Background: High level of NO and TNF-α can induce diverse effects on host survival.
Results: Lentinan inhibits NO and TNF-α secretion and phosphorylation of MAP kinases JNK1/2 and ERK1/2.
Conclusion: Inhibition of NO and TNF-α is partially through suppression of JNK1/2 and ERK1/2 activation.
Significance: A novel pharmacological molecule is discovered to control the diseases associated with NO and TNF-α overproduction.
Keywords: ERK, Jun N-terminal kinase (JNK), NF-κB, Nitric Oxide, Tumor Necrosis Factor (TNF), β-Glucan
Abstract
Lentinan (LNT), a β-glucan from the fruiting bodies of Lentinus edodes, is well known to have immunomodulatory activity. NO and TNF-α are associated with many inflammatory diseases. In this study, we investigated the effects of LNT extracted by sonication (LNT-S) on the NO and TNF-α production in LPS-stimulated murine RAW 264.7 macrophages. The results suggested that treatment with LNT-S not only resulted in the striking inhibition of TNF-α and NO production in LPS-activated macrophage RAW 264.7 cells, but also the protein expression of inducible NOS (iNOS) and the gene expression of iNOS mRNA and TNF-α mRNA. It is surprising that LNT-S enhanced LPS-induced NF-κB p65 nuclear translocation and NF-κB luciferase activity, but severely inhibited the phosphorylation of JNK1/2 and ERK1/2. The neutralizing antibodies of anti-Dectin-1 and anti-TLR2 hardly affected the inhibition of NO production. All of these results suggested that the suppression of LPS-induced NO and TNF-α production was at least partially attributable to the inhibition of JNK1/2 and ERK1/2 activation. This work discovered a promising molecule to control the diseases associated with overproduction of NO and TNF-α.
Introduction
Nitric oxide (NO) and tumor necrosis factor-α (TNF-α) are two key molecules in the immunopharmacology which have beneficial biological effects on a variety of normal cells mostly related to immunomodulatory or inflammatory or physiological processes (1–4). Both of them are also two important cytotoxic mediators contributing to the antimicrobial and tumoricidal activity of the macrophages (5–7) and exert a key role in the pathogenesis of many infectious and inflammatory diseases (3, 8, 9). However, high levels of NO and TNF-α generated by activated macrophages can induce diverse effects on host survival, ranging from direct cellular cytotoxicity (2, 10) to damage of components leading to mutagenesis (6, 11, 12). It has been reported that NO inhibition could markedly increase the survival rate in the cecal ligation and puncture model (13). Therefore, identification of novel pharmacological reagents which can suppress NO and TNF-α overproduction is of considerable medical interest.
LPS is known to activate multiple signaling pathways in macrophages, leading to production of proinflammatory mediators and cytokines such as NO, TNF-α, and interleukins. Some researchers have reported that certain compounds such as quercetin and taurine chloramine can effectively inhibit NO and TNF-α production in LPS-stimulated macrophages. As we know, β-glucans possess anti-infective and antitumorigenic activity (14–16) by activating leukocytes and then producing reactive oxygen intermediates, inflammatory mediators, and cytokines such as NO and TNF-α (14, 17). It is well known that the particulate zymosan containing β-glucan as the main component from Saccharomyces cerevisiae is especially used widely to examine the proinflammatory responses of immune cells such as phagocytes and macrophages (18–20). Zymosan has been confirmed to be able to activate NF-κB and induce production of TNF-α in RAW 264.7 macrophages (21). Currently, another two important β-glucans, Schizophyllan (SPG)3 and Lentinan (LNT), having the main chain of (1,3)-β-glucan with one β-(1,6)-glucose branch at every five main-chain glucose residues are clinically used as antitumor agents (22, 23). Yadomae and co-workers have demonstrated that SPG could not enhance NO or TNF-α secretion, but the alkaline-treated SPG (SPG-OH) could induce NO and TNF-α synthesis in vitro and in vivo (24, 25). Moreover, SPG-OH augmented NO production in the presence of cytokines such as TNF-α, IL-1α, IL-6, GM-CSF, especially in the presence of interferon-γ (IFN-γ) in vitro. As for LNT, it has been shown that it can stimulate NK cell activity (26–28) and several macrophage/monocyte functions (secreting IL-1 and superoxide anion), phagocytosis, and cytotoxicity (29–34). The cytotoxic activity and TNF secretion of macrophages were found to be elevated by LNT in vitro and in vivo (35, 36). Moreover, pretreatment of bone marrow macrophages with LNT resulted in increased production of NO in vitro (36). However, Noel et al. demonstrated that pretreatment of LNT before LPS administration induced a striking inhibition of up to 89% circulating TNF-α in bacillus Calmette-Guérin-primed mice (37). In a word, LNT exhibits various immunomodulatory behaviors under different conditions. To our knowledge, there are few publications involved in the immunomodulating effects of combined application of LPS with LNT. Therefore, in this study, we investigated the effects of LNT on LPS-induced NO and TNF-α secretion from murine macrophage RAW 264.7 cells and on the signal transduction induced by LPS.
EXPERIMENTAL PROCEDURES
Sample
LNT used in this study was extracted from the fruiting bodies of Lentinus edodes according to the previously described procedures (38), which was coded as LNT-S. LNT-S was dissolved in PBS and sterilized at 121 °C for 20 min before use.
Reagents and Antibodies
DMEM (glutamine, high glucose), penicillin, streptomycin, and LPS from Escherichia coli 0111:B4 were purchased from Sigma. Antibodies were obtained from the following sources: mouse monoclonal anti-inducible NOS (iNOS), BD Transduction Laboratories; rabbit polyclonal anti-NF-κB p65, rabbit polyclonal anti-phospho-ERK1/2, rabbit polyclonal anti-phospho-JNK1/2, and rabbit polyclonal anti-phospho-p38, Cell Signaling Technology (Beverly, MA); mouse monoclonal anti-β-actin, Santa Cruz Biotechnology (Santa Cruz, CA); monoclonal antibody against TLR2/CD282, Imgenex (San Diego, CA); and mouse monoclonal antibody against Dectin-1/CLEC7A (R&D Systems), Wako Pure Chemical Industries (Osaka, Japan).
Cell Culture
RAW 264.7 cells (a murine macrophage/monocyte-like cell line, American Type Culture Collection) were maintained in DMEM (glutamine, high glucose) supplemented with penicillin (100 units/ml), streptomycin (100 μg/ml), and 10% heat-inactivated FBS (Sekisui Medical Co., Ltd., Tokyo, Japan) at 56 °C for 30 min. Subculturing was done by dislodging the cells with trypsin (0.25%) and EDTA·2Na·2H2O (0.02%), followed by centrifugation and seeding into the culture flask or dish, which was incubated at 37 °C under a humidified atmosphere of 95% air and 5% CO2.
Nitrite Determination
NO production was determined based on the amount of nitrite present in the conditioned medium, a stable end product of NO, by Griess reaction. Briefly, RAW 264.7 cells were seeded (2 × 105 cells/well) in 48-well plates and incubated for 24 h. The cells were rinsed with PBS and exposed to LPS (100 ng/ml) and LNT-S in DMEM without FBS. After a 48-h stimulation, each supernatant (100 μl) was mixed with an equal volume of Griess reagent (50 μl of 1% sulfanilamide in 5% phosphoric acid and 50 μl of 0.1% N-(1-naphthyl)ethylenediamine dihydrochloride in distilled water) at room temperature. The absorbance was measured at 570 nm using a Wallac microplate reader (1420 ARVO Sx), and the concentration of NO was quantified with a standard curve generated with sodium nitrite in the range of 0–100 μm.
TNF-α Assays by ELISA
RAW 264.7 cells (5 × 105 cells/well) were seeded into 24-well plates and incubated for 24 h before stimulation. At the end of preincubation, cells were rinsed with PBS and, the medium was exchanged to DMEM without FBS. The cells were exposed to LPS (100 ng/ml) with and without LNT-S (200 μg/ml). After a 24-h stimulation, the conditioned medium was collected and centrifuged, and TNF-α levels in the supernatant were assessed using an ELISA kit (Thermo Scientific) according to the manufacturer's instructions.
Western Blot Analysis
The cytoplasmic and nuclear proteins were extracted according to procedures described previously (21) with a slight modification. Briefly, RAW 264.7 cells were seeded in a 60-mm culture dish at a density of 5 × 106 cells/dish in DMEM with 10% FBS for 24 h. The cells were then rinsed with sterilized PBS and exposed to LPS (100 ng/ml) and LNT-S (0–200 μg/ml) in DMEM without FBS. At the end of treatment, the cells were harvested by scraping and treated with 200 μl of lysis buffer (50 mm KCl, 0.5% Nonidet P-40, 25 mm HEPES (pH 7.8), 1 mm PMSF, 10 μg/ml leupeptin, 20 μg/ml aprotinin, and 100 mm dithiothreitol) on ice for 10 min. After a 5-min centrifugation at 14,000 rpm, the supernatant was saved as a cytoplasmic extract. The left nuclear pellet was washed once with the same volume of buffer without Nonidet P-40. The nuclear pellet was then treated with 40 μl of extraction buffer (500 mm KCl, 10% glycerol with the same concentrations of HEPES, PMSF, leupeptin, aprotinin, and dithiothreitol as the lysis buffer) on ice for 40 min with pipetting every 10 min. After centrifugation at 14,000 rpm for 10 min, the supernatant was harvested as the nuclear protein extract. Both the cytoplastic and nuclear protein extracts were preserved at −70 °C for Western blot assay. The protein concentration was determined using a Bradford protein assay reagent (Bio-Rad) and BSA (Nacalai tesque, Kyoto, Japan) as the standard.
Both cytoplasmic and nuclear lysates were mixed with 4×SDS sample buffer and denatured in boiling water for 5 min. Aliquots of 10–15 μg of denatured cytoplasmic proteins and 15–25 μg of denatured nuclear proteins were respectively separated by SDS-PAGE on a 10% polyacrylamide gel and then electrically transferred to a PVDF membrane (BioTrace; Pall Corp., Port Washington, NY). After blocking with 5% (w/v) BSA in TBS (10 mm Tris-HCl (pH 8.0), 150 mm NaCl) containing 0.1% Tween 20 at room temperature for 1 h, the membranes were then incubated with appropriate specific primary antibody (anti-iNOS, 1:8,000; anti-ERK1/2, 1:1,000; anti-JNK1/2, 1:1,000; anti-p38, 1:1,000; anti-NF-κB p65, 1:1,000; anti-β-actin, 1:20,000) overnight at 4 °C. The reactive bands were visualized with an HRP-conjugated secondary antibody (1:20,000; Santa Cruz Biotechnology) via ECL Western blot detection reagents on a LightCapture II (ATTO, Tokyo) according to the manufacturer's instructions.
RNA Isolation and Quantitative Real Time RT-PCR Assay
RAW 264.7 cells (5 × 105 cells/well) were seeded into 24-well plates and incubated for 24 h before stimulation. At the end of preincubation, the cells were rinsed with PBS, and the medium was exchanged to DMEM without FBS. The cells were then exposed to LPS (100 ng/ml) and LNT-S (200 μg/ml) for the desired time. Total cellular RNA was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. RNA concentrations and purity were determined by spectrophotometer (GeneQuant Pro RNA/DNA calculator; Amersham Biosciences) measuring absorbance at 260 and 280 nm. RNA was temporarily stored at −20 °C in nuclease-free water. The reverse transcription (RT) was performed with 20 ng of total RNA to synthesize cDNA using PrimeScript PLUS RTase obtained from Takara (Takara, Shiga, Japan) according to the manufacturer's protocol. RT-generated cDNA with Takara Ex TaqHS was amplified using Expand High Fidelity PCR system from Roche Dignostics according to manufacturer's instructions. Thermal cycling was performed on Thermal Cycler Dice Real Time System TP860 (Takara, Kyoto, Japan) using One Step SYBR PrimerScript Plus RT-PCR Kit (Takara) according to the manufacturer's protocol. The primers used were purchased from Invitrogen, and PCR conditions were as follows: hypoxanthine guanine phosphoribosyl transferase (hprt), as a control (5′-gTAATgATCATTCAACgggggAC-3′ and 5′-CCAgCAAgCTTgCAACCTTAACCA-3′), at 94 °C for 30 s, 55 °C for 25 s, and 72 °C for 45 s; iNOS (5′-CTgCAgCACTTggATCAggAACCTg-3′ and 5′-gggAgTAgCCTgTgTgCACCTggAA-3′), 27 cycles at 95 °C for 30 s, 55 °C for 25 s, and 72 °C for 45 s; TNF-α (forward, GACAGTGACCTGGACTGTGG and reverse, TGAGACAGAGGCAACCTGAC), at 95 °C for 5 s, 60 °C for 30 s, and 72 °C for 30 s. The resultant iNOS mRNA and TNF-α mRNA were normalized by hprt mRNA and expressed relative to LPS.
NF-κB-Luciferase Assay
RAW 264.7 cells in 24-well plates (3 × 105 cells/well) were preincubated for 12 h and then transfected with 0.8 μl of NF-κB-driven luciferase reporter vectors using 1.56 μl of Lipofectamine (Invitrogen) in 26 μl of OPTI-MEM (Invitrogen) per well. After a 6-h transfection, the cells were rinsed with PBS and incubated for additional 6 h in DMEM with 10% FBS. The cells were then stimulated with LPS (100 ng/ml) and LNT-S (200 μg/ml) in DMEM without FBS. Following 12-h stimulations, cells were lysed with Passive Lysis buffer, and firefly and Renilla luciferase activities were assessed using a Dual-Luciferase Reporter assay (Promega, Madison, WI) and a Wallac microplate luminometer (1420 ARVO Sx, PerkinElmer Life Sciences), according to the manufacturer's instructions. NF-κB activity was expressed as -fold induction relative to the control cells without LPS treatment.
SPR Analysis
Surface Plasma Resonance (SPR) analysis was performed using Biacore 3000 (GE Healthcare) according to the manufacturer's instruction. Before the analysis, anti-Dectin-1 antibody was immobilized on sensor chip CM5 using the amine-coupling method. The resultant sensor chip was equilibrated with running buffer (HEPES-EP: 10 mm HEPES (pH 7.4), containing 150 mm NaCl, 3 mm EDTA and 0.005% Surfactant P20) at a flow rate of 20 μl/min at 4 °C. LNT-S dissolved in running buffer at a concentration of 100 μg/ml was loaded onto sensor chip, and the signal data were collected with the Biacore program (GE Healthcare). After each analysis, the sensor chip was regenerated by 50 mm NaOH. Affinity constants were calculated using BIA evaluation 4.1 software by globally fitting the association and dissociation phases of the overlay plots to a 1:1 Langmuir binding model.
Anti-Dectin-1 and Anti-TLR2 Antibody Neutralization Assay
RAW 264.7 cells were seeded at a density of 5 × 105 cells/ml in 24-well plate. After a 24-h preincubation, the cells were incubated with 5–20 μg/ml monoclonal antibodies against murine Dectin-1/CLEC7A and TLR2/CD282 for 30 min at 37 °C. LPS (100 ng/ml) and LNT-S (100 μg/ml) were then added into the medium free of FBS and incubated for another 24 h. The supernatants were finally collected, and the concentration of NO was measured by the method described above.
Statistical Analysis
Data are given as the mean ± S.E. from at least three independent experiments, unless specified otherwise. Student's t test was performed, and the differences were considered statistically significant at p < 0.05.
RESULTS
LNT-S Suppresses NO and TNF-α Production in LPS-Stimulated RAW 264.7
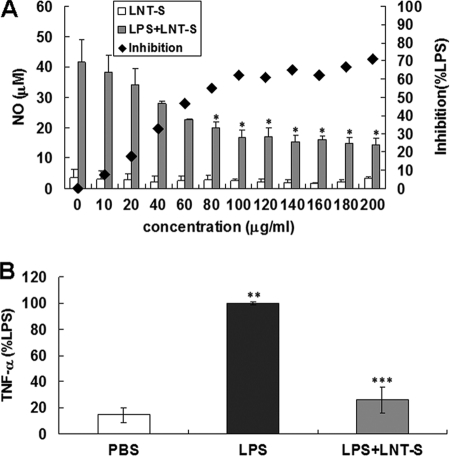
As described in the Introduction, NO and TNF-α are two important inflammatory mediators produced from macrophages corresponding to inflammation. Therefore, NO and TNF-α were used here as an indicator of macrophage response to LPS and LNT-S. As shown in Fig. 1A, LPS-induced NO production was suppressed by LNT-S in a dose-dependent manner, and ∼70% inhibition was observed at LNT-S concentration of 200 μg/ml. A significant decrease (∼60%) in NO production was observed at 80 μg/ml and then changed slightly with increasing concentration. Similarly, compared with control, LPS induced substantial TNF-α production in RAW 264.7 cells, and LNT-S suppressed it with inhibition of ∼75% at a concentration of 200 μg/ml and a 20-h stimulation (Fig. 1B).
FIGURE 1.
Inhibition of LPS-induced NO and TNF-α production in RAW 264.7 macrophages. A, RAW 264.7 cells (2 ×105 cells/well) were seeded into 48-well plates and incubated for 24 h, and the cells were then exposed to LNT-S with different concentrations in the absence and presence of LPS (100 ng/ml). After a 48-h stimulation, NO production in the supernatant was measured by Griess reaction as described under “Experimental Procedures.” B, RAW 264.7 cells (5 ×105 cells/well) were seeded into 24-well plates and incubated for 24 h, and the cells were then exposed to LPS (100 ng/ml) in the absence and presence of LNT-S (200 g/ml). After a 20-h stimulation, TNF-α production in the supernatant was measured by ELISA kit as described under “Experimental Procedures.” Each value represents the mean ± S.E. of three independent experiments. *, p < 0.05 versus LPS; **, p < 0.001 versus PBS; ***, p < 0.005 versus LPS.
LNT-S Inhibits LPS-induced iNOS Protein Expression
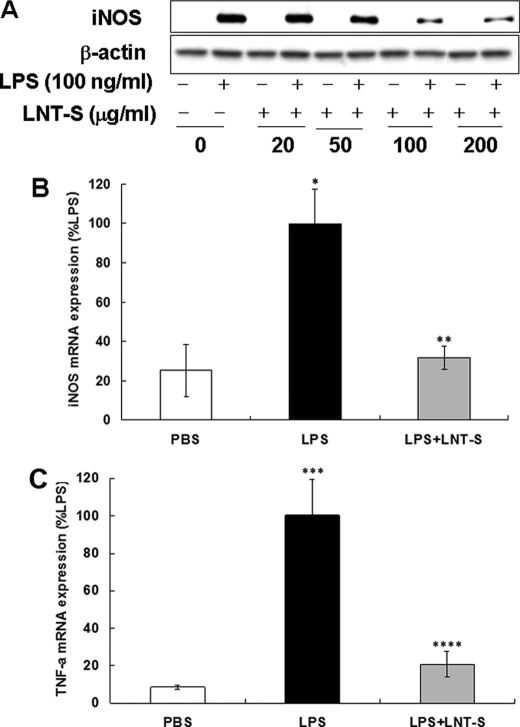
NO is synthesized by a family of enzymes known as NOS. To date, three main isoforms of NOS have been identified as brain constitutive NOS (nNOS), endothelial constitutive NOS (eNOS), and inducible NOS (iNOS) (39, 40). In contrast, the iNOS protein is not expressed constitutively (41, 42), but it can be up-regulated by proinflammatory stimuli (43, 44) and is the most important proinflammatory enzyme responsible for increasing the levels of NO (45). Nathan's laboratory has purified and cloned iNOS that makes NO in activated macrophage (46). Therefore, in this study, we checked the iNOS expression in LPS-stimulated RAW 264.7 cells. The results indicated that iNOS protein in LPS-stimulated RAW 264.7 cells exhibited a strong time dependence (data not shown). At the 24-h time point, a strong iNOS protein band induced by LPS appeared, and significant inhibition by LNT-S was observed. In accord with the results of NO assay (Fig. 1A), the substantial iNOS protein expression showed a strong concentration dependence (Fig. 2A). LNT-S itself was not effective in inducing notable iNOS production. This result indicates that the inhibitory activity of LNT-S toward NO production is through the inhibition of iNOS enzyme expression.
FIGURE 2.
LNT-S inhibited LPS-induced iNOS protein, iNOS mRNA, and TNF-α mRNA expression. A, RAW 264.7 cells (5 × 106 cells) were seeded in a φ60 cell culture dish and incubated for 24 h, and the cells were then exposed to LNT-S with different concentration in the absence and presence of LPS (100 ng/ml). After a 24-h stimulation, the cytoplasmic proteins were extracted and measured by Western blotting using the specific antibodies against iNOS and β-actin as described under “Experimental Procedures.” Data are representative of three independent experiments. B and C, RAW 264.7 cells (5 ×105 cells/well) were seeded into 24-well plates and incubated for 24 h, and the cells were then exposed to LPS (100 ng/ml) in the absence and presence of LNT-S (200 μg/ml). After a 20-h stimulation, total RNA was isolated from cells and measured as described under “Experimental Procedures.” The iNOS mRNA and TNF-α mRNA are expressed relative to LPS. Each value represents the mean ± S.E. of at least three independent experiments. *, p < 0.05 versus PBS; **, p < 0.05 versus LPS; ***, p < 0.001 versus PBS; ****, p < 0.001 versus LPS.
LNT-S Suppressed LPS-induced iNOS mRNA and TNF-α mRNA Expression
As we know, iNOS and TNF-α are synthesized by translation of the corresponding mRNA in the cytoplasm. To clarify how LNT-S attenuates NO and TNF-α production, iNOS mRNA and TNF-α mRNA expression was examined by RT-PCR analysis. The time dependence of iNOS mRNA and TNF-α mRNA showed that after co-stimulation for 20 h, LNT-S exhibited the maximum inhibition (data not shown here). As shown in Fig. 2, B and C, a significant inhibition of iNOS mRNA (∼70%) and TNF-α mRNA (∼80%) by LNT-S was observed, suggesting that inhibition of NO and TNF-α is through suppression of their respective mRNA expression.
LNT-S Inhibits LPS-induced Phosphorylation of ERK1/2 and JNK1/2
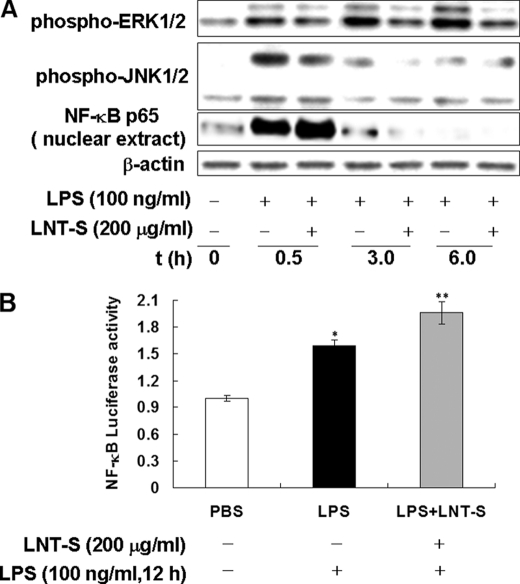
It is now generally accepted that LPS from Gram-negative bacterial stimulates inflammatory responses through TLR4-initiated several distinct signaling pathways including MAPKs and NF-κB (47–49). Specially, ERK and JNK are thought to regulate production of inflammatory cytokines and mediators such as TNF-α and NO (50–53). To determine whether LNT-S interfered with MAPK signaling pathways, we analyzed the cytoplasmic cell lysates by Western blotting with the specific phospho-antibodies against ERK1/2, JNK1/2, and p38 MAP kinases. Fig. 3A demonstrates that LPS stimulation induced rapid phosphorylation of JNK1/2. The maximal JNK1/2 phosphorylation induced by LPS occurred at a time point of 0.5 h and then attenuated with increasing incubation time, similar to the result reported by Okugawa et al. previously (54). However, the marked suppression of phospho-JNK1/2 expression by LNT-S at 0.5 h and 3.0 h after LPS stimulation was clearly observed. Similarly, rapid phosphorylation of ERK1/2 at 0.5 h after LPS stimulation was also observed and increased with extending stimulation time, showing a different time-dependent manner from JNK1/2. Co-treatment of LPS with LNT-S also resulted in strong inhibition of ERK1/2 activation (Fig. 3A). However, LPS stimulation resulted in no detectable phosphorylation of p38 MAPK in this study (data not shown). These results indicated that LNT-S interfered with MAPK signaling pathways in response to LPS, resulting in inhibition of NO and TNF-α.
FIGURE 3.
LNT-S interfered with MAPK signaling pathways and enhanced NF-κB p65 nuclear translocation and NF-κB transcription activity. A, RAW 264.7 cells (5 ×106 cells) were seeded into a φ60 dish and incubated for 24 h, and the cells were then exposed to LPS in the presence of LNT-S. After stimulation for desired time, the cytoplasmic and nuclear proteins were extracted and measured by Western blotting using the specific antibodies against phospho-ERK1/2, phospho-JNK1/2, NF-κB p65, and β-actin as described under “Experimental Procedures.” Data are representative of three independent experiments. B, RAW 264.7 macrophages were transfected with an NF-κB-luciferase reporter vector using Lipofectamine. Transiently transfected cells were stimulated LPS (100 ng/ml) in the absence and presence of LNT-S (200 μg/ml) for 12 h. NF-κB-luciferase activity was measured as described under “Experimental Procedures” and is expressed relative to the control (PBS). Each value represents the mean ± S.E. of three independent experiments. *, p < 0.002 versus PBS; **, p < 0.001 versus PBS.
LNT-S Enhances Nuclear Translocation and Transactivation of NF-κB p65
The transcription factor NF-κB is largely involved in the immune responses and expression of proinflammation elicited by a variety of mediators including LPS. To clarify whether the inhibition of NO and TNF-α production is associated with NF-κB, the nuclear translocation of NF-κB p65 was examined in the nuclear extracts of RAW 264.7 cells which were stimulated with LPS plus LNT-S. The results show that there was a strong protein expression of NF-κB p65 in LPS-stimulated RAW 264.7 cells within 30 min compared with the unstimulated cells (Fig. 3A). At a longer time point, the protein bands became very weak and even disappeared, which might be attributable to degradation of the protein. It is surprising that LNT-S could not attenuate LPS-induced nuclear translocation of NF-κB p65, but enhanced it slightly.
It has been reported that NO productivity was strongly dependent on some cytokines such as TNF-α, IL-1α, IL-6, or GM-CSF (24). NF-κB is a major activator for TNF-α transcription in macrophages. To investigate whether NF-κB was transactivated, transcriptional activity of NF-κB was detected by analyzing the gene reporter activity. As shown in Fig. 3B, a significant increase of luciferase activity was observed in LPS-stimulated microphages and slight augmentation by LNT-S, consistent with the enhancement of nuclear translocation of NF-κB p65 (Fig. 3B).
Effect of Anti-Dectin-1 or Anti-TLR2 Antibodies on Inhibition of NO and TNF-α by LNT-S
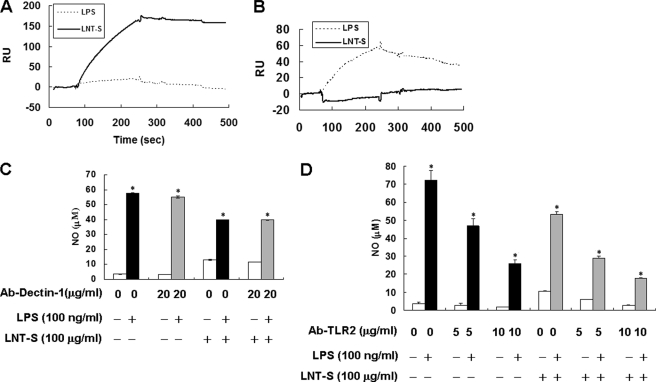
Dectin-1 has been extensively identified as a zymosan β-glucan receptor (20, 57–59), which is primarily expressed by cells of myeloid origin, including macrophages, dendritic cells, and neutrophils in mice. Rogers et al. reported that both Dectin-1 and TLR2 are activated by zymosan, but the TLR2 ligand is not yet known (60). To determine whether LNT-S binds to Dectin-1 or TLR2 receptors, we first determined the interactions between LNT-S and the two receptors by SPR analysis. Fig. 4A demonstrates high affinity interaction between LNT-S and Dectin-1, but little binding to TLR2 was observed as shown in Fig. 4B. In contrast, LPS could not bind to Dectin-1 but interacted partly with TLR2. These results indicated that Dectin-1 but not TLR2 can also recognize water-soluble β-glucan LNT-S, and LPS may bind to TLR2 although the major receptor is TLR4 (61).
FIGURE 4.
A, affinity constant of LNT-S and anti-Dectin-1 antibody by SPR analysis. B, affinity constant of LNT-S and anti-TLR2 antibody by SPR analysis. C, effect of anti-Dectin-1 antibody on inhibition of inhibition of NO by LNT-S in LPS-stimulated RAW 264.7 cells. D, effect of anti-TLR 2 antibody on inhibition of NO production by LNT-S in LPS-stimulated RAW 264.7 cells. RAW 264.7 cells (5 × 105 cells/well) were seeded in 24-well plates. After 24-h preincubation, the cells were incubated with 5–20 μg/ml monoclonal antibodies against murine Dectin-1/CLEC7A and TLR2/CD282 for 30 min. The cells were then exposed to LPS (100 ng/ml) and LNT-S (100 μg/ml) for 24 h, and NO in the supernatant was measured as described under “Experimental Procedures.” Each value represents the mean ± S.E. of at least three independent experiments.*, p < 0.001 versus respective controls.
Because they are expressed on macrophages, we used anti-Dectin-1 mAb and anti-TLR2 mAb to neutralize Dectin-1 and TLR2, respectively, and then detected NO production. As shown in Fig. 4, C and D, addition of anti-Decin-1 mAb hardly affected the inhibition of LPS-induced NO production, but anti-TLR2 mAb suppressed NO production, which could be explained by the results of SPR analysis. Namely, anti-TLR2 mAb bound to TLR2, leading to a decrease of LPS-induced NO production. Although anti-TLR2 mAb inhibited LPS-induced NO production, it had little effect on the inhibition effect induced by LNT-S.
DISCUSSION
It is well known that NO and TNF-α are two very important inflammatory mediators, which are associated with microbial and tumor cell killing (3, 62). However, overproduction of NO and TNF-α is harmful to the body as described in the Introduction. In this study, we studied the effect of LNT-S on LPS-induced activation of MAP kinases that lead to activation of NF-κB and production of NO and TNF-α. All of the results shown in Figs. 1 and 2 proved that LNT-S showed strong inhibition of LPS-induced proinflammatory mediators TNF-α, NO, and iNOS by suppressing the expression of TNF-α mRNA and iNOS mRNA. Clearly, it has a completely different effect from SPG-OH, which augments cytokine production and subsequent NO synthesis in the presence of cytokines such as TNF-α, IL-1α, IL-6, GM-CSF, especially in the presence of IFN-γ in vitro (24).
LPS has been documented to induce activation of MAP kinases, JAK2, and PI3K, hereby activating multiple transcription factors such as NF-κB and AP-1 and so forth to translocate to the nucleus facilitating DNA binding activity and leading to up-regulation of iNOS and TNF-α mRNA expression in RAW 264.7 cells (54, 63). Our results obtained from Western blotting documented that LNT-S severely suppressed LPS-induced JNK1/2 and ERK1/2 activation in RAW 264.7 cells (Fig. 3A). These observations confirmed that LNT-S inhibited LPS-induced iNOS and TNF-α mRNA expression and subsequent NO and TNF-α production through selective suppression of JNK1/2 and ERK1/2 in the MAPK signaling pathways. It is very interesting that LNT-S can activate MAP kinases JNK1/2 and ERK1/2 and enhance the nuclear translocation of NF-κB p65 and its transactivation activity but without production of NO and TNF-α in RAW 264.7 macrophages, which was reported in our very recent work (38). When it co-stimulates RAW 264.7 cells with LPS, LNT-S shows completely different immunomodulatory behavior, that is, it did not augment the activation of ERK1/2 and JNK1/2 (Fig. 3A), but suppressed them and thus the proinflammatory mediators such as TNF-α and NO (Fig. 1). These results indicate that LNT-S can be used as not only an immunomodulatory agent but also a molecule controlling the diseases associated with NO and TNF-α overproduction.
The promoter region of the mouse iNOS gene contains several binding sites for transcription factors such as NF-κB, AP-1, CCAAT-enhancer box-binding protein (C/EBP), cAMP-responsive element-binding protein (CREB), interferon regulatory factor-1 (IRF-1), nuclear factor-IL6 (NF-IL6), octamer factor-1 (Oct-1), serum response factor (SRF), and the signal transducers and activators of transcription (STAT)1α (64). Of these, NF-κB, STAT1, and AP-1 are required for iNOS gene expression in RAW 264.7 macrophages induced by LPS and proinflammatory cytokines (65–67) Especially, NF-κB is a central target for activators or inhibitors of iNOS induction (68). The inhibition of iNOS expression by some agents such as glucocorticoids, TGF-β1, and antioxidants have been reported to result from direct capture of NF-κB by protein-protein interactions (69, 70), inhibition of nuclear translocation of NF-κB (71), inhibition of NF-κB transactivation activity, or from enhancement of the expression of the specific inhibitor of NF-κB, I-κB (72, 73). In the present study, we observed that LNT-S could not inhibit the nuclear translocation of NF-κB p65 (Fig. 3A) or the transactivation activity of NF-κB (Fig. 3B), but slightly enhanced them, respectively. It is possible that inhibition of NO and TNF-α production by LNT-S was not through NF-κB signaling pathway, and that other transcription factors such as AP-1, C/EBP, CREB and so on might be involved in the inhibition mechanism, which will be investigated further in future study.
It has been extensively accepted that Dectin-1 is a key receptor on the surface of macrophages, which is required for β-glucan recognition and even can collaborate with TLR2 mediating the biological effects of β-glucan (55, 56, 74). The SPR analysis demonstrates that LNT-S bound Dectin-1 strongly (Fig. 4A) but hardly to TLR2 (Fig. 4B). However, as shown in Fig. 4C, after neutralization by anti-Dectin-1 mAb, LNT-S still exhibited almost the same inhibition of NO production in LPS-stimulated RAW 264.7 cells (Fig. 4C). This result suggested that the inhibition of NO and TNF-α production by LNT-S might be not through Dectin-1 signaling pathways. Apart from Dectin-1, other receptors of β-glucans might be involved in this process.
In summary, our results suggest that LNT-S inhibits LPS-induced NO and TNF-α production in RAW 264. 7 macrophages partially via suppression of MAP kinases JNK1/2 and ERK1/2, and the resultant decrease in the expression of iNOS mRNA and TNF-α mRNA. Do other β-glucans show similar results? We thus treated LPS-stimulated RAW 264.7 macrophages with another β-glucan named BBG from S. cerevisiae (Bakery). The results showed that BBG could inhibit secretion of proinflammatory mediators including NO, TNF-α, IL-1α, and IL-1γα. The phosphorylation of MAP kinases ERK1/2 and JNK1/2 was also confirmed to be suppressed by BBG. It is thus supposed that β-glucans besides LNT-S and BBG have the bioactivity of inhibiting proinflammatory mediators through MAPK signaling pathways. However, more and more β-glucans should be chosen to confirm it, and the upstreaming MAPK signaling pathways, transcription factor(s), and the receptor(s) involved in the inhibition mechanism will be further studied in detail in our future work.
This work was supported by Japan Society for the Promotion of Science Postdoctoral Fellowship 20.08431 (to X. X.), National Natural Science Foundation Grant 20874078, and Youth Technology Chenguang Project of Wuhan 200950431193.
- SPG
- Schizophyllan
- iNOS
- inducible NOS
- LNT
- Lentinan
- LNT-S
- LNT extracted by sonication
- SPG-OH
- alkaline-treated SPG
- SPR
- surface plasma resonance
- TLR
- toll-like receptor.
REFERENCES
- 1. Casale T. B., Costa J. J., Galli S. J. (1996) Am. J. Respir. Cell Mol. Biol. 15, 35–44 [DOI] [PubMed] [Google Scholar]
- 2. Manjeet K. R., Ghosh B. (1999) Int. J. Immunopharmacol. 21, 435–443 [DOI] [PubMed] [Google Scholar]
- 3. Nathan C. (1992) FASEB J. 6, 3051–3064 [PubMed] [Google Scholar]
- 4. Davis K. L., Martin E., Turko I. V., Murad F. (2001) Annu. Rev. Pharmacol. Toxicol. 41, 203–236 [DOI] [PubMed] [Google Scholar]
- 5. Liew F. Y. (1995) Curr. Opin. Immunol. 7, 396–399 [DOI] [PubMed] [Google Scholar]
- 6. Bogdan C. (2001) Nat. Immunol. 2, 907–916 [DOI] [PubMed] [Google Scholar]
- 7. MacMicking J., Xie Q. W., Nathan C. (1997) Annu. Rev. Immunol. 15, 323–350 [DOI] [PubMed] [Google Scholar]
- 8. Kindler V., Sappino A. P., Grau G. E., Piguet P. F., Vassalli P. (1989) Cell 56, 731–740 [DOI] [PubMed] [Google Scholar]
- 9. Lehmann V., Freudenberg M. A., Galanos C. (1987) J. Exp. Med. 165, 657–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fehsel K., Kröncke K. D., Meyer K. L., Huber H., Wahn V., Kolb-Bachofen V. (1995) J. Immunol. 155, 2858–2865 [PubMed] [Google Scholar]
- 11. Kröncke K. D., Fehsel K., Kolb-Bachofen V. (1998) Clin. Exp. Immunol. 113, 147–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wink D. A., Kasprzak K. S., Maragos C. M., Elespuru R. K., Misra M., Dunams T. M., Cebula T. A., Koch W. H., Andrews A. W., Allen J. S. (1991) Science 254, 1001–1003 [DOI] [PubMed] [Google Scholar]
- 13. Hogaboam C. M., Steinhauser M. L., Schock H., Lukacs N., Strieter R. M., Standiford T., Kunkel S. L. (1998) Infect. Immun. 66, 650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Czop J. K. (1986) Pathol. Immunopathol. Res. 5, 286–296 [DOI] [PubMed] [Google Scholar]
- 15. Williams D. L. (1997) Mediators Inflamm. 6, 247–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ross G. D., Vetvicka V., Yan J., Xia Y., Vetvicková J. (1999) Immunopharmacology 42, 61–74 [DOI] [PubMed] [Google Scholar]
- 17. Williams D. L., Mueller A., Browder W. (1996) Clin. Immunother. 5, 392–399 [Google Scholar]
- 18. Riggi S. J., Di Luzio N. R. (1961) Am. J. Physiol. 200, 297–300 [DOI] [PubMed] [Google Scholar]
- 19. Kankkunen P., Teirilä L., Rintahaka J., Alenius H., Wolff H., Matikainen S. (2010) J. Immunol. 184, 6335–6342 [DOI] [PubMed] [Google Scholar]
- 20. Brown G. D., Herre J., Williams D. L., Willment J. A., Marshall A. S., Gordon S. (2003) J. Exp. Med. 197, 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Young S. H., Ye J., Frazer D. G., Shi X., Castranova V. (2001) J. Biol. Chem. 276, 20781–20787 [DOI] [PubMed] [Google Scholar]
- 22. Suga T., Shiio T., Maeda Y. Y., Chihara G. (1984) Cancer Res. 44, 5132–5137 [PubMed] [Google Scholar]
- 23. Okamura K., Suzuki M., Chihara T., Fujiwara A., Fukuda T., Goto S., Ichinohe K., Jimi S., Kasamatsu T., Kawai N. (1986) Cancer 58, 865–872 [DOI] [PubMed] [Google Scholar]
- 24. Ohno N., Hashimoto T., Adachi Y., Yadomae T. (1996) Immunol. Lett. 52, 1–7 [DOI] [PubMed] [Google Scholar]
- 25. Hashimoto T., Ohno N., Adachi Y., Yadomae T. (1997) FEMS Immunol. Med. Microbiol. 19, 131–135 [DOI] [PubMed] [Google Scholar]
- 26. Taguchi T., Kaneko Y. (1986) in Host Defense Mechanisms against Cancer (Urushizaki I., Aoki T., Tsubura E., eds) p. 221, Excerpta Medica, Amsterdam [Google Scholar]
- 27. Nanba H., Kuroda H. (1987) Chem. Pharm. Bull. 35, 2459–2464 [DOI] [PubMed] [Google Scholar]
- 28. Péter G., Károly V., Imre B., János F., Kaneko Y. (1988) Immunopharmacol. Immunotoxicol. 10, 157–163 [DOI] [PubMed] [Google Scholar]
- 29. Fruehauf J. P., Bonnard G. D., Herberman R. B. (1982) Immunopharmacology 5, 65–74 [DOI] [PubMed] [Google Scholar]
- 30. Akiyama T., Kashima S., Hayami T., Izawa M., Mitsugi K., Hamuro J. (1987) in Manipulation of Host Defence Mechanism (Acki T., Uneshizaki I., Tsubura E., eds) pp. 227–238, Excerpta Medica, Amsterdam [Google Scholar]
- 31. Abel G., Szöllösi J., Chihara G., Fachet J. (1989) Int. J. Immunopharmacol. 11, 615–621 [DOI] [PubMed] [Google Scholar]
- 32. Herlyn D., Kaneko Y., Powe J., Aoki T., Koprowski H. (1985) Gann 76, 37–42 [PubMed] [Google Scholar]
- 33. Nanba H., Mori K., Toyomasu T., Kuroda H. (1987) Chem. Pharm. Bull. 35, 2453–2458 [DOI] [PubMed] [Google Scholar]
- 34. Ladányi A., Tímár J., Lapis K. (1993) Cancer Immunol. Immunother. 36, 123–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kerékgyártó C., Virág L., Tankó L., Chihara G., Fachet J. (1996) Int. J. Immunopharmacol. 18, 347–353 [DOI] [PubMed] [Google Scholar]
- 36. Kupfahl C., Geginat G., Hof H. (2006) Int. Immunopharmacol. 6, 686–696 [DOI] [PubMed] [Google Scholar]
- 37. Noel M., Madaj K., Hintelmann H., Gast G., Kaneko Y. (1997) Int. J. Immunopharmacol. 19, 463–468 [DOI] [PubMed] [Google Scholar]
- 38. Xu X., Pan C., Zhang L., Ashida H. (2011) J. Biol. Chem. 286, 31194–31198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Miralles C., Busquets X., Santos C., Togores B., Hussain S., Rahman I., MacNee W., Agustí A. G. (2000) FEBS Lett. 476, 253–257 [DOI] [PubMed] [Google Scholar]
- 40. Moncada S., Higgs A. (1993) N. Engl. J. Med. 329, 2002–2012 [DOI] [PubMed] [Google Scholar]
- 41. Gess B., Schricker K., Pfeifer M., Kurtz A. (1997) Am. J. Physiol. 273, R905–910 [DOI] [PubMed] [Google Scholar]
- 42. Osei S. Y., Ahima R. S., Fabry M. E., Nagel R. L., Bank N. (1996) Blood 88, 3583–3588 [PubMed] [Google Scholar]
- 43. Taylor B. S., Alarcon L. H., Billiar T. R. (1998) Biochemistry 63, 766–781 [PubMed] [Google Scholar]
- 44. Taylor B. S., de Vera M. E., Ganster R. W., Wang Q., Shapiro R. A., Morris S. M., Jr., Billiar T. R., Geller D. A. (1998) J. Biol. Chem. 273, 15148–15156 [DOI] [PubMed] [Google Scholar]
- 45. Yang E. J., Yim E. Y., Song G., Kim G. O., Hyun C. G. (2009) Interdiscip. Toxicol. 2, 245–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ruttimann J. (2007) J. Exp. Med. 204, 3057 [Google Scholar]
- 47. Arbour N. C., Lorenz E., Schutte B. C., Zabner J., Kline J. N., Jones M., Frees K., Watt J. L., Schwartz D. A. (2000) Nat. Genet. 25, 187–191 [DOI] [PubMed] [Google Scholar]
- 48. Muzio M., Ni J., Feng P., Dixit V. M. (1997) Science 278, 1612–1615 [DOI] [PubMed] [Google Scholar]
- 49. Takeda K., Akira S. (2001) Genes Cells 6, 733–742 [DOI] [PubMed] [Google Scholar]
- 50. Hambleton J., Weinstein S. L., Lem L., DeFranco A. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2774–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Han J., Lee J. D., Bibbs L., Ulevitch R. J. (1994) Science 25, 808–811 [DOI] [PubMed] [Google Scholar]
- 52. Lee J. C., Laydon J. T., McDonnell P. C., Gallagher T. F., Kumar S., Green D., McNulty D., Blumenthal M. J., Heys J. R., Landvatter S. W. (1994) Nature 372, 739–746 [DOI] [PubMed] [Google Scholar]
- 53. Kim J. W., Kim C. (2005) Biochem. Pharmacol. 70, 1352–1360 [DOI] [PubMed] [Google Scholar]
- 54. Okugawa S., Ota Y., Kitazawa T., Nakayama K., Yanagimoto S., Tsukada K., Kawada M., Kimura S. (2003) Am. J. Physiol. Cell Physiol. 285, C399–408 [DOI] [PubMed] [Google Scholar]
- 55. Brown G. D. (2006) Nat. Rev. Immunol. 6, 33–43 [DOI] [PubMed] [Google Scholar]
- 56. Gantner B. N., Simmons R. M., Canavera S. J., Akira S., Underhill D. M. (2003) J. Exp. Med. 197, 1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brown G. D., Gordon S. (2003) Immunity 19, 311–315 [DOI] [PubMed] [Google Scholar]
- 58. Gantner B. N., Simmons R. M., Underhill D. M. (2005) EMBO J. 24, 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Brown G. D., Gordon S. (2005) Cell Microbiol. 7, 471–479 [DOI] [PubMed] [Google Scholar]
- 60. Rogers N. C., Slack E. C., Edwards A. D., Nolte M. A., Schulz O., Schweighoffer E., Williams D. L., Gordon S., Tybulewicz V. L., Brown G. D., Reis e Sousa C. (2005) Immunity 22, 507–517 [DOI] [PubMed] [Google Scholar]
- 61. Verstrepen L., Bekaert T., Chau T. L., Tavernier J., Chariot A., Beyaert R. (2008) Cell. Mol. Life Sci. 65, 2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lorsbach R. B., Murphy W. J., Lowenstein C. J., Snyder S. H., Russell S. W. (1993) J. Biol. Chem. 268, 1908–1913 [PubMed] [Google Scholar]
- 63. Tebo J. M., Chaoqun W., Ohmori Y., Hamilton T. A. (1994) J. Immunol. 153, 4713–4720 [PubMed] [Google Scholar]
- 64. Kleinert H., Paultz A., Linker K., Schwarz P. M. (2004) Eur. J. Pharmacol. 500, 255–266 [DOI] [PubMed] [Google Scholar]
- 65. Xie Q. W., Kashiwabara Y., Nathan C. (1994) J. Biol. Chem. 269, 4705–4708 [PubMed] [Google Scholar]
- 66. Gao J., Morrison D. C., Parmely T. J., Russell S. W., Murphy W. J. (1997) J. Biol. Chem. 272, 1226–1230 [DOI] [PubMed] [Google Scholar]
- 67. Xie Q. W., Whisnant R., Nathan C. (1993) J. Exp. Med. 177, 1779–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ghosh S., May M. J., Kopp E. B. (1998) Annu. Rev. Immunol. 16, 225–260 [DOI] [PubMed] [Google Scholar]
- 69. Kleinert H., Euchenhofer C., Ihrig-Biedert I., Förstermann U. (1996) Mol. Pharmacol. 49, 15–21 [PubMed] [Google Scholar]
- 70. Mukaida N., Morita M., Ishikawa Y., Rice N., Okamoto S., Kasahara T., Matsushima K. (1994) J. Biol. Chem. 269, 13289–13295 [PubMed] [Google Scholar]
- 71. Jeon Y. J., Han S. H., Lee Y. W., Yea S. S., Yang K. H. (1998) Biochem. Mol. Biol. Int. 45, 435–441 [DOI] [PubMed] [Google Scholar]
- 72. De Vera M. E., Taylor B. S., Wang Q., Shapiro R. A., Billiar T. R., Geller D. A. (1997) Am. J. Physiol. 273, G1290–1296 [DOI] [PubMed] [Google Scholar]
- 73. Saura M., Zaragoza C., Diaz-Cazorla M., Hernandez-Perera O., Eng E., Lowenstein C. J, Perez-Sala D., Lamas S. (1998) Kideny Int. 53, 38–49 [DOI] [PubMed] [Google Scholar]
- 74. Taylor P. R., Tsoni S. V., Willment J. A., Dennehy K. M., Rosas M., Findon H., Haynes K., Steele C., Botto M., Gordon S., Brown G. D. (2007) Nat. Immunol. 8, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]