Background: Human protein-disulfide isomerase (hPDI) is a key enzyme and chaperone for protein folding.
Results: Oxidation of domain a′ releases the compact conformation of hPDI and exposes the buried substrate-binding sites facilitating its high chaperone activity.
Conclusion: Oxidation of hPDI activates its chaperone activity.
Significance: This study provides the first structural evidence of and mechanistic insights into the redox-regulated chaperone activity of hPDI.
Keywords: Endoplasmic Reticulum (ER), Protein Dynamics, Protein Folding, Protein Structure, Redox Regulation, Chaperone, Protein-disulfide Isomerase
Abstract
Protein-disulfide isomerase (PDI), with domains arranged as abb′xa′c, is a key enzyme and chaperone localized in the endoplasmic reticulum (ER) catalyzing oxidative folding and preventing misfolding/aggregation of proteins. It has been controversial whether the chaperone activity of PDI is redox-regulated, and the molecular basis is unclear. Here, we show that both the chaperone activity and the overall conformation of human PDI are redox-regulated. We further demonstrate that the conformational changes are triggered by the active site of domain a′, and the minimum redox-regulated cassette is located in b′xa′. The structure of the reduced bb′xa′ reveals for the first time that domain a′ packs tightly with both domain b′ and linker x to form one compact structural module. Oxidation of domain a′ releases the compact conformation and exposes the shielded hydrophobic areas to facilitate its high chaperone activity. Thus, the study unequivocally provides mechanistic insights into the redox-regulated chaperone activity of human PDI.
Introduction
The ER5 is a eukaryotic organelle containing many chaperones and enzymes and provides an optimized environment for the folding and maturation of membrane and secretory proteins (1). PDI is a key enzyme (2) and chaperone (3, 4) localized in the ER with versatile functions (e.g. catalyzing formation, breakage, and rearrangement of the disulfide bonds in protein substrates (5), facilitating ER-associated degradation of misfolded proteins (6, 7), and serving as a chaperone-like subunit of prolyl-4-hydroxylase (8) and microsomal triglyceride transfer protein (9)). Additionally, PDI has non-ER locations (10); for example, on the cell surface, it participates in the activation of the fusion of HIV virus (11) and mediates the adhesion, secretion, and aggregation of platelet (12). Recently, it has been recognized that PDI plays a critical role in the control of neurodegenerative diseases related to protein misfolding and aggregation (e.g. Alzheimer disease, Parkinson disease, amyotrophic lateral sclerosis, and Huntington disease (13–16)).
PDI is composed of four thioredoxin-like domains (a, b, b′, and a′) plus an x linker and a C-terminal acidic tail (Fig. 1A) (17–19). Both domains of a and a′ contain a CGHC active site responsible for the oxidoreductase activity (20). The b′ domain possesses the main peptide binding site, whereas binding of large substrates or partner proteins requires all four domains (21, 22). The crystal structures of yeast PDI (yPDI) at 4 and 22 °C revealed that four domains are arranged in a “twisted U shape” and a “boat shape,” respectively, with domains a and a′ representing two mobile arms connected to the rigid bb′ base, and the a arm was demonstrated to be more flexible than the a′ arm (23, 24). The full-length structure of hPDI has not been reported so far; however, the major flexible sites are located in the b′xa′ region (25). PDI can shuttle between the oxidized and reduced states with abilities to bind to various targets and catalyze different reactions (i.e. the oxidized PDI preferentially binds to reduced/unfolded polypeptide to introduce disulfide bonds, whereas the reduced PDI mainly interacts with misoxidized substrates functioning as a reductase or isomerase (26)). In mammalian cells, only the reduced and not the oxidized PDI interacts with its oxidase Ero1 (27) or peroxiredoxin 4 (28). Hence, it is elusive how reduced/oxidized PDI distinguishes its substrates/partner proteins or selectively binds to the same substrate at different stages in the protein-folding pathway (26).
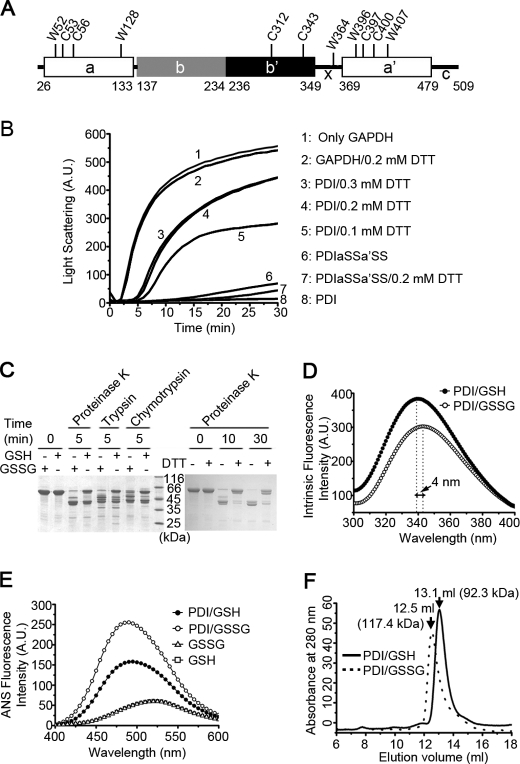
FIGURE 1.
The chaperone activity and overall conformation of hPDI are redox-regulated. A, a schematic representation of hPDI with domain organization in abb′xa′c. The positions of all of the tryptophan and cysteine residues are marked, and the residue numbering is for hPDI with signal sequence. B, hPDI is a redox-regulated chaperone. Guanidine hydrochloride-denatured GAPDH at 0.14 mm was 50-fold diluted into refolding buffer in the absence or presence of 28 μm hPDI proteins with various concentrations of DTT as indicated. Aggregation produced during the refolding was monitored by recording the light scattering at 488 nm, and the suppression of aggregation was used to measure the chaperone activity of hPDI proteins. A.U., arbitrary units. C, protease digestion profiles of hPDI at different redox states. hPDI at 1 mg/ml was incubated with 1 mm GSSG, GSH, or DTT at 25 °C for 30 min and then digested by 2 μg/ml proteinase K, trypsin, or chymotrypsin for various time as indicated. The reactions were terminated by adding PMSF to a final concentration of 0.5 mm and analyzed by reducing SDS-PAGE. The oxidized hPDI is more accessible for protease digestion than the reduced hPDI. D and E, fluorescence spectra of hPDI at different redox states. hPDI at 5 μm was incubated with 1 mm GSSG or GSH at 25 °C for 30 min, and the intrinsic (D) and ANS (E) fluorescence spectra were recorded. A.U., arbitrary units. F, size exclusion chromatography profiles of hPDI treated with 1 mm GSH (solid curve) or GSSG (dashed curve) at 25 °C for 30 min. The molecular weight of hPDI in different redox states calculated according to the elution volume is marked.
About 10 years ago, PDI was reported to be a redox-dependent chaperone based on the finding that PDI binds and unfolds the cholera toxin A1 subunit in the reduced form (29) and releases it after being oxidized by Ero1, thus facilitating the retrograde transport of A1 subunit from the ER into the cytosol (30). However, this redox-dependent chaperone activity of PDI was subsequently challenged by arguing that the release of the A1 subunit is mediated by substrate competition rather than by a change in the redox state of PDI (31). Therefore, whether the chaperone activity of PDI is redox-regulated is controversial (32). Moreover, the putative redox-dependent conformational changes of PDI are much less characterized, and only very recently, studies of PDI from a thermophilic fungus showed a redox-dependent intramolecular rearrangement of the b′ and a′ domains by using NMR spectroscopy and small-angle x-ray scattering (33, 34). Nevertheless, the underlying molecular mechanism for redox-regulated conformational changes and chaperone activity of PDI remains largely unknown.
In this study, we extensively explored different conformations and the chaperone activity of hPDI in different redox states. We demonstrate that hPDI is a redox-regulated chaperone undergoing large redox-dependent conformational changes. We further map out b′xa′ to be the minimal region of hPDI enduring the conformational rearrangement and identify the active site in the a′ domain to be the key trigger. The crystal structure of the reduced bb′xa′ was determined and reveals that the a′ domain packs tightly with both the b′ domain and the x linker to form one compact structural module. Oxidation of the a′ domain releases this compact conformation and subsequently exposes the shielded substrate binding areas for higher chaperone activity of hPDI. We finally demonstrate that the redox-dependent conformational changes are essential for regulation of the chaperone activity of hPDI.
EXPERIMENTAL PROCEDURES
Plasmid Construction and Protein Purification
The genes encoding hPDI, abb′x and bb′xa′, were cloned into pQE30 (Qiagen), and the resulting proteins contained N-terminal (MRGSH6GS) tags. The bb′xa′ gene was also cloned into a modified pET32a for structural studies, and the resulting protein contained an N-terminal (H6SSGLEVLFQGSEF) tag. hPDI mutants were created by the overlap extension PCR method, and the sequences of all of the constructs were verified by DNA sequencing. Proteins were expressed and purified as described previously (35). Protein concentrations were determined by the Bradford method with bovine serum albumin as a standard.
Chaperone Activity
Complete denaturation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was carried out by incubation of 0.14 mm GAPDH in 3 m guanidine hydrochloride with 1 mm DTT overnight at 4 °C. Refolding was initiated by 50-fold dilution of the denatured GAPDH into 100 mm sodium phosphate buffer (pH 7.4) containing 2.5 mm EDTA in the absence or presence of hPDI proteins and different concentrations of DTT at 25 °C. Aggregation produced during the refolding was monitored by recording the light scattering at 488 nm, and the suppression of aggregation was used to measure the chaperone activity of hPDI proteins (3).
Limited Proteolysis Assay
hPDI proteins at 1 mg/ml were incubated with or without 1 mm GSH, GSSG, or DTT in 50 mm Tris-HCl buffer (pH 7.6) containing 5 mm CaCl2 at 25 °C for 30 min, and then different proteases were added for digestion. The reactions were terminated at different times by phenylmethanesulfonyl fluoride (PMSF) and analyzed by reducing SDS-PAGE.
Fluorescence Measurements
hPDI proteins of 5 μm were incubated with or without 1 mm GSH, GSSG, or DTT in 50 mm Tris-HCl buffer (pH 7.6) containing 150 mm NaCl at 25 °C for 30 min. Intrinsic fluorescence spectra were recorded at 310–400 nm at 25 °C with excitation at 290 nm. 1-Anilino-8-naphthalene sulfonate (ANS) was added in protein samples to 50 μm and incubated for 20 min at 25 °C in the dark before the ANS fluorescence emission spectra (400–600 nm) were determined with excitation at 370 nm. The concentration of ANS was determined using an extinction coefficient at 350 nm of 5000 m−1 cm−1.
Crystallization, Data Collection, and Process
The purified bb′xa′ at 10.0 mg/ml in 20 mm Tris-HCl buffer, 100 mm NaCl, pH 7.5, was crystallized at 16 °C using the hanging drop method by mixing 1 μl of protein sample containing 50 mm DTT with an equal volume of 25% (w/v) PEG 3350 in 0.1 m Tris-HCl, 0.2 m ammonium acetate, pH 8.5. The crystals were flash-cooled with liquid nitrogen. The diffraction data were collected at Shanghai Synchrotron Radiation Facility beam line BL17U at a wavelength of 0.979 Å at 100 K. The diffraction data were processed and scaled using HKL2000 package (36).
Structure Determination and Refinement
The bb′xa′ structure was solved by the molecular replacement method with the program Phaser (37) using the structure of yPDI (Protein Data Bank code 2B5E) (23) as the search model. Structures were fitted and rebuilt with the program Coot (38) and refined with REFMAC5 (37) and CNS (40). At the final refinement stage, TLS refinement was used with groups defined by the TLSMD server (41). The overall quality of the final structural model was assessed by PROCHECK (42). The atomic coordinates of bb′xa′ have been deposited in the Protein Data Bank (PDB code 3UEM).
Mass Spectrometry
Protein samples were purified by size exclusion chromatography with 0.1% formic acid/water for digests by proteinase K and by reverse-phase HPLC on a C8 chip (G4240-63001) with a water-ACN system containing 0.1% formic acid for digests by trypsin or chymotrypsin. The samples were introduced into the spectrometer by JetStream electrospray ionization using a spray voltage of 3.7 kV for digests by proteinase K and by HPLC-chip using 1.88 kV for digests by trypsin or chymotrypsin. Spectra were acquired on a calibrated Q-TOF (Agilent 6530) instrument operated in positive ion mode over the m/z range 400–1600 for digests by proteinase K and 400–3200 for digests by trypsin or chymotrypsin. Data were processed via Mass Hunter qualitative analysis software.
RESULTS
Chaperone Activity of hPDI Is Regulated by Its Redox States
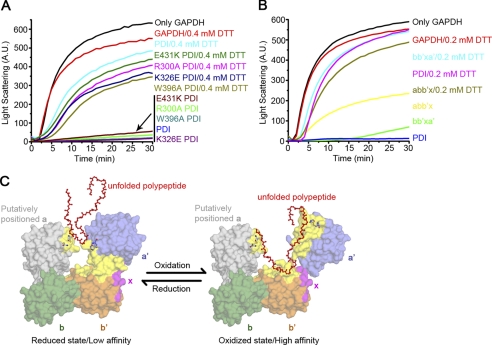
To reconcile the controversy on redox-regulated chaperone activity of hPDI, we first tested whether the chaperone activity of hPDI is regulated by its redox states. A well established system was used to characterize the chaperone activity of hPDI (3) (i.e. the presence of hPDI in the refolding buffer strongly suppressed the aggregation of guanidine hydrochloride-denatured GAPDH during dilution-induced refolding) (Fig. 1B). The purified hPDI contains about 2.5 free thiols as determined by 5,5′-dithiobis(2-nitrobenzoic acid), indicating the active sites in domains a and a′ are mostly oxidized because the two cysteines in the b′ domain do not form a disulfide bond (43). Adding DTT in the refolding buffer significantly decreased the chaperone activity of hPDI in a concentration-dependent manner (i.e. the more reduced hPDI was, the lower chaperone activity it showed) (Fig. 1B), demonstrating that the redox states of hPDI indeed control its chaperone activity. Because both active sites of domains a and a′ are redox state sensors, we next made double Cys/Ser point mutations on both sites (PDIaSSa′SS) to impair its redox responses. Notably, PDIaSSa′SS showed chaperone activity similar to that of the oxidized hPDI, and the DTT-induced chaperone activity decrease was completely abolished (Fig. 1B), indicating that redox-regulated hPDI chaperone activity is dependent on its active sites.
hPDI Undergoes Redox-dependent Conformational Changes
We next looked for the potential redox-dependent conformational changes of hPDI by direct digestion with proteinase K, trypsin, and chymotrypsin under various redox conditions (Fig. 1C). After 5 min of digestion by proteinase K, the GSSG-incubated hPDI almost disappeared, whereas about half of the GSH-incubated hPDI was intact. Similar digestion profiles were produced by trypsin and chymotrypsin. To avoid S-glutathionylation-induced conformational change, hPDI was next digested in the presence or absence of DTT. After a 10-min digestion by proteinase K, most of hPDI but little of the DTT-incubated hPDI was digested (Fig. 1C). Hence, the oxidized hPDI adopts a loose conformation, and the reduced one is very compact.
We next carried out fluorescence studies of hPDI under different redox conditions to further probe redox-regulated conformational changes. Consistent with a compact conformation of the reduced hPDI, the intrinsic fluorescence spectrum of the GSH-incubated hPDI showed a blue shift (∼4 nm) of the maximum excited wavelength compared with that of the GSSG-incubated hPDI (Fig. 1D). The GSSG-incubated hPDI showed higher ANS fluorescence intensity than that of the GSH-incubated hPDI, again indicating the oxidized hPDI being much looser and exposing more hydrophobic areas (Fig. 1E). Additionally, the intrinsic and ANS fluorescence spectra of DTT-treated and -untreated hPDI showed similar results (supplemental Fig. S1). Finally, size exclusion chromatography analysis also demonstrated that the reduced hPDI was eluted with a larger volume, confirming its compact conformation (Fig. 1F). Taken together, hPDI shows significant conformational changes induced by different redox conditions.
Redox States of a′ Domain Regulate Interdomain Conformations of b′xa′
To identify the critical cysteine responsible for the redox-regulated conformational changes of hPDI, we made cysteine mutations in a, b′, or a′ domain, respectively. The digestion profiles of the mutants (Fig. 2A) showed that the redox-regulated susceptibility to hydrolysis disappeared only for the a′ domain mutant (PDIaCCa′SS), indicating that the active site of the a′ domain is the key position. Moreover, mutation of either cysteine in the a′ domain resulted in loss of the redox-regulated conformation changes (supplemental Fig. S2). Furthermore, the proteinase digestion profiles (Fig. 2B) and the intrinsic (Fig. 2C) and ANS (Fig. 2D) fluorescence spectra of abb′x and bb′xa′ in different redox conditions were determined, showing little redox-dependent difference for abb′x but significant difference for bb′xa′. All of the above data demonstrated that the active site of the a′ domain plays a predominant role in the regulation of hPDI conformation under different redox conditions.
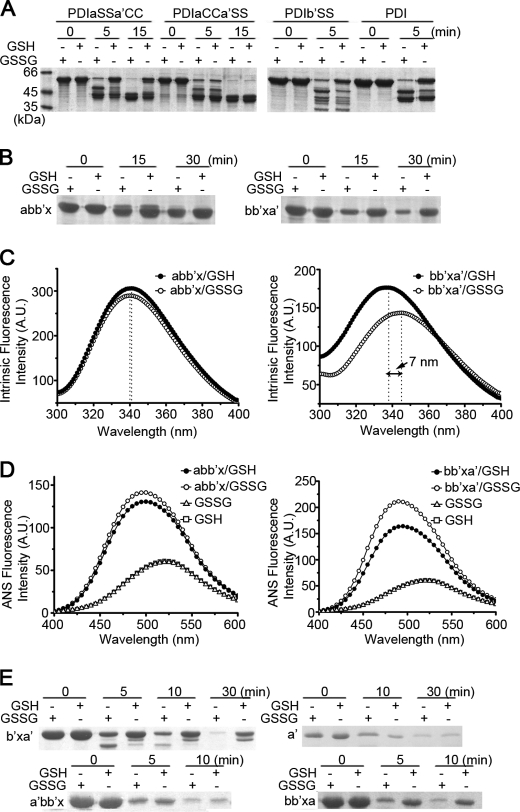
FIGURE 2.
The active site of domain a′ regulates the redox-dependent conformational change of hPDI. A, B, and E, protease digestion profiles of various hPDI mutants with mutations on cysteine residues (A), different truncations, and domain swapping (B and E) at different redox states. hPDI and its mutants at 1 mg/ml were incubated with 1 mm GSSG or GSH at 25 °C for 30 min and then digested by proteinase K for different times as indicated. The reactions were terminated by adding PMSF to a final concentration of 0.5 mm and analyzed by reducing SDS-PAGE. Intrinsic (C) and ANS fluorescence spectra (D) of abb′x and bb′xa′ at 5 μm in different redox states. A.U., arbitrary units.
Next, to map out the corresponding minimal structural element undergoing the redox-induced conformational changes, we made truncations on the bb′xa′. Further deletion of domain b still showed different susceptibilities to proteinase digestion under different redox conditions, whereas domain a′ alone did not show any difference (Fig. 2E), indicating that the minimum region is b′xa′. We further made domain-swapped mutants by exchanging the position of domain a and a′ in the abb′x and bb′xa′, respectively. Interestingly, limited proteolysis studies showed that a′bb′x did not undergo the redox-regulated conformational changes, whereas bb′xa did (Fig. 2E). Hence, the presence of an active thioredoxin-like domain (either a or a′) following the b′x is a most critical element for the redox-regulated interdomain conformational changes of hPDI.
Overall Structure of Reduced bb′xa′
After mapping out b′xa′ to be the minimal structural element undergoing the redox-induced interdomain conformational changes, we next tried to solve the structure of this fragment to explore the molecular mechanism. Due to the fact that b and b′ extensively contact with each other to form one structural unit (44), we included b to b′xa′ for further structural studies. After extensive crystallization trails, high quality crystals of the reduced bb′xa′ were successfully achieved and finally diffracted to 2.29 Å for the structure determination (Table 1). From the overall topology, the structure of the reduced bb′xa′ demonstrated that, consistent with the above biochemical characterizations, the bb′ tandem tightly packs with the a′ domain to form one compact structural entity integrated by the x linker. (Fig. 3A) (see below for details).
TABLE 1.
Data collection and refinement statistics
| Diffraction data | |
| Space group | P212121 |
| Unit cell (Å) | a = 63.585, b = 76.076, c = 79.656, α = β = γ = 90° |
| Resolution (Å) | 50–2.29 (2.38–2.29)a |
| Unique reflections | 17,802 |
| Rmergeb (%) | 4.7 (41.2) |
| I/σ | 30.3 (3.9) |
| Average redundancy | 5.1 (5.3) |
| Completeness (%) | 99.5 (100.0) |
| Refinement | |
| Resolution (Å) | 50–2.29 (2.35–2.29) |
| Rworkc (last shell) | 22.3 (30.9) |
| Rfreed (last shell) | 28.0 (39.2) |
| Mean B factors (Å2) | 55.4 |
| Bond lengthe (Å) | 0.007 |
| Bond angles (degrees) | 1.044 |
| Ramachandran plot (residues, %) | |
| Most favored | 93.0 |
| Additionally allowed | 6.6 |
| Generously allowed | 0.3 |
a The values in parentheses refer to the highest resolution shell.
b Rmerge = ΣhΣi|lih ln|ΣhΣi lh, where lh is the mean intensity of the i observation of reflection h.
c Rfactor = Σh‖Fo| − |Fc‖/Σ|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively. Summation includes all reflections used in the refinement.
d Rfree = Σ‖Fo| − |Fc‖/Σ|Fo|, evaluated for a randomly chosen subset of 5% of the diffraction data not included in the refinement.
e Root mean square deviation from ideal values.
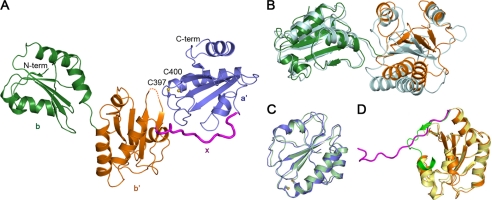
FIGURE 3.
Overall structure of the reduced bb′xa′. A, a ribbon diagram of the crystal structure of the reduced bb′xa′. The b, b′, x, and a′ domains are colored in green, orange, magenta, and blue, respectively. The side chains of the active site cysteines in domain a′ are shown as sticks. Both the N and C termini are also labeled. B, superimposed overlay of the bb′ tandem in the solution structure (44) (cyan) and that in the crystal structure of bb′xa′ based on domain b. C, superposition of the a′ domain in the solution structure (Protein Data Bank entry 1X5C) (pale green) and that in the crystal structure of bb′xa′. D, superposition of the crystal structure of I289A b′x (43) (the b′ domain in yellow and the x linker in forest green) and b′x in the crystal structure of bb′xa′.
Superposition of the bb′xa′ structure and the bb′ solution structure (44) revealed obvious interdomain motion (Fig. 3B), although single domains b and b′ in the two structures are similar. The a′ domain in the bb′xa′ structure is similar to the isolated a′ solution structure (Protein Data Bank entry 1X5C) (Fig. 3C) and exists in the reduced form with a distance of 3.62 Å between two sulfur atoms in the active site. Although the b′ domains in bb′xa′ and in I289A b′x (43) are also highly similar (Fig. 3D), differences were observed around the x linker. In I289A b′x, the first α-helix shifts toward the bound x linker with a tilt of ∼20°; the N-terminal half of the x linker forms an anti-parallel β-strand with the fifth β-strand of b′, and the C-terminal half of the x linker twists and folds back to directly interact with the b′ domain (Fig. 3D). In contrast, the x linker in bb′xa′ orients perpendicularly to the β-sheet of b′ and extends to a′, packing against both the fourth β-strand of b′ and the first α-helix of a′, thus to mediate the compact interactions between them (Fig. 3A).
Reduced bb′xa′ Structure Reveals Compact Structural Module
We next compared the reduced bb′xa′ structure with two different crystal structures of yeast PDI (yPDI22 °C (24) and yPDI4 °C (23)) and found significant interdomain motions of the a′ domain relative to the bb′ base in the reduced bb′xa′ (Fig. 4A). Based on the structural superposition analysis, the a′ domain in the reduced bb′xa′ exhibits a ∼8–9-Å dislocation of the center of the a′ domain due to a ∼35–45° rotation around the x linker toward the hydrophobic surface of the b′ domain. Furthermore, compared with the recently solved structure of human ERp57 (45), which is a PDI family member with similar domain architecture to PDI, the a′ domain endures more significant changes with an ∼11 Å dislocation through a ∼50° rotation (Fig. 4A).
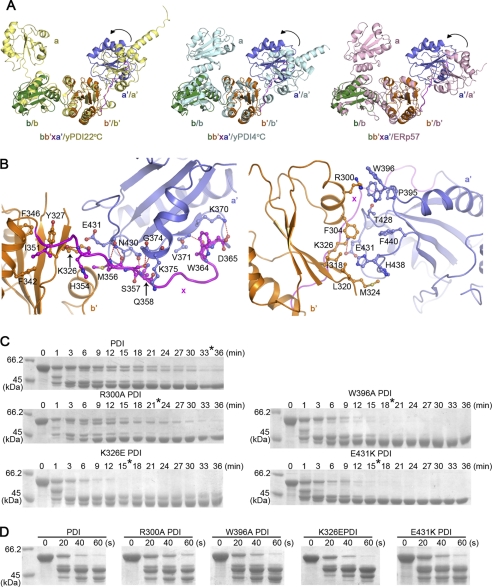
FIGURE 4.
Reduced bb′xa′ adopts a compact conformation. A, superpositions of the reduced bb′xa′ with yPDI structure at 22 °C (24) (yPDI22 °C) (left) or 4 °C (23) (yPDI4 °C) (middle) and ERp57 structure (45) (right) based on the bb′ domains. The interdomain motions of the a′ domain relative to the bb′ domains are indicated by arrows. B, left, the interface of the x linker with domains of b′ and a′; right, the interface between domains of b′ and a′. Residues involved in the interaction are labeled and shown in sticks. Hydrogen bonds (left) and the salt bridge (right) are indicated with red dashed lines. The b′, x, and a′ are colored in orange, magenta, and blue, respectively. C and D, protease digestion profiles of hPDI with various point mutations on the interface of the compact bb′xa′ structure. hPDI and the mutants at 1 mg/ml were incubated with 1 mm GSH (C) and GSSG (D) at 25 °C for 30 min and then digested by 2 μg/ml proteinase K for different times as indicated. The reactions were terminated by adding PMSF to a final concentration of 0.5 mm and analyzed by reducing SDS-PAGE. The time for complete digestion of hPDI and its mutants under GSH conditions are indicated by asterisks. All hPDI mutants showed increased accessibility for protease digestion at reduced conditions.
The x linker in the reduced bb′xa′ connects the b′ and a′ domains with intensive interactions, mediating their interdomain conformation (Fig. 4B, left). The interaction of the N-terminal x linker was contributed by the side chain of Ile-351, which inserts into the hydrophobic pocket formed by side chains of Tyr-327, Phe-342, and Phe-346 of the b′ domain. The central and C-terminal x linker interact with the b′ and a′ domains via forming intensive hydrogen bonds (i.e. His-354–Lys-326, Met-356–Glu-431, Ser-357–Gly-374 and –Lys-375, Gln-358–Asn-430, Trp-364–Val-371, and Asp-365–Lys-370) (Fig. 4B, left). Although the relative positions of domains b′ and a′ are different among yPDI, ERp57, and the reduced bb′xa′, the interaction pattern between the x linker and the b′ or a′ domain is similar (i.e. hydrophobic interactions in the N-terminal x linker and intensive hydrogen bonds in the central and C-terminal x linker).
Surprisingly, direct interactions between domains b′ and a′ were observed in the reduced bb′xa′ (Fig. 4B, right), whereas little was found in yPDI22 °C, yPDI4 °C, and ERp57. The side chain of Trp-396 in the a′ domain exists in two alternative conformations; in one, it forms a cation-π interaction with the side chain of Arg-300 in the b′ domain, and in the other, it contacts with Pro-395 and Thr-428. Interestingly, the side chain of His-438 in the a′ domain also adopts two alternative conformations with the major conformation, in which it is buried in the hydrophobic pocket formed by Phe-304, Ile-318, Leu-320, and Met-324 in the b′ domain. In addition, Lys-326 in domain b′ forms a salt bridge with Glu-431 in domain a′, and Phe-304 in domain b′ interacts with Thr-428, Phe-440, and His-438 in domain a′ (Fig. 4B, right). It is noteworthy that most of residues in the interface of domains b′ and a′ (Arg-300, Phe-304, Ile-318, Leu-320, Met-324, Trp-396, Thr-428, His-438, and Phe-440) coincidentally have been previously characterized for ligand binding (34, 43, 44). Therefore, the reduced a′ domain packs extensively with both the b′ domain and the x linker to form a compact structural module that shields parts of the substrate binding areas, explaining why reduced hPDI has much lower chaperone activity (Fig. 1B). To test whether such direct interactions between the b′ and a′ domains identified in the crystal structure also exist in reduced hPDI in solution, we selectively made R300A, W396A, K326E, and E431K mutants. After incubation with GSH, these mutants are more susceptible to proteinase K than wild-type PDI (Fig. 4C). As the control, the mutants and wild-type PDI showed similar susceptibility to protease digestion after incubation with GSSG (Fig. 4D). These results indicated that the interdomain interactions in b′xa′ stabilize the compact conformation of the reduced hPDI.
Oxidation of Active Site in a′ Domain Exposes Shielded Interfaces
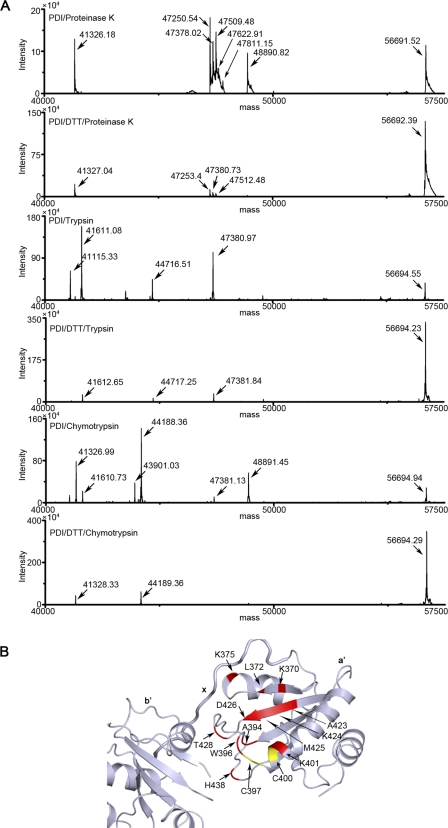
To further understand how oxidation of the a′ domain opens the compact conformation of hPDI, we next tried to identify oxidation-induced exposed regions in detail. The SDS-polyacrylamide gel mobility analysis showed that the major digested products (indicated by asterisks) of proteinase K, trypsin, or chymotrypsin are largely comparable but with different intensity between the oxidized and reduced hPDI, indicating that the major digestion sites are similar and more exposed in the oxidized form (supplemental Fig. S3A). These digests were then further analyzed by mass spectrometry (Fig. 5A). Our previous work has shown that the C-terminal half of hPDI is more flexible to be digested than the N-terminal half (25). Indeed, all of the observed molecular mass of major products corresponds to the fragment extending from the N-terminal tag to a residue in the a′ domain (supplemental Table S1). Although three proteases have different preferences for attacking amino acid residues, they target similar regions in the oxidized hPDI, including the beginning part (Lys-370, Leu-372, and Lys-375), a region around the active sites (Ala-394, Trp-396, and Lys-401), the β2 sheet (Ala-423, Lys-424, Met-425, and Asp-426), and the β2-β3 loop (Thr-428 and His-438) of the a′ domain (Fig. 5B). However, the corresponding products in the mass spectra of the reduced hPDI are either in very low intensity or not observed (Fig. 5A), indicating that these residues are protected from digestion. Interestingly, most of them are located at the interfaces between domains b′ and a′ (Ala-394, Trp-396, Thr-428, and His-438) or between domain a′ and the x linker (Lys-370 and Lys-375). Hence, the mass spectrometry analysis firmly demonstrated that the oxidation of the active site in the a′ domain releases the compact module formed by domains b′, a′, and the x linker and exposes shielded sites for digestion.
FIGURE 5.
Oxidation of the a′ domain release the compact structure formed by domains b′, a′ and the x linker. A, mass spectrum of the digestion products of hPDI at different redox states. hPDI at 1 mg/ml was incubated with or without 1 mm DTT at 25 °C for 30 min and then digested by 0.3 μg/ml proteinase K, 2 μg/ml trypsin or chymotrypsin for 5 min. The reactions were terminated by adding PMSF to a final concentration of 0.5 mm and analyzed by mass spectrometry. B, mapping the identified redox-dependent digestion sites of hPDI onto the three-dimensional structure of bb′xa′. The digestions sites are shown in red, and the active sites are shown in yellow.
Conformational Transfer Is Essential for Redox-regulated hPDI Chaperone Activity
As indicated above, significant interdomain conformational changes of hPDI occur in response to different redox conditions. We envision that this kind of compact/loose conformation transfer could directly regulate hPDI chaperone activity by shielding/exposing the substrate binding areas. We next tested the chaperone activity of various hPDI mutants with point mutations on the domain interfaces observed in the compact reduced bb′xa′ (such as R300A, W396A, K326E, and E431K) to interfere with the ability of conformational transfer. These mutants showed chaperone activity similar to that of the wild-type hPDI in the oxidized form but higher chaperone activity in the presence of DTT (Fig. 6A), indicating that disruption of the interdomain rearrangement can interfere with redox-dependent chaperone activity changes. To further verify the importance of the a′ domain in the redox-regulated chaperone activity of hPDI, we finally tested different hPDI truncations. Oxidized bb′xa′ suppressed the aggregation of GAPDH more efficiently than oxidized abb′x, and the presence of DTT decreased the chaperone activity of bb′xa′ remarkably (Fig. 6B). Because we and others have previously indicated that the four domains of the PDI molecule are all required for binding to large substrates (21, 22), such as the GAPDH folding intermediate (22), chaperone activity of abb′x still showed some, but much less, redox regulation (Fig. 6B). Taken together, all of the above results demonstrated that the conformational transfer, based on interdomain rearrangement and trigged by the active site of the a′ domain, is essential for the redox-regulated chaperone activity of hPDI.
FIGURE 6.
Redox-regulated chaperone activity of hPDI. A and B, guanidine hydrochloride-denatured GAPDH at 0.14 mm was 50-fold diluted into refolding buffer in the absence or presence of 28 μm (A) or 5.6 μm (B) hPDI proteins with various concentrations of DTT as indicated. Aggregation produced during the refolding was monitored by recording the light scattering at 488 nm, and the suppression of aggregation was used to measure the chaperone activity of hPDI proteins. C, a schematic model of redox-regulated chaperone activity of hPDI. The b, b′, x, and a′ domains are colored in green, orange, magenta, and blue, respectively. The position of the a domain (gray) in this model is achieved by superposition of the bb′xa′ structure with that of yPDI at 4 °C (23). Residues involved in ligand binding (23, 34, 43, 44) are colored in yellow. Active sites of both the a and a′ domains are shown in stick representations. The reduced hPDI adopts the compact conformation with small hydrophobic areas exposed for substrate binding, and oxidation of the a′ domain of hPDI results in exposure of more extended hydrophobic areas for its substrate binding and chaperone activity.
DISCUSSION
PDI is a multidomain protein that functions as both a dithiol-disulfide oxidoreductase and a chaperone to promote the oxidative folding or reductive unfolding of various protein substrates. Conformational changes of PDI are indicated to be a key factor for its promiscuous binding nature (24, 25, 43). The results presented here provide structural insights into the molecular mechanism of redox-regulated interdomain conformational changes and chaperone activity of hPDI.
We found that the redox-regulated conformational changes of hPDI are mainly located in the b′xa′ region and triggered by redox switch of the active site in the a′ domain. Interestingly, this location is in line with the major flexible region in hPDI revealed by our previous work (25), indicating that the large flexibility in the b′xa′ region is required for the interdomain conformational change. Furthermore, the redox-regulated conformational change happened only when an active thioredoxin-like domain (either a or a′) connected to the C termini of b′x, clearly demonstrating that the b′ domain and the x linker are the necessary components for the conformational change of hPDI regulated by redox. Hence, the domain organization integrity of b′xa′ determines its redox-dependent regulation in nature.
The crystal structure of the reduced bb′xa′, which is the longest fragment of hPDI with structure determined so far, allowed us to understand the underlying molecular mechanism. Particularly, it is the first time that the b′xa′ region of hPDI has been observed at the atomic level; this is the most critical region for the versatile activities of hPDI (21, 46–48). The structure revealed a novel domain organization (i.e. a′ domain packs tightly with both the b′ domain and the x linker to form one compact structural module). Previous 1H-15N HSQC spectra comparison of b′-a′oxi and b′-a′red of PDI from thermophilic fungus showed that the redox states of the a′ domain influence the dynamic properties of the b′ domain (34), and the small angle x-ray scattering studies revealed a redox-regulated interdomain motion of the a′ domain (33). Here we uncovered multiple interactions between the b′ and a′ domains in detail according to the reduced bb′xa′ structure. We further identified the regions and residues exposed upon the oxidation of the a′ domain by mass spectrometry analysis, which were reversely shielded in the reduced form of bb′xa′, further demonstrating that the oxidation of the a′ domain releases the compact structure module.
How does the redox switch of the a′ domain active site trigger the large interdomain conformational change? It is found that the residue Trp-396, adjacent to the active site (supplemental Fig. S3B), interacts with domain b′ in the reduced state but is exposed upon oxidation of the a′ domain active site, indicating its important role in sensing the redox change and triggering the redox-regulated interdomain conformational changes. Another region involved in this conformational change is the β2-β3 loop (supplemental Fig. S3B), which extensively interacts with domain b′ in the reduced state and is exposed upon oxidation. In addition, this loop, although far away from the active site in the primary sequence, contains an extremely well conserved (in thioredoxin-like proteins) cis-proline (Pro-441) located adjacent to the active site in the three-dimensional structure (supplemental Fig. S3B) that has been shown to be involved in substrate binding/release based on the studies of thioredoxin (49), DsbA (50), and yPDI (23). Therefore, Pro-441 may also play a critical role in sensing the redox change. Interestingly, both Trp-396 (adjacent to the active site) and His-438 (near the cis-proline) were found in alternative conformations in the reduced bb′xa′ structure (supplemental Fig. S3B). The high mobility of these two residues may contribute to sensing redox microenvironment and triggering conformational changes.
The redox-regulated interdomain conformational changes of the b′ and a′ domain can be further supported by the flexibility of the x linker. It has been reported that the N-terminal part of the x linker can stabilize the b′ domain (18), whereas the C-terminal part loosely interacts with the a′ domain and does not contribute to its structural integrity (51). Actually, the C-terminal portion of the x linker has been reported to adopt multiple conformations (25, 43), which may further increase interdomain mobility of the b′xa′ region and underpin the redox-regulated conformational changes. Besides PDI, a number of other PDI family members also contain a linker connecting the b′ and a′ domains (17), and it is worthwhile to further explore whether such redox-regulated conformational changes exist in these members.
PDI displays its chaperone activity by temporarily binding with unfolded polypeptides/proteins or folding intermediates through hydrophobic interactions, thereby preventing the misfolding and aggregation of substrates (52). ANS fluorescence studies showed that the reduced hPDI exposed smaller hydrophobic area. The crystal structure of reduced bb′xa′ also revealed that several residues in both the b′ and a′ domains, supposed to be involved in substrate binding, are shielded in the compact structure and released to be exposed to solvent upon the oxidation of the a′ domain active site. It is logical to suppose that such prominent conformational changes induced by different redox states may lead to potential regulation of hPDI chaperone activity. Indeed, we showed in this study that the chaperone activity of hPDI in suppressing the aggregation of GAPDH folding intermediates is redox-regulated. A schematic model of redox-regulated chaperone activity of PDI is shown in Fig. 6C.
Previously, PDI was shown as a redox-regulated chaperone to unfold the cholera toxin A1 subunit (29), and bb′xa′ was shown to be necessary and sufficient to trigger the unfolding (48). Therefore, the redox-regulated conformational changes in the b′xa′ region identified here may also be involved in this process. In their toxin unfolding assay, the reduced PDI showed higher binding affinity to bind with the folded toxin A1 subunit and then unfold and release it after being oxidized (29), whereas in our GAPDH refolding assay, the oxidized PDI preferentially associates with partially folded GAPDH folding intermediates to facilitate its folding. There are some other cases: the reduced PDI showed higher binding affinities with Ero1-Lα (53) and antigen peptides (54), and the oxidized PDI displayed higher affinities with mastoparan (34) and α-subunit of prolyl-4-hydroxylase (31). These observations suggested that PDI at different redox states may recognize different substrates; however, the general mechanism is still unknown.
A well studied redox-regulated chaperone, Hsp33, is a conserved heat shock protein in prokaryotes (55). Induced by oxidation along with unfolding, the chaperone activity of Hsp33 is activated with exposure of its buried hydrophobic surfaces in the reduced form. Although the redox-regulated activation of chaperone activity of PDI was previously considered distinct from Hsp33 (29, 56), our results showed that it is similar to Hsp33 (i.e. oxidation of PDI significantly increases its chaperone activity). This feature thereby may endow PDI with the first characterized redox-regulated chaperone in the ER for sensing and protecting protein damage from reactive oxygen species (consistently generated in the ER (39)), further explaining why PDI plays a critical role in protecting neurodegenerative disease induced by oxidative stress and/or protein misfolding (39).
Supplementary Material
Acknowledgments
We thank Prof. Mingjie Zhang for stimulating discussion and helpful advice on this work, Prof. Fuquan Yang and Dr. Zhensheng Xie for mass spectrometry analysis, Wenqi Li for some preliminary work, and Prof. Paul Curmi for kindly reading the paper.
This work was supported by National Major Basic Research Program of China Grants 2011CB910303 and 2011CB910503, National Natural Science Foundation of China Grants 31000351 and 31070657, and Knowledge Innovation Program of the Chinese Academy of Sciences Grants KSCX2-YW-R-154 and KSCX2-EW-J-3.

This article contains supplemental Table S1 and Figs. S1–S3.
The atomic coordinates and structure factors (code 3UEM) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ER
- endoplasmic reticulum
- PDI
- protein-disulfide isomerase
- hPDI
- human PDI
- yPDI
- yeast PDI
- ANS
- 1-anilino-8-naphthalene sulfonate.
REFERENCES
- 1. Sitia R., Braakman I. (2003) Nature 426, 891–894 [DOI] [PubMed] [Google Scholar]
- 2. Goldberger R. F., Epstein C. J., Anfinsen C. B. (1963) J. Biol. Chem. 238, 628–635 [PubMed] [Google Scholar]
- 3. Cai H., Wang C. C., Tsou C. L. (1994) J. Biol. Chem. 269, 24550–24552 [PubMed] [Google Scholar]
- 4. Wilson R., Lees J. F., Bulleid N. J. (1998) J. Biol. Chem. 273, 9637–9643 [DOI] [PubMed] [Google Scholar]
- 5. Freedman R. B., Klappa P., Ruddock L. W. (2002) EMBO Rep. 3, 136–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Molinari M., Galli C., Piccaluga V., Pieren M., Paganetti P. (2002) J. Cell Biol. 158, 247–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee S. O., Cho K., Cho S., Kim I., Oh C., Ahn K. (2010) EMBO J. 29, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pihlajaniemi T., Helaakoski T., Tasanen K., Myllylä R., Huhtala M. L., Koivu J., Kivirikko K. I. (1987) EMBO J. 6, 643–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wetterau J. R., Combs K. A., Spinner S. N., Joiner B. J. (1990) J. Biol. Chem. 265, 9800–9807 [PubMed] [Google Scholar]
- 10. Turano C., Coppari S., Altieri F., Ferraro A. (2002) J. Cell. Physiol. 193, 154–163 [DOI] [PubMed] [Google Scholar]
- 11. Fenouillet E., Barbouche R., Jones I. M. (2007) Antioxid. Redox Signal. 9, 1009–1034 [DOI] [PubMed] [Google Scholar]
- 12. Essex D. W. (2009) Antioxid. Redox Signal. 11, 1191–1225 [DOI] [PubMed] [Google Scholar]
- 13. Hoffstrom B. G., Kaplan A., Letso R., Schmid R. S., Turmel G. J., Lo D. C., Stockwell B. R. (2010) Nat. Chem. Biol. 6, 900–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Uehara T., Nakamura T., Yao D., Shi Z. Q., Gu Z., Ma Y., Masliah E., Nomura Y., Lipton S. A. (2006) Nature 441, 513–517 [DOI] [PubMed] [Google Scholar]
- 15. Walker A. K., Farg M. A., Bye C. R., McLean C. A., Horne M. K., Atkin J. D. (2010) Brain 133, 105–116 [DOI] [PubMed] [Google Scholar]
- 16. Walker A. K., Atkin J. D. (2011) Neurol. Res. Int. 2011, 317340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alanen H. I., Salo K. E., Pekkala M., Siekkinen H. M., Pirneskoski A., Ruddock L. W. (2003) Antioxid. Redox Signal. 5, 367–374 [DOI] [PubMed] [Google Scholar]
- 18. Pirneskoski A., Klappa P., Lobell M., Williamson R. A., Byrne L., Alanen H. I., Salo K. E., Kivirikko K. I., Freedman R. B., Ruddock L. W. (2004) J. Biol. Chem. 279, 10374–10381 [DOI] [PubMed] [Google Scholar]
- 19. Freedman R. B., Gane P. J., Hawkins H. C., Hlodan R., McLaughlin S. H., Parry J. W. (1998) Biol. Chem. 379, 321–328 [DOI] [PubMed] [Google Scholar]
- 20. Darby N. J., Creighton T. E. (1995) Biochemistry 34, 11725–11735 [DOI] [PubMed] [Google Scholar]
- 21. Klappa P., Ruddock L. W., Darby N. J., Freedman R. B. (1998) EMBO J. 17, 927–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun X. X., Dai Y., Liu H. P., Chen S. M., Wang C. C. (2000) Biochim. Biophys. Acta 1481, 45–54 [DOI] [PubMed] [Google Scholar]
- 23. Tian G., Xiang S., Noiva R., Lennarz W. J., Schindelin H. (2006) Cell 124, 61–73 [DOI] [PubMed] [Google Scholar]
- 24. Tian G., Kober F. X., Lewandrowski U., Sickmann A., Lennarz W. J., Schindelin H. (2008) J. Biol. Chem. 283, 33630–33640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang C., Chen S., Wang X., Wang L., Wallis A. K., Freedman R. B., Wang C. C. (2010) J. Biol. Chem. 285, 26788–26797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatahet F., Ruddock L. W. (2009) Antioxid. Redox Signal. 11, 2807–2850 [DOI] [PubMed] [Google Scholar]
- 27. Mezghrani A., Fassio A., Benham A., Simmen T., Braakman I., Sitia R. (2001) EMBO J. 20, 6288–6296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zito E., Melo E. P., Yang Y., Wahlander Å., Neubert T. A., Ron D. (2010) Mol. Cell 40, 787–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsai B., Rodighiero C., Lencer W. I., Rapoport T. A. (2001) Cell 104, 937–948 [DOI] [PubMed] [Google Scholar]
- 30. Tsai B., Rapoport T. A. (2002) J. Cell Biol. 159, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lumb R. A., Bulleid N. J. (2002) EMBO J. 21, 6763–6770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Winter J., Jakob U. (2004) Crit. Rev. Biochem. Mol. Biol. 39, 297–317 [DOI] [PubMed] [Google Scholar]
- 33. Nakasako M., Maeno A., Kurimoto E., Harada T., Yamaguchi Y., Oka T., Takayama Y., Iwata A., Kato K. (2010) Biochemistry 49, 6953–6962 [DOI] [PubMed] [Google Scholar]
- 34. Serve O., Kamiya Y., Maeno A., Nakano M., Murakami C., Sasakawa H., Yamaguchi Y., Harada T., Kurimoto E., Yagi-Utsumi M., Iguchi T., Inaba K., Kikuchi J., Asami O., Kajino T., Oka T., Nakasako M., Kato K. (2010) J. Mol. Biol. 396, 361–374 [DOI] [PubMed] [Google Scholar]
- 35. Li S. J., Hong X. G., Shi Y. Y., Li H., Wang C. C. (2006) J. Biol. Chem. 281, 6581–6588 [DOI] [PubMed] [Google Scholar]
- 36. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Malhotra J. D., Kaufman R. J. (2007) Antioxid. Redox Signal. 9, 2277–2293 [DOI] [PubMed] [Google Scholar]
- 40. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 41. Painter J., Merritt E. A. (2006) Acta Crystallogr. D 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 42. Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 43. Nguyen V. D., Wallis K., Howard M. J., Haapalainen A. M., Salo K. E., Saaranen M. J., Sidhu A., Wierenga R. K., Freedman R. B., Ruddock L. W., Williamson R. A. (2008) J. Mol. Biol. 383, 1144–1155 [DOI] [PubMed] [Google Scholar]
- 44. Denisov A. Y., Määttänen P., Dabrowski C., Kozlov G., Thomas D. Y., Gehring K. (2009) FEBS J. 276, 1440–1449 [DOI] [PubMed] [Google Scholar]
- 45. Dong G., Wearsch P. A., Peaper D. R., Cresswell P., Reinisch K. M. (2009) Immunity 30, 21–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pirneskoski A., Ruddock L. W., Klappa P., Freedman R. B., Kivirikko K. I., Koivunen P. (2001) J. Biol. Chem. 276, 11287–11293 [DOI] [PubMed] [Google Scholar]
- 47. Wang L., Li S. J., Sidhu A., Zhu L., Liang Y., Freedman R. B., Wang C. C. (2009) J. Biol. Chem. 284, 199–206 [DOI] [PubMed] [Google Scholar]
- 48. Forster M. L., Mahn J. J., Tsai B. (2009) J. Biol. Chem. 284, 13045–13056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maeda K., Hägglund P., Finnie C., Svensson B., Henriksen A. (2006) Structure 14, 1701–1710 [DOI] [PubMed] [Google Scholar]
- 50. Kadokura H., Tian H., Zander T., Bardwell J. C., Beckwith J. (2004) Science 303, 534–537 [DOI] [PubMed] [Google Scholar]
- 51. Kozlov G., Määttänen P., Thomas D. Y., Gehring K. (2010) FEBS J. 277, 3924–3936 [DOI] [PubMed] [Google Scholar]
- 52. Wilkinson B., Gilbert H. F. (2004) Biochim. Biophys. Acta 1699, 35–44 [DOI] [PubMed] [Google Scholar]
- 53. Masui S., Vavassori S., Fagioli C., Sitia R., Inaba K. (2011) J. Biol. Chem. 286, 16261–16271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cho K., Cho S., Lee S. O., Oh C., Kang K., Ryoo J., Lee S., Kang S., Ahn K. (2011) Antioxid. Redox Signal. 15, 621–633 [DOI] [PubMed] [Google Scholar]
- 55. Kumsta C., Jakob U. (2009) Biochemistry 48, 4666–4676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sitia R., Molteni S. N. (2004) Sci. STKE 2004, pe27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.