrCel5A | 0.71 ± 0.05 | 0.18 | 0.44 | 0.29 | 0.09 | ND | ND |
| 100 nmTrCel12A | 0.82 ± 0.06 | 0.09 | 0.45 | 0.20 | 0.25 | ND | ND |
Calibration
It may be seen from Fig. 1 that there is a direct proportionality between the area under the ITC signal and the moles of reducing ends produced for each enzyme. The total activity, measured as the sum of both soluble and insoluble reducing sugar equivalents, is thus measured continuously by the calorimeter. The slopes of the calibration plots, expressed in kJ/mol as the molar enthalpy of the reaction (ΔappH), are different for each enzyme, with −1.7 kJ/mol measured for TrCel7B, −3.0 kJ/mol for TrCel5A, and −5.4 kJ/mol TrCel12A. Clearly, the covalent change (hydrolysis of β-1,4 glycosidic bonds) is the same in all cases, and the difference in the apparent enthalpy must rely on non-covalent interactions. Such interaction energies may be sizable for the breakdown of cellulose. Thus, the lattice energy of crystalline cellulose is about 25 kJ/(mol glucosyl unit) (61), and the energy changes associated with both the creation of a cavity in water that can accommodate a small oligosaccharide and the hydration of the dissolved sugar is in the 10–20 kJ/mol range and dependent on the size of the oligosaccharide (62, 63). The three EGs tested here produce different profiles of products (Table 1 and Ref. 45), and this most likely accounts for the observed differences of a few kJ/mol in ΔappH. Comparisons of the apparent enthalpies in Fig. 1 and the product profile in Table 1 suggest that the enthalpy becomes less negative the smaller the average product size. This relationship may be used in future calorimetric product profiling, but it also calls for caution in prolonged calorimetric measurements with high degrees of conversion, where the product profile may shift as the process progresses.
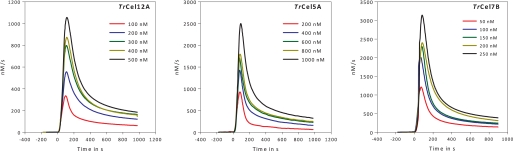
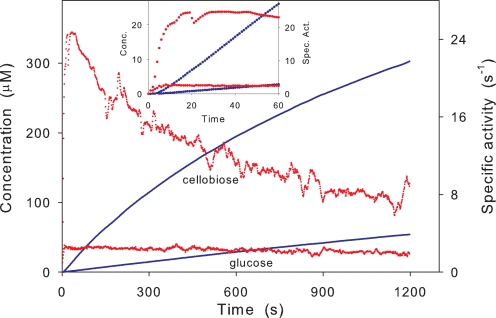
The calorimetric data (Fig. 2) immediately shows that the rate of hydrolysis changes conspicuously even at very low conversion. Thus, if the results for TrCel7B (Fig. 2C) are normalized with respect to the enzyme concentration, we find maximal a turnover of 15–20 s−1. This value falls 5–10-fold (depending on the enzyme concentration) over the following 5 min, although the total conversion at this point is <3–5% for the maximum enzyme dose. TrCel5A also showed an initial burst with a 5–10-fold reduction in activity, but the maximal turnover for this enzyme was significantly lower (maximal rate 2.5–4 s−1). For TrCel12A the burst was weaker, and the maximal reaction rate in the early part of the hydrolysis was only 3–4 times larger than the rate after 15 min. The enzyme also had the lowest absolute rate, with a maximal turnover of 2.2–2.8 s−1. Changes in reaction rate near the maximum are too rapid to be fully resolved by the ITC instrument. This smearing is due to the thermal inertia of the calorimetric vessel, and it is conventionally corrected on the basis of the instrument time constants (64). The slightly sharper changes produced by this dynamic correction showed a maximum for TrCel7B at 30–40 s and a peak value that was 15–20% higher than in the raw calorimetric data. This matches the results from measurements with biosensors (inset of Fig. 3), which has about an order of magnitude better time resolution than the ITC, and we conclude that a maximal turnover of 24 ± 3 s−1 occurs after about a half-minute for TrCel7B. We note that the delay of the ITC signal only affects the results in the initial phase (<∼60 s), when the hydrolytic rate changes rapidly. In Fig. 3 it is particularly interesting to note the different kinetics of glucose and oligosaccharides (which according to Table 1 is almost exclusively cellobiose). Hence, cellobiose shows an initial burst that parallels the total activity detected by calorimetry (Fig. 2), whereas the rate of glucose production raises quickly (within ∼8 s) and monotonically to a value that remains constant (to within the experimental resolution) for over 20 min. In other words, there is no sign of a burst in glucose production in Fig. 3, and this behavior was confirmed in other trials with variable enzyme loads up to 500 nm (data not shown). This implies that the ratio of production rates for cellobiose and glucose changes quite markedly over the first minutes; it is 10 after 30 s and less than 4 after 20 min (Fig. 3). This behavior could arise if the EG hydrolyzed cellobiose or if the enzyme preparation was contaminated with β-glucosidase (producing increasing amounts of glucose as the concentration of cellobiose rises). However, control experiments in which cellobiose was added to EG solutions (without substrate) showed constant concentrations over the relevant times (20 min), and we conclude that secondary hydrolysis of cellobiose is not the cause for the decoupling of glucose and cellobiose production.
FIGURE 2.
ITC data. Varied doses of TrCel12A, TrCel5A, and TrCel7B added to 8 g/liter RAC at 30 °C, pH 5. 0 are shown. The raw data (μJ/s) has been converted to nm/s using the ΔappH determined from Fig. 1, and the distinctive burst of all three EGs is evident. The enzyme was added under vigorous (400 rpm) stirring at time = 0. The stirring was stopped after the injection was complete at time = 60 s, and the hydrolysis was monitored for 15 min. For a more detailed account see the supplemental material Fig. S1.
FIGURE 3.
Hydrolysis of RAC (8 g/liter) by 20 nmTrCel7B as monitored by the biosensors. The blue curves show the directly measured concentrations (left ordinate) of cellobiose (upper curve) and glucose (lower curve). The red data points are the specific rates (right ordinate); i.e. the slope of the concentration traces normalized with respect to the enzyme concentration. The inset shows an enlargement of the first 60 s (symbols and units as in the main figure). The differentiated values were calculated by a function in OriginPro 8.0, which assigns the derivative at a given data point as the average of the slopes of lines connecting the point and its two closest neighbors.
We suggest that the characteristic initial kinetics (the burst phase) illustrated in Fig. 2 is the origin of pronounced variation in kinetic parameters (particularly kcat) found for EGs in the literature (cf. supplemental Table S1). Thus, the rate may decrease strongly as the hydrolysis progresses, and end-point measurements obtained at different stages of the hydrolysis cannot be meaningfully converted to turnover numbers or kcat values. This conclusion is substantiated by further analysis of the data in supplemental Table S1. If the the turnover number from different works using filter paper as the substrate are plotted as a function of the experimental time, the course is close to a single-exponential decay (supplemental Fig. S5, R2 = 0.93), and this correlation occurs despite differences in temperature and experimental set-up. We conclude that kinetic analyses of EGs must include rapid and frequent data sampling over the first few minutes and preferably be based on a continuous assay technology.
The kinetic behavior illustrated in Fig. 2 may be interpreted as an initial burst phase that subsequently falls off toward a nearly constant reaction rate, which reflects a (near) steady-state situation with (nearly) constant concentrations of free enzyme and enzyme intermediates. This behavior is in contrast to simple enzyme kinetics, where the rate of product formation increases monotonously in the presteady-state regime. Hence, the key question that arises from Fig. 2 is why this pronounced overshoot in reaction rate occurs. We suggest that the possible reasons for this can in principle be arranged into three groups. These are as follows. (i) The burst may be linked to the degree of conversion. If, for example, the EG has a high activity toward certain structures in the insoluble substrate and these good attack points are scarce, the rate of hydrolysis may decrease quickly as they are depleted as a result of topological changes in the substrate surface (surface erosion). Other conversion-dependent causes for a slowdown include inhibition by accumulated product (65–69). (ii) The burst may rely on the fact that both the insoluble substrate (in this case RAC) and some of the products (soluble COS of a certain degree of polymerization) are substrates to the EGs. This may generate a complex interplay between the release (or initial presence) of COS and its rapid breakdown in the aqueous phase, where kcat values may reach up to 50–100 s−1 (45). (iii) Finally, the burst could depend on inactivation of enzyme. Irreversible inactivation, dependent on e.g. conventional protein stability issues, would be expected to give a single exponential decay unlike the biphasic course in Fig. 2. A more likely interpretation is a reversible inactivation model where rapid hydrolysis prevails until the enzyme encounters some structure in the substrate (an “obstacle” or “trap”), which dictates a protracted (but finite) inactive period (13, 14). In this case the activity is high over some initial period, but it will decrease as more and more enzyme gets stalled by obstacles. Eventually, when the population of inactivated enzyme approaches a constant (steady-state) level, the reaction rate will reach a constant (low) value.
To assess the importance of (i), we first note that the highest concentration of soluble sugars in the current experiments is about 1–3 mm. This is much lower that reported product inhibition constants for the investigated enzymes (13, 70, 71), and it, therefore, seems unlikely that the slowdown depends on product inhibition. This reasoning is supported in both Figs. 3 and 4. The former shows that the burst (and slowdown) can be readily detected by the biosensors even at very low enzyme load (20 nm), where the concentration of products and the degree of conversion are extremely low at the end of the burst (a fraction of a mm and <0.5%, respectively). Fig. 4 shows that the addition of a second enzyme dose to a sample with about 0.5 mm product generates a second burst phase. The initial kinetic is re-established at ∼95% of the original activity, and there is 1% conversion at this time; that is to say the two enzyme doses are kinetically identical within experimental boundaries. More importantly, this figure also shows that the slowdown cannot depend on surface erosion or the depletion of good attack points on the substrate. If there was erosion (or other factors making the substrate less reactive), adding more enzyme could not reestablish an equivalent burst for the second enzyme dosage. Taken together, we did not find evidence for a link between the slowdown after about 1 min and the degree of conversion (that is indeed very low at this stage).
FIGURE 4.
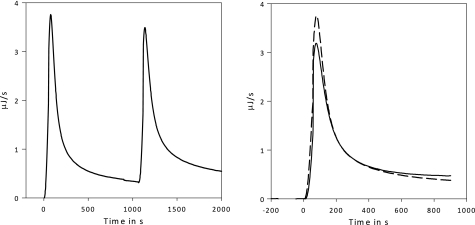
ITC data (exothermic up). Left, a double injection analysis is shown. 75 nm TrCel7B is injected to 8 g/liter RAC at 30 °C, pH 5.0, at t = 0 s and again at t = 1100 s. Right, shown is the extrapolation of the base lines and a plot of the curves on top of each other for comparison. Dashed line = injection at t = 0 s; solid line = injection at t = 1100 s. The relative areas have been calculated by integrating the peaks.
With respect to (ii), we first note that no background COS could be detected in the supernatant of centrifuged substrates, either by BCA analysis or calorimetric measurements. We, therefore, rule out soluble COS in the substrate samples. The current results also speak against an initial release of hydrolysable COS (e.g. COS 4–6) and their subsequent rapid hydrolysis as the origin of the burst. This is seen, for example, from Table 1, which shows that TrCel7B (that shows the strongest burst) generates almost exclusively (97%) soluble reducing ends. If the enzyme released COS as a “true endo-glucanase” with hydrolysis of random glycosidic bonds, there would be a more pronounced production of insoluble reducing ends. The depletion experiments shown in Fig. 5 show that COS breakdown, even at concentrations far in excess of those potentially found in the burst phase, is not fast enough to account for the peaks in Fig. 2. The COS hydrolyses also produce significant amounts of glucose (measured by high performance anion exchange chromatography and the biosensor). The latter would imply a coupling of the glucose production and the total enzyme activity. However, the biosensor data in Fig. 3 showed that the production rates of glucose and cellobiose are not coupled but in fact developed quite differently over the first 20 min. In conclusion, the current results do not suggest a link between the initial burst and rapid hydrolysis of soluble COS.
This leaves us with the reversible inactivation model sketched out under point (iii). To evaluate this, we first assess the magnitude of the burst using two independent approaches (see the supplemental material Fig. S4). This showed that the product concentration generated during the burst scaled proportionally to the enzyme concentration with increments of 1239 ± 200 (n = 18) for TrCel7B, 242 ± 38 (n = 20) for TrCel5A, and 280 ± 29 (n = 5) for TrCel12A. These figures specify the number of catalytic cycles conducted by each enzyme molecule during the burst. If instead we varied the substrate concentration from 4 to 6 and 8 g/liter and maintained the concentration of TrCel7B at 200 nm, no significant changes in the magnitude of the burst could be detected (see supplemental material Fig. S4C). We conclude that under the investigated conditions the extent of the burst is proportional to the enzyme concentration and independent of the substrate concentration. This behavior is in perfect accord with the reversible inactivation model. Hence, in this picture each enzyme is predicted to perform a certain number of rapid cycles determined by the frequency of traps and not the concentration of substrate. For TrCel5A and TrCel12A this frequency is about 1:250 hydrolysable bonds, and if the slowdown depends on reversible inactivation, changing the substrate concentration (as long as it is in excess) would not alter the average number of hydrolytic steps preceding the encounter with a trap. For TrCel7B, the extent of the burst is significantly larger. This suggests that this enzyme is either less susceptible to reversible inactivation or that it has a preferential affinity for substrate regions with fewer traps. The reversible inactivation model is also in line with the reestablished burst upon a second enzyme injection (Fig. 4) inasmuch as a new population of enzymes is predicted to run through the same initial phase as long as the substrate is in excess. We conclude that the observations concurrently support (iii).
A similar course of burst and inactivation has previously been suggested for processive exo-glucanases (11, 12, 14, 70, 72). TrCel6A, formerly cellobiohydrolase II (36), has two loops enclosing the active site, one of which can move, allowing for endo-attack initiation on insoluble substrates (73, 74). TrCel6A is classified as an endo-processive enzyme (12), being able to degrade CMC, hydroxyethylcellulose, and bacterial cellulose in an endo fashion (75, 76). Similar claims have been made for TrCel7A (formerly cellobiohydrolase I (77)), with a recorded 90% endo-attack initiation on amorphous substrates recently being recorded (14). There are four loops enclosing the catalytic domain of TrCel7A that are shortened on TrCel7B (14, 78). The processivity of the endo-glucanases investigated here remains to be fully elucidated, but under the conventional definition (3, 4) EGs hydrolyze randomly and are as such non-processive. However, strict boundaries between endo- and (processive) exo-cellulases are becoming blurred as their mode of action is better elucidated.
The processivity under the current experimental conditions may be approximated from the ratio of soluble and insoluble reducing ends (79–82). This analysis suggests (Table 1) that TrCel7B shows pronounced processivity on RAC (33 catalytic events), whereas the other enzymes appear to perform only a few (4–6 catalytic events) sequential steps on the same strand. A recent analysis of endo-processivity on TrCel5A and TrCel12A has reported apparent processivity values very close to this, 3–5 (14). Our analysis should be considered an indication only, as the product profiles change over time, and the concentration of soluble reducing ends may rely on both processive enzyme action and the hydrolysis of soluble COS produced non-processively. Nevertheless, these numbers demonstrate that all three EGs (TrCel7B in particular) show some degree of processivity and hence do not attack accessible glycosidic bonds randomly.
In conclusion we have found that the three most common endo-glucanases from T. reesei exhibit a pronounced non-monotonic initial activity on amorphous cellulose with a maximum after about 0.5–1 min. Subsequently, the enzymatic turnover decreases 2–10-fold over a few minutes as the reaction appears to enter a (near) steady-state condition. This maximum (or burst) must be taken into account in kinetic analysis of EG, and we suggest that the pronounced variation in the reported values of e.g. kcat based on end-point measurements could depend on the failure to do so. This aspect is particularly relevant when estimates of kinetic constants are used in functional models for the breakdown of cellulose (cf. Refs. 83–85), and the results in Fig. 2 and supplemental Table S1 show that parameters estimated at different time scales may vary by orders of magnitude. On the basis of complementary experimental data, we suggest that the burst reflects a balance between rapid hydrolysis with moderate processivity on one hand and the transient inactivation of enzyme on the other (Fig. 6). The mechanism of the temporary inactivation cannot be assessed, but we note that the interpretation is in line with the notion of non-productively bound enzyme, which has been repeatedly discussed for cellulases (11, 12, 14, 70, 72). The number of catalytic cycles in the burst phase was much higher (about 2 orders of magnitude) than processivity for all three enzymes. This implies that upon addition to the substrate, the EG will perform many (10s or 100s) endo-processive runs at a turnover rate that is over 20 s−1 for TrCel7B. It follows that unlike earlier interpretations for exo-cellulases (11, 13, 14, 66), the early slowdown for the enzymes studied here is not necessarily linked to processivity. As the process progresses, an increasing population of the enzyme gets randomly trapped in unproductive positions for extended periods, and the turnover rates consequently begins to fall. After several minutes the trapped population approaches a constant level, and the turnover rate levels off toward a constant value well below the maximum. At this stage the rate-limiting step is the release from the non-productively bound position. We stress that this interpretation only pertains to the initial reduction in hydrolytic rate, which is studied here. Longer trials with significant changes in the structure and amounts of substrate and concentration of soluble products are likely to be affected by a range of other inhibitory mechanisms. In a wider perspective, this interpretation is analogous to the original description of burst-phase kinetics over 50 years ago (86). This work pointed out that an ordered two-step (bi-bi) enzyme reaction could produce an initial burst if the first product is released from the enzyme complex faster than the second. Obviously the EG reactions studied here are more complex, but the basic course for the initial maximum in the turnover rate may result from an ordered sequence of a rapid (endo-processive hydrolysis) and a slower (release from non-productively bound positions) process.
FIGURE 6.
Schematic presentation of the proposed mechanism for the initial burst of T. reesei endo-glucanases. The enzymes make random attacks on the cellulose strands followed by short processive sequences (A) probably involving about five steps for TrCel5A and TrCel12A (14) and somewhat more for TrCel7B (Table 1). This leaves small gaps in the strand. Some structures on the cellulose surface are postulated to act as traps for the enzymes (B). This implies that enzymes interacting with these structures are released much more slowly than otherwise and, hence, that they experience prolonged inactive periods. The results suggest that traps for TrCel5A and TrCel12A occur at a frequency of about 1 for each 200–300 hydrolyzable bonds, and for TrCel7B this number was >1000. This implies that the enzymes conduct (on the average) 10s or 100s of short processive sequences with a specific rate up to 24 s−1 before encountering a trap, and this is suggested to be the origin of the observed initial burst. Subsequently, an increasing enzyme population is rendered inactive on the surface (B), and the reaction rate decreases toward a lower, pseudosteady-state level where the rate-limiting step is the release of trapped enzyme. We emphasize that the substrate was in large excess in the current experiments, and hence that temporary inactivation was a stochastic process governed by the bond:trap ratio and not the depletion of hydrolysable substrate (cf. Fig. 4).
Supplementary Material
Acknowledgment
We thank Erik Lumby Rasmusssen for excellent technical assistance.
This work was supported by Danish Agency for Science, Technology, and Innovation Grant 2104-07-0028 (to P. W.) and the Carlsberg Foundation.

This article contains supplemental Tables S1 and S2 and Figs. S1–S5.
- EG
- endo-glucanase
- CMC
- carboxymethyl cellulose
- ITC
- isothermal titration calorimetry
- BCA
- 2,2′-bicinchoninic acid
- RAC
- regenerated amorphous cellulose
- COS
- cello-oligosaccharide
- Tr
- T. reesei.
REFERENCES
- 1. Henrissat B. (1991) A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rabinovich M. L., Melnik M. S., Boloboba A. V. (2002) Appl. Biochem. Microbiol. 38, 305–321 [Google Scholar]
- 3. Lynd L. R., Weimer P. J., van Zyl W. H., Pretorius I. S. (2002) Microbial cellulose utilization: fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 66, 506–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang Y. H., Lynd L. R. (2004) Toward an aggregated understanding of enzymatic hydrolysis of cellulose: noncomplexed cellulase systems. Biotechnol. Bioeng. 88, 797–824 [DOI] [PubMed] [Google Scholar]
- 5. Barr B. K., Hsieh Y. L., Ganem B., Wilson D. B. (1996) Identification of two functionally different classes of exocellulases. Biochemistry 35, 586–592 [DOI] [PubMed] [Google Scholar]
- 6. Cao Y., Tan H. (2002) Effects of cellulase on the modification of cellulose. Carbohydr. Res. 337, 1291–1296 [DOI] [PubMed] [Google Scholar]
- 7. Väljamäe P., Sild V., Pettersson G., Johansson G. (1998) The initial kinetics of hydrolysis by cellobiohydrolases I and II is consistent with a cellulose surface-erosion model. Eur. J. Biochem. 253, 469–475 [DOI] [PubMed] [Google Scholar]
- 8. Lee Y. H., Fan L. T. (1983) Kinetic studies of enzymatic hydrolysis of insoluble cellulose: (II). Analysis of extended hydrolysis times. Biotechnol. Bioeng. 25, 939–966 [DOI] [PubMed] [Google Scholar]
- 9. Xiao Z.., Zhang X., Gregg D. J., Saddler J. N. (2004) Appl. Biochem. Biotechnol. 113–116, 1115–1126 [DOI] [PubMed] [Google Scholar]
- 10. Eriksson T., Borjesson J., Tjerneld F. (2002) Enzyme Microb. Technol. 31, 353–364 [Google Scholar]
- 11. Jalak J., Väljamäe P. (2010) Mechanism of initial rapid rate retardation in cellobiohydrolase-catalyzed cellulose hydrolysis. Biotechnol. Bioeng. 106, 871–883 [DOI] [PubMed] [Google Scholar]
- 12. Kipper K., Väljamäe P., Johansson G. (2005) Processive action of cellobiohydrolase Cel7A from Trichoderma reesei is revealed as “burst” kinetics on fluorescent polymeric model substrates. Biochem. J. 385, 527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Praestgaard E., Elmerdahl J., Murphy L., Nymand S., McFarland K. C., Borch K., Westh P. (2011) A kinetic model for the burst phase of processive cellulases. FEBS J. 278, 1547–1560 [DOI] [PubMed] [Google Scholar]
- 14. Kurasin M., Väljamäe P. (2011) Processivity of cellobiohydrolases is limited by the substrate. J. Biol. Chem. 286, 169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Penttilä M., Lehtovaara P., Nevalainen H., Bhikhabhai R., Knowles J. (1986) Homology between cellulase genes of Trichoderma reesei: complete nucleotide sequence of the endoglucanase I gene. Gene 45, 253–263 [DOI] [PubMed] [Google Scholar]
- 16. Saloheimo M., Lehtovaara P., Penttilä M., Teeri T. T., Ståhlberg J., Johansson G., Pettersson G., Claeyssens M., Tomme P., Knowles J. K. (1988) EGIII, a new endoglucanase from Trichoderma reesei: the characterization of both gene and enzyme. Gene 63, 11–22 [DOI] [PubMed] [Google Scholar]
- 17. Sandgren M., Shaw A., Ropp T. H., Wu S., Bott R., Cameron A. D., Ståhlberg J., Mitchinson C., Jones T. A. (2001) The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 A resolution. J. Mol. Biol. 308, 295–310 [DOI] [PubMed] [Google Scholar]
- 18. Divne C., Sinning I., Ståhlberg J., Pettersson G., Bailey M., Siika-aho M., Margolles-Clark E., Teeri T., Jones T. A. (1993) Crystallization and preliminary X-ray studies on the core proteins of cellobiohydrolase I and endoglucanase I from Trichoderma reesei. J. Mol. Biol. 234, 905–907 [DOI] [PubMed] [Google Scholar]
- 19. Ståhlberg J., Johansson G., Pettersson G. (1988) A binding-site-deficient, catalytically active, core protein of endoglucanase III from the culture filtrate of Trichoderma reesei. Eur. J. Biochem. 173, 179–183 [DOI] [PubMed] [Google Scholar]
- 20. Nakazawa H., Okada K., Onodera T., Ogasawara W., Okada H., Morikawa Y. (2009) Directed evolution of endoglucanase III (Cel12A) from Trichoderma reesei. Appl. Microbiol. Biotechnol. 83, 649–657 [DOI] [PubMed] [Google Scholar]
- 21. Okada H., Mori K., Tada K., Nogawa M., Morikawa Y. (2000) J. Mol. Catal. B Enzym. 10, 249–255 [Google Scholar]
- 22. Bayer E. A., Chanzy H., Lamed R., Shoham Y. (1998) Cellulose, cellulases, and cellulosomes. Curr. Opin. Struct. Biol. 8, 548–557 [DOI] [PubMed] [Google Scholar]
- 23. Miller G. L. (1959) Anal. Chem. 31, 426–428 [Google Scholar]
- 24. Lever M. (1972) A new reaction for colorimetric determination of carbohydrates. Anal. Biochem. 47, 273–279 [DOI] [PubMed] [Google Scholar]
- 25. Lever M. (1973) Colorimetric and fluorometric carbohydrate determination with p-hydroxybenzoic acid hydrazide. Biochem. Med. 7, 274–281 [DOI] [PubMed] [Google Scholar]
- 26. Nelson N. (1944) J. Biol. Chem. 153, 375–380 [Google Scholar]
- 27. Somgyi M. (1952) Notes on sugar determination. J. Biol. Chem. 195, 19–23 [PubMed] [Google Scholar]
- 28. Anthon G. E., Barrett D. M. (2002) Determination of reducing sugars with 3-methyl-2-benzothiazolinonehydrazone. Anal. Biochem. 305, 287–289 [DOI] [PubMed] [Google Scholar]
- 29. Doner L. W., Irwin P. L. (1992) Assay of reducing end groups in oligosaccharide homologues with 2,2′-bicinchoninate. Anal. Biochem. 202, 50–53 [DOI] [PubMed] [Google Scholar]
- 30. Sharrock K. R. (1988) Cellulase assay methods: a review. J. Biochem. Biophys. Methods 17, 81–105 [DOI] [PubMed] [Google Scholar]
- 31. Coward-Kelly G., Aiello-Mazzari C., Kim S., Granda C., Holtzapple M. (2003) Suggested improvements to the standard filter paper assay used to measure cellulase activity. Biotechnol. Bioeng. 82, 745–749 [DOI] [PubMed] [Google Scholar]
- 32. Sengupta S., Jana M. L., Sengupta D., Naskar A. K. (2000) A note on the estimation of microbial glycosidase activities by dinitrosalicylic acid reagent. Appl. Microbiol. Biotechnol. 53, 732–735 [DOI] [PubMed] [Google Scholar]
- 33. Miller G. L., Blum R., Glennon W. E., Burton A. L. (1960) Anal. Biochem. 1, 127–132 [Google Scholar]
- 34. Lee J. M., Heitmann J. A., Pawlak J. J. (2007) BioResources 2, 20–33 [Google Scholar]
- 35. Joos P., Sierens W., Ruyssen R. (1969) The determination of cellulase activity by viscometry. J. Pharm. Pharmacol. 21, 848–853 [DOI] [PubMed] [Google Scholar]
- 36. Rouvinen J., Bergfors T., Teeri T., Knowles J. K., Jones T. A. (1990) Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science 249, 380–386 [DOI] [PubMed] [Google Scholar]
- 37. Vlasenko E. Y., Ryan A. I., Shoemaker C. F., Shoemaker S. P. (1998) Enzyme Microb. Technol. 23, 350–359 [Google Scholar]
- 38. Mullings R. (1985) Enzyme. Microb. Technol. 7, 586–591 [Google Scholar]
- 39. Percival, Zhang Y. H., Himmel M. E., Mielenz J. R. (2006) Outlook for cellulase improvement: screening and selection strategies. Biotechnol. Adv. 24, 452–481 [DOI] [PubMed] [Google Scholar]
- 40. Eveleigh D. E., Mandels M., Andreotti R., Roche C. (2009) Prefermentation improves xylose utilization in simultaneous saccharification and co-fermentation of pretreated spruce. Biotechnol. Biofuels 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Child J. J., Eveleigh D. E., Sieben A. S. (1973) Determination of cellulase activity using hydroxyethylcellulose as substrate. Can. J. Biochem. 51, 39–43 [DOI] [PubMed] [Google Scholar]
- 42. Todd M. J., Gomez J. (2001) Enzyme kinetics determined using calorimetry: a general assay for enzyme activity? Anal. Biochem. 296, 179–187 [DOI] [PubMed] [Google Scholar]
- 43. Olsen S. N. (2006) Thermochim. Acta 448, 12–18 [Google Scholar]
- 44. Murphy L., Borch K., McFarland K. C., Bohlin C., Westh P. (2010) Enzyme Microb. Technol. 46, 141–146 [DOI] [PubMed] [Google Scholar]
- 45. Karlsson J., Siika-aho M., Tenkanen M., Tjerneld F. (2002) Enzymatic properties of the low molecular mass endoglucanases Cel12A (EG III) and Cel45A (EG V) of Trichoderma reesei. J. Biotechnol. 99, 63–78 [DOI] [PubMed] [Google Scholar]
- 46. Murphy L., Baumann M. J., Borch K., Sweeney M., Westh P. (2010) An enzymatic signal amplification system for calorimetric studies of cellobiohydrolases. Anal. Biochem. 404, 140–148 [DOI] [PubMed] [Google Scholar]
- 47. Dubois M., Gilles K. A., Hamilton J. K., Rebers P. A., Smith F. (1956) Anal. Chem. 28, 350–356 [Google Scholar]
- 48. Zhang Y. H., Lynd L. R. (2005) Determination of the number-average degree of polymerization of cellodextrins and cellulose with application to enzymatic hydrolysis. Biomacromolecules 6, 1510–1515 [DOI] [PubMed] [Google Scholar]
- 49. Matulova M., Nouaille R., Capek P., Péan M., Delort A. M., Forano E. (2008) NMR study of cellulose and wheat straw degradation by Ruminococcus albus 20. FEBS J. 275, 3503–3511 [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y. H., Lynd L. R. (2003) Cellodextrin preparation by mixed-acid hydrolysis and chromatographic separation. Anal. Biochem. 322, 225–232 [DOI] [PubMed] [Google Scholar]
- 51. Briggner L. E., Wadsö I. (1991) Test and calibration processes for microcalorimeters, with special reference to heat conduction instruments used with aqueous systems. J. Biochem. Biophys. Methods 22, 101–118 [DOI] [PubMed] [Google Scholar]
- 52. Jeoh T., Baker J. O., Ali M. K., Himmel M. E., Adney W. S. (2005) β-d-Glucosidase reaction kinetics from isothermal titration microcalorimetry. Anal. Biochem. 347, 244–253 [DOI] [PubMed] [Google Scholar]
- 53. Bohlin C., Olsen S. N., Morant M. D., Patkar S., Borch K., Westh P. (2010) A comparative study of activity and apparent inhibition of fungal β-glucosidases. Biotechnol. Bioeng. 107, 943–952 [DOI] [PubMed] [Google Scholar]
- 54. Karim N., Okada H., Kidokoro S. (2005) Thermochim. Acta 431, 9–20 [Google Scholar]
- 55. Karim N., Kidokoro S. (2004) Thermochim. Acta 412, 91–96 [Google Scholar]
- 56. Ludwig R., Harreither W., Tasca F., Gorton L. (2010) Cellobiose dehydrogenase: a versatile catalyst for electrochemical applications. Chemphyschem 11, 2674–2697 [DOI] [PubMed] [Google Scholar]
- 57. Hildén L., Eng L., Johansson G., Lindqvist S. E., Pettersson G. (2001) An amperometric cellobiose dehydrogenase-based biosensor can be used for measurement of cellulase activity. Anal. Biochem. 290, 245–250 [DOI] [PubMed] [Google Scholar]
- 58. Tatsumi H., Katano H., Ikeda T. (2006) Kinetic analysis of enzymatic hydrolysis of crystalline cellulose by cellobiohydrolase using an amperometric biosensor. Anal. Biochem. 357, 257–261 [DOI] [PubMed] [Google Scholar]
- 59. Tatsumi H., Katano H. (2005) Kinetics of the surface hydrolysis of raw starch by glucoamylase. J. Agric. Food Chem. 53, 8123–8127 [DOI] [PubMed] [Google Scholar]
- 60. Vanderhart D. L., Atalla R. H. (1984) Macromolecules 17, 1465–1472 [Google Scholar]
- 61. Calvet E., Hermans P. H. (1951) J. Polym. Sci. 6, 33–38 [Google Scholar]
- 62. Dale B. E., Tsao G. T. (1982) J. Appl. Polym. Sci. 27, 1233–1241 [Google Scholar]
- 63. Taylor J. B. (1957) Transactions of the Faraday Society 53, 1198–1203 [Google Scholar]
- 64. Loblich K. R. (1994) Thermochim. Acta 231, 7–20 [Google Scholar]
- 65. Igarashi K., Wada M., Hori R., Samejima M. (2006) Surface density of cellobiohydrolase on crystalline celluloses. A critical parameter to evaluate enzymatic kinetics at a solid-liquid interface. FEBS J. 273, 2869–2878 [DOI] [PubMed] [Google Scholar]
- 66. Eriksson T., Karlsson J., Tjerneld F. (2002) A model explaining declining rate in hydrolysis of lignocellulose substrates with cellobiohydrolase I (cel7A) and endoglucanase I (cel7B) of Trichoderma reesei. Appl. Biochem. Biotechnol. 101, 41–60 [DOI] [PubMed] [Google Scholar]
- 67. Nidetzky B., Steiner W. (1993) A new approach for modeling cellulase-cellulose adsorption and the kinetics of the enzymatic hydrolysis of microcrystalline cellulose. Biotechnol. Bioeng. 42, 469–479 [DOI] [PubMed] [Google Scholar]
- 68. Zhang S., Wolfgang D. E., Wilson D. B. (1999) Substrate heterogeneity causes the nonlinear kinetics of insoluble cellulose hydrolysis. Biotechnol. Bioeng. 66, 35–41 [DOI] [PubMed] [Google Scholar]
- 69. Yang B., Willies D. M., Wyman C. E. (2006) Changes in the enzymatic hydrolysis rate of Avicel cellulose with conversion. Biotechnol. Bioeng. 94, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 70. Gruno M., Väljamäe P., Pettersson G., Johansson G. (2004) Inhibition of the Trichoderma reesei cellulases by cellobiose is strongly dependent on the nature of the substrate. Biotechnol. Bioeng. 86, 503–511 [DOI] [PubMed] [Google Scholar]
- 71. Holtzapple M. T., Caram H. S., Humphrey A. E. (1984) Determining the inhibition constants in the HCH-1 model of cellulose hydrolysis. Biotechnol. Bioeng. 26, 753–757 [DOI] [PubMed] [Google Scholar]
- 72. Väljamäe P., Pettersson G., Johansson G. (2001) Mechanism of substrate inhibition in cellulose synergistic degradation. Eur. J. Biochem. 268, 4520–4526 [DOI] [PubMed] [Google Scholar]
- 73. Breyer W. A., Matthews B. W. (2001) A structural basis for processivity. Protein Sci. 10, 1699–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zou J., Kleywegt G. J., Ståhlberg J., Driguez H., Nerinckx W., Claeyssens M., Koivula A., Teeri T. T., Jones T. A. (1999) Crystallographic evidence for substrate ring distortion and protein conformational changes during catalysis in cellobiohydrolase Ce16A from Trichoderma reesei. Structure Fold. Des. 7, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 75. Kleman-Leyer K. M., Siika-Aho M., Teeri T. T., Kirk T. K. (1996) The cellulases endoglucanase I and cellobiohydrolase II of Trichoderma reesei act synergistically to solubilize native cotton cellulose but not to decrease its molecular size. Appl. Environ. Microbiol. 62, 2883–2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bailey M. J., Siika-aho M., Valkeajärvi A., Penttilä M. E. (1993) Hydrolytic properties of two cellulases of Trichoderma reesei expressed in yeast. Biotechnol. Appl. Biochem. 17, 65–76 [PubMed] [Google Scholar]
- 77. Shoemaker S., Watt K., Tsitovsky G., Cox R. (1983) Biotechnology 1, 687–690 [Google Scholar]
- 78. MacKenzie L. F., Sulzenbacher G., Divne C., Jones T. A., Wöldike H. F., Schülein M., Withers S. G., Davies G. J. (1998) Crystal structure of the family 7 endoglucanase I (Cel7B) from Humicola insolens at 2.2 A resolution and identification of the catalytic nucleophile by trapping of the covalent glycosyl-enzyme intermediate. Biochem. J. 335, 409–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang S., Irwin D. C., Wilson D. B. (2000) Site-directed mutation of noncatalytic residues of Thermobifida fusca exocellulase Cel6B. Eur. J. Biochem. 267, 3101–3115 [DOI] [PubMed] [Google Scholar]
- 80. Irwin D. C., Spezio M., Walker L. P., Wilson D. B. (1993) Activity studies of eight purified cellulases: Specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 42, 1002–1013 [DOI] [PubMed] [Google Scholar]
- 81. Zhou W., Irwin D. C., Escovar-Kousen J., Wilson D. B. (2004) Kinetic studies of Thermobifida fusca Cel9A active site mutant enzymes. Biochemistry 43, 9655–9663 [DOI] [PubMed] [Google Scholar]
- 82. Watson B. J., Zhang H., Longmire A. G., Moon Y. H., Hutcheson S. W. (2009) Processive endoglucanases mediate degradation of cellulose by Saccharophagus degradans. J. Bacteriol. 191, 5697–5705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Levine S. E., Fox J. M., Blanch H. W., Clark D. S. (2010) A mechanistic model of the enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 107, 37–51 [DOI] [PubMed] [Google Scholar]
- 84. Peri S., Karra S., Lee Y. Y., Karim M. N. (2007) Modeling intrinsic kinetics of enzymatic cellulose hydrolysis. Biotechnol. Prog. 23, 626–637 [DOI] [PubMed] [Google Scholar]
- 85. Zhang Y. H., Lynd L. R. (2006) A functionally based model for hydrolysis of cellulose by fungal cellulase. Biotechnol. Bioeng. 94, 888–898 [DOI] [PubMed] [Google Scholar]
- 86. Hartley B. S., Kilby B. A. (1954) The reaction of p-nitrophenyl esters with chymotrypsin and insulin. Biochem. J. 56, 288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vlasenko E., Cherry J., Xu F. (July 28, 2005) PCT International Patent Appl. WO/2005/067531
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.