Background: We identified target transcripts of the RNA-binding protein CUGBP1 in resting and activated T cells.
Results: T cell activation induced CUGBP1 phosphorylation, causing decreased CUGBP1 binding to target transcripts.
Conclusion: CUGBP1 binding to a network of target transcripts is regulated by CUGBP1 phosphorylation following T cell activation.
Significance: CUGBP1 target transcripts are coordinately regulated during T cell activation.
Keywords: Cellular Immune Response, MRNA Decay, Protein Phosphorylation, RNA-binding Protein, T Cell, Translation Control, CELF1, CUGBP1, mRNA Degradation, mRNA Stability, RNA Immunoprecipitation
Abstract
The RNA-binding protein, CUG-binding protein 1 (CUGBP1), regulates gene expression at the levels of alternative splicing, mRNA degradation, and translation. We used RNA immunoprecipitation followed by microarray analysis to identify the cytoplasmic mRNA targets of CUGBP1 in resting and activated primary human T cells and found that CUGBP1 targets were highly enriched for the presence of GU-rich elements (GREs) in their 3′-untranslated regions. The number of CUGBP1 target transcripts decreased dramatically following T cell activation as a result of activation-dependent phosphorylation of CUGBP1 and decreased ability of CUGBP1 to bind to GRE-containing RNA. A large percentage of CUGBP1 target transcripts exhibited rapid and transient up-regulation, and a smaller percentage exhibited transient down-regulation following T cell activation. Many of the transiently up-regulated CUGBP1 target transcripts encode important regulatory proteins necessary for transition from a quiescent state to a state of cellular activation and proliferation. Overall, our results show that CUGBP1 binding to certain GRE-containing target transcripts decreased following T cell activation through activation-dependent phosphorylation of CUGBP1.
Introduction
The activation and clonal expansion of human T cells during an immune response requires rapid and precise changes in gene expression that are regulated at multiple levels through transcriptional and posttranscriptional mechanisms (1). The molecular events leading to T cell activation must be tightly controlled to prevent the development of disease states, such as autoimmunity or malignancy (2–4), and it is becoming increasingly clear that posttranscriptional gene regulation at the level of mRNA degradation is critical for normal cellular activation, proliferation, and immune effector function (5, 6). Indeed, over half of the gene expression changes in early T cell activation are a result of changes in mRNA half-life (7). The significance of mRNA decay regulation is highlighted by the fact that T cell malignancies display abnormal stabilization of numerous transcripts that encode proteins that promote cellular growth and proliferation (8).
RNA-binding proteins or microRNAs bind to specific recognition motifs in mRNA and coordinately regulate the posttranscriptional fate of networks of genes involved in cellular responses (9). The best characterized example of a posttranscriptional regulatory network that coordinately regulates gene expression during immune responses is AU-rich element (ARE)5-mediated mRNA decay. AREs are conserved sequence elements found in the 3′-untranslated region (UTR) of transcripts encoding numerous cytokine transcripts and other inflammatory mediators, and AREs function to coordinately regulate mRNA decay during immune responses by interacting with cytoplasmic ARE-binding proteins (10, 11). Another recently described posttranscriptional regulatory network involves the RNA-binding protein CUG-binding protein 1 (CUGBP1), also referred to as CUGBP- and ELAV-like family member 1 (CELF1), which binds to a GU-rich element (GRE) residing in the 3′-UTR of target transcripts and mediates coordinate degradation of GRE-containing transcripts (12). The GRE was originally identified as a sequence that was highly enriched in the 3′-UTR of transcripts that decayed rapidly in primary human T cells and was shown to function as a regulator of mRNA decay (12). Based on a bioinformatic analysis of mRNA targets of CUGBP1 in HeLa cells, the GRE was defined to be the consensus sequence UGU(G/U)UGU(G/U)UGU (13). Binding by CUGBP1 to certain GRE-containing transcripts has been shown to promote their rapid degradation, but how this process is regulated during cellular activation is poorly understood.
In addition to regulating mRNA degradation in the cytoplasm, CUGBP1 has other functions as a regulator of alternative splicing and translation (14). In the nucleus, CUGBP1 regulates the alternative splicing of a number of transcripts (15, 16), whereas in the cytoplasm, CUGBP1 binds to the untranslated regions of transcripts and regulates their translation efficiency or stability (14). In addition to binding to GREs in the 3′-UTR of mRNA, CUGBP1 can bind to some transcripts that contain a GC-rich element in their 5′-UTR and promote their translation (17, 18). The function of CUGBP1 is regulated by phosphorylation. In a mouse model of myotonic dystrophy, it has been shown that CUGBP1 is phosphorylated by protein kinase C (PKC), resulting in altered cellular distribution and stability of the CUGBP1 protein, correlating with altered splicing patterns (19, 20). The CUGBP1 target transcript TNF-α was stabilized upon chemical activation of the PKC pathway (21). In addition, the RNA binding specificity of CUGBP1 has been shown to be altered via phosphorylation by cyclin D3-Cdk4/6 (22). These results suggest that CUGBP1 is regulated by phosphorylation, which could provide a mechanism for activation-induced changes in CUGBP1 function during cellular processes, such as T cell activation.
Here, we used RNA immunoprecipitation (RNA-IP) followed by microarray analysis (23) to investigate the cytoplasmic target transcripts of CUGBP1 in resting and activated primary human T cells. We found that CUGBP1 target transcripts in resting and activated T cells were highly enriched for the presence of the GRE in their 3′-UTRs, but the number of CUGBP1 target transcripts decreased dramatically following T cell activation. The decrease in the number of CUGBP1 targets upon T cell activation was caused by activation-dependent phosphorylation of CUGBP1 and decreased ability of CUGBP1 to bind to GRE-containing RNA. A large percentage of CUGBP1 target transcripts exhibited rapid and transient up-regulation, and a smaller percentage exhibited transient down-regulation following T cell activation. Many of the transiently up-regulated CUGBP1 target transcripts encode important regulators necessary for transition from a quiescent state to a state of cellular activation and proliferation. Our data support a model whereby CUGBP1 phosphorylation regulates a network of transcripts involved in T cell activation. Overall, our results verify that CUGBP1 binds to GRE-containing target transcripts in primary human T cells and that its ability to bind to mRNA is altered following T cell activation.
EXPERIMENTAL PROCEDURES
Purification of Human T Cells
Primary human T cells were purified from peripheral blood mononuclear cells by negative selection using the CD3+ Rosette-Sep antibody mixture from Stem Cell Technologies as described previously (10). CD3+ lymphocytes were then isolated through a Ficoll-Hypaque cushion (GE Healthcare).
T Cell Stimulation
Purified human T cells were cultured overnight in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 units/ml penicillin G, and 100 μg/ml streptomycin. Cells were then incubated for 6 h in 15-cm dishes (5 × 107 cells/dish) with medium alone or with a combination of immobilized monoclonal antibodies (1 μg/ml) directed against the CD3 component of the TCR complex (R&D Systems) and the CD28 co-stimulatory molecule (R&D Systems) as described previously (12).
RNA-IP and Microarray Analysis
Cytoplasmic extracts were prepared from resting and stimulated T cells, and RNA-IP reactions were performed as described previously, using antibodies targeting hemagglutinin (HA) (F7, Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)), CUGBP1 (3B1, Santa Cruz Biotechnology, Inc.) or poly(A)-binding protein (PABP) (ImmunoQuest) (13). Three RNA-IP experiments were performed using resting or activated T cells isolated from three individual anonymous donors. For each experiment, RNA was purified from the input and immunoprecipitated material from equal numbers of resting and stimulated T cells using the RNeasy kit (Qiagen) following the manufacturer's recommendations. For the input RNA, 5 μg was used to prepare labeled cRNA for microarray hybridizations. SF9 insect cell RNA was added to the immunoprecipitated RNA such that the total amount of RNA was 5 μg. This RNA was then used to prepare cRNA using the MessageAMP III RNA amplification kit (Ambion). The cRNA from the input and RNA-IP samples from resting and stimulated lysates were hybridized to Affymetrix U133a Plus-2 microarrays. Microarrays were normalized using the gene content robust multiarray average algorithm using Genespring 11.0 software (Agilent Technologies Inc). Transcripts were determined to be present if the log2 normalized signal from the input microarray was greater than the log2 normalized signal from the HA microarray with p < 0.05 as determined by Welch's t test (R) in either the resting or the stimulated conditions.
Target Transcript Identification
Transcripts were determined to be CUGBP1 targets if the difference between the log2 normalized signal from the microarrays hybridized with the cRNA from the anti-CUGBP1 RNA-IP and the anti-PABP RNA-IP was greater than the same value derived from the difference between the anti-HA RNA-IP and anti-PABP RNA-IP, with p < 0.005, as determined by Welch's t test (R), in either resting or stimulated conditions.
Two-dimensional Western Blotting
100 μg of cytoplasmic lysates were diluted 1:1 with rehydration buffer from Bio-Rad and dialyzed against two-dimensional gel buffer (7 m urea, 2 m thiourea, 2% CHAPS, 10 mm Tris) overnight at room temperature. The samples were then loaded onto 10-cm pH 3–10 IPG strips. Samples were focused using a Bio-Rad Protean IEF cell. Subsequently, IPG strips were loaded onto BisTris 4–12% precast gels and run at 175 V using MOPS-SDS buffer. Gels were then blotted onto charged PVDF, and Western blots were performed by probing with an anti-CUGBP1 antibody (3B1) from Santa Cruz Biotechnology, Inc. For λ-phosphatase treatment, 100 μg of cytoplasmic lysate was incubated with CUGBP1 antibody for 90 min at 4 °C and subsequently incubated with protein A/G-Sepharose beads from Pierce for 90 min at 4 °C. Beads were washed three times with RNA-IP lysis buffer and resuspended in 100 μl of λ-phosphatase buffer. Samples were then treated with λ-phosphatase (New England Biolabs) or were mock-treated, and subsequently, CUGBP1 was eluted from the beads by the addition of 1% SDS and heating at 65 °C for 15 min. Samples were then diluted 1:1 with rehydration buffer and were separated by two-dimensional electrophoresis.
EMSA and UV Cross-linking Assay Using T Cell Cytoplasmic Lysates
Biotinylated GRE RNA was ordered from Sigma-Aldrich containing the sequence 5′-biotin-GAGUGUGUGUGUGUGUGUGUGUUGUUU-3′. Mutated, biotinylated RNA was also purchased from Sigma-Aldrich and had the sequence 5′-biotin-GACACAGUGUCACAGUGUCACAUGUUU-3′. Binding reactions were the same as described previously (10) using 10 μg of cytoplasmic lysate and 25 fmol of biotinylated RNA. For EMSA, electrophoresis was performed using 0.5× TBE buffer and 5% polyacrylamide gels under non-denaturing conditions. For the UV cross-linking assay, reactions were exposed to UV light and were separated by SDS-PAGE on a 10% polyacrylamide gel. In the indicated reactions, CUGBP1 was immunoprecipitated by the addition of an anti-CUGBP1 antibody for 90 min at 4 °C, followed by the addition of protein A/G beads for 90 min at 4 °C. Beads were washed three times, and the material on the beads was separated by SDS-PAGE. Following electrophoresis, gels were blotted onto nylon membranes and UV-cross-linked. The blots were then exposed using the Chemiluminescent Nucleic Acid Detection Module from Thermo Scientific, following the manufacturer's instructions.
Reverse Transcription-Polymerase Chain Reaction
cDNA used for reverse transcription-PCR (RT-PCR) amplification was synthesized from total cellular RNA using the StrataScript reverse transcriptase (Stratagene) using oligo(dT). PCR amplifications were then performed as described previously (24). Quantitative real-time PCR assays were performed using Roche Applied Science Universal Probe Library technology, and transcript abundance was normalized to the level of the β2-microglobulin transcript. Oligonucleotide primers used for all assays are shown in supplemental Table 2.
UV Cross-linking Assay Using Immunopurified CUGBP1
CUGBP1 was immunoprecipitated from 20 μg of cytoplasmic lysate, and samples were treated with λ-phosphatase or were mock-treated as described under “Two-dimensional Western Blotting.” The immunoprecipitated material was then incubated for 30 min with 25 fmol of either the biotinylated GRE or mutant GRE probe as well as a 10-fold excess of unlabeled poly(U) RNA and 5 mg/ml heparan sulfate in 20 μl of RBB buffer (10). Following incubation, the reactions were treated with UV light, and material was eluted from the beads by incubation in SDS loading buffer for 5 min at 95 °C. The eluted material was separated by SDS-PAGE on a 10% acrylamide gel, and the gel was blotted onto nylon membranes. Subsequently, the biotinylated RNA probe was visualized with the Chemiluminescent Nucleic Acid Detection Module from Thermo Scientific, following the manufacturer's instructions. The blots were then stripped and probed by Western blot with an anti-CUGBP1 antibody. Blots were then quantified with ImageJ to determine the RNA/CUGBP1 ratio.
RESULTS
GREs Are Enriched in CUGBP1 Target Transcripts in Primary Human T Cells
We recently showed that the GRE was highly enriched in short lived transcripts expressed in primary human T cells (12) and that CUGBP1 bound to the GRE and mediated the decay of GRE-containing transcripts (13). The transcriptional network targeted by CUGBP1 and the regulation of CUGBP1 function in T cells remain poorly understood. In order to better understand the role of CUGBP1 in T cell function, we sought to identify the cytoplasmic mRNA targets of CUGBP1 in primary human T cells using RNA-IP, followed by analysis of targets using Affymetrix U133A Plus 2.0 microarrays. Immunoprecipitation experiments were performed in triplicate using antibodies targeting CUGBP1, PABP, or HA. For each experiment, transcript abundance in input RNA and immunoprecipitated RNA was analyzed using Affymetrix U133A Plus 2.0 microarrays. The results of these analyses revealed that CUGBP1 in resting primary human T cells was associated with 1245 unique transcripts, corresponding to 1309 Affymetrix probe sets. A subset of these target transcripts were verified to be CUGBP1 targets using traditional RT-PCR, because an anti-CUGBP1 antibody specifically co-immunoprecipitated JunD, topoisomerase I, and ubiquitin-protein ligase E3A transcripts in cytoplasmic extracts from resting T cells (Fig. 1, lane 5). GAPDH, which is not predicted to be a CUGBP1 target transcript, was not copurified with CUGBP1. Immunoprecipitation with an antibody against HA did not copurify any of these transcripts (lane 3), whereas an antibody against PABP copurified all of these transcripts (lane 1).
FIGURE 1.
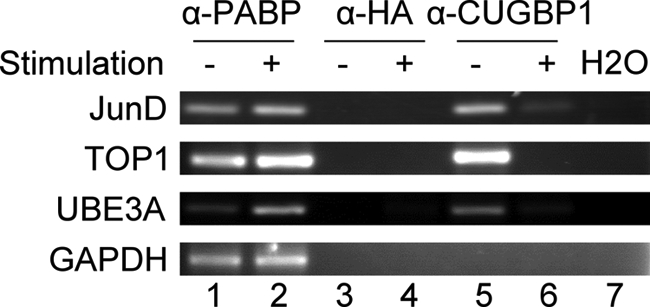
Validation of CUGBP1 target transcripts by RT-PCR. RNA-IP was performed using cytoplasmic lysates from unstimulated (−) and stimulated (+) T cells and antibodies targeting PABP (α-PABP), HA (α-HA), and CUGBP1 (α-CUGBP1). The levels of Topoisomerase I (TOP1), JunD proto-oncogene (JunD), ubiquitin-protein ligase E3A (UB3EA), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) transcripts were measured in the immunoprecipitated material by RT-PCR.
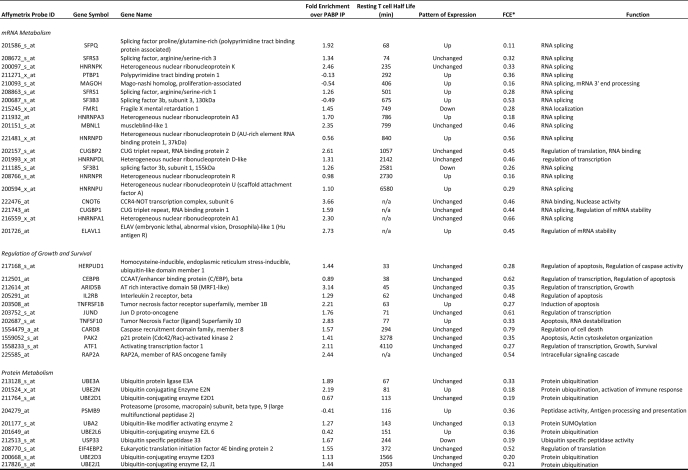
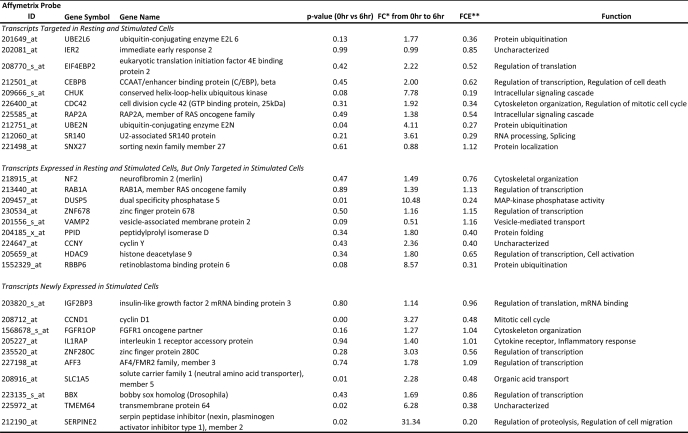
Transcripts determined to be targets of CUGBP1 encoded proteins serving various functions, including mRNA processing, metabolism of RNA, protein turnover, as well as regulators of cell death and proliferation. Transcripts encoding RNA-binding proteins were particularly enriched in the CUGBP1 targets, suggesting that in resting T cells, CUGBP1 may be functioning as a posttranscriptional “regulator of regulators” whereby CUGBP1 influences the expression of a network of target transcripts encoding RNA-binding proteins, which in turn regulate individual subnetworks of transcripts. A partial list of important CUGBP1 target transcripts is shown in Table 1, and a full list can be found in supplemental Table 1.
TABLE 1.
CUGBP1 target transcripts (resting T cells)
* -Fold change in enrichment as defined under “Results.”
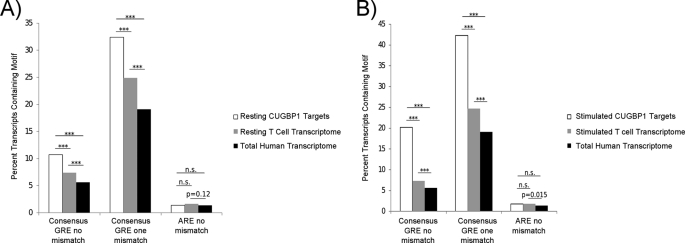
CUGBP1 binds preferentially to a GU-rich RNA motif, UGU(G/U)UGU(G/U)UGU, found in the 3′-UTR of target transcripts (13). We sought to determine whether our experimentally determined CUGBP1 target transcripts in T cells were enriched for this motif. GenBankTM files were downloaded from the NCBI database, and custom C++ scripts were written to extract the 5′-UTRs, coding sequences, and 3′-UTRs from CUGBP1 target transcripts as well as all transcripts present in the genome (total human transcriptome) and all transcripts present in the resting human T cell transcriptome, determined as described under “Experimental Procedures.” A bioinformatic search of these sequences for the GRE, allowing one or two mismatches, was performed, and no enrichment of the GRE was found in the 5′-UTR or coding region of the target transcripts compared with either the total human transcriptome or the resting T cell transcriptome. In contrast, a significant enrichment of the GRE was found in the 3′-UTR of CUGBP1 target transcripts compared with both the total human transcriptome and the T cell transcriptome (Fig. 2A). These differences were highly statistically significant (p < 0.001 for both zero mismatches and one mismatch, χ2 test). Interestingly, the resting T cell transcriptome exhibited a small but significant enrichment in GRE-containing transcripts (p < 0.001 for both zero mismatches and one mismatch, χ2 test) when compared with the total human transcriptome frequency, suggesting that GRE-containing transcripts may be expressed preferentially in T cells. To control for specificity of the CUGBP1-GRE interaction, we investigated the prevalence of the 11-mer ARE (UAUUUAUUUAU), allowing for no mismatches, and found no enrichment in the CUGBP1 IP compared with the resting T cell transcriptome or the total human transcriptome. Similar to the GRE, we observed a trend toward enrichment of the ARE in the T cell transcriptome compared with the total human transcriptome. It has been reported previously that CUGBP1 also binds to a CUG/CCG sequence, such as that found in the 5′-UTR of CCAAT/enhancer-binding protein β (17). We did not observe enrichment of this motif in the 5′-UTR, coding region, or 3′-UTR of CUGBP1 target transcripts.
FIGURE 2.
GREs were enriched in the 3′-UTRs of CUGBP1 target transcripts expressed in primary human T cells. The 3′-UTRs from the total human transcriptome were extracted from NCBI refseq records. A, the prevalence of the consensus GRE or ARE sequence in the 3′-UTRs of CUGBP1 target transcripts from resting T cells (Resting CUGBP1 Targets) was compared with the presence of these sequences in the total human transcriptome or the resting T cell transcriptome. B, the prevalence of the consensus GRE or ARE sequences in the 3′-UTRs of CUGBP1 target transcripts from T cells that were stimulated for 6 h with anti-CD3 and anti-CD28 antibodies (Stimulated CUGBP1 Targets) was compared with the presence of these sequences in the total human transcriptome or the stimulated T cell transcriptome. For the GRE, we allowed zero mismatches or one mismatch, and for the ARE, we allowed zero mismatches. ***, p < 0.001; n.s., not significant; χ2 test).
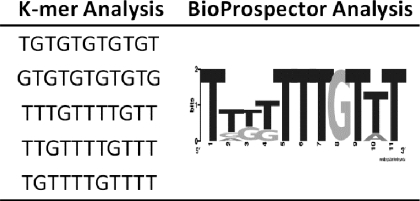
As an independent approach to identify CUGBP1 target sequences, we performed de novo motif searches on the 3′-UTRs of the experimentally determined CUGBP1 target transcripts. First, the 3′-UTRs were submitted to a simple overrepresentation analysis, where the frequency of all oligonucleotide sequences of length k (k-mers) in the target 3′UTRs was compared with the frequency of the same k-mers in the total human transcriptome for k values from 8 to 13. Using custom C++ scripts, k-mers were then sorted by the difference of the two frequencies to identify the most highly enriched k-mers. We found that several GRE-like sequences were the most highly enriched motifs (Table 2, left). These results corroborated previous reports showing that both (UGUU)n motifs and (UG)n motifs were enriched in the 3′-UTRs of CUGBP1 target transcripts (13, 25). Additionally, the 3′-UTRs from CUGBP1 target transcripts were submitted to a BioProspector motif search (26) to evaluate enriched sequences. The top result from this search was also a GRE-like sequence (Table 2, right). Overall, these results confirm that CUGBP1 has preference for binding to GREs.
TABLE 2.
Sequence analysis of CUGBP1 target transcripts
Only a Subset of CUGBP1 Target Transcripts Exhibit Short Half-lives
CUGBP1 has been shown to promote the instability of reporter transcripts that contain the GRE in their 3′-UTRs (13). Furthermore, short lived transcripts in primary human T cells were significantly enriched for transcripts containing a GRE in their 3′-UTR (12). We sought to determine whether CUGBP1 target transcripts in primary human T cells exhibited short half-lives. To answer this question, we utilized the results from a previously published global assessment of mRNA half-lives in resting and activated primary human T lymphocytes (27). Briefly, primary human T cells were either stimulated for 3 h with anti-CD3 and anti-CD28 antibodies or allowed to rest, and transcription was halted by administration of actinomycin D. Total cellular RNA was then isolated at 0, 1.5, and 3 h. Transcript abundance was assessed using U95 Affymetrix microarrays, and half-lives were calculated based on a first order decay model. The U133A+2.0 Affymetrix probe set IDs of the CUGBP1 targets were converted to the corresponding U95 Affymetrix probe set ID using the ID converter tool available from Babylomics. We identified 198 short lived GRE-containing CUGBP1 target transcripts in resting T cells with half-lives less than 180 min. Ingenuity Pathway Analyst was used to investigate the functions of the proteins encoded by these short lived target transcripts, and this analysis revealed an enrichment of transcripts involved in cell death (p = 1.4 × 10−4), protein degradation/ubiquitination (p = 0.02), and proliferation (p = 0.002). All of these cellular functions must be turned on upon T cell stimulation and play a crucial role in the acquisition of an activated phenotype. CUGBP1 may be functioning to maintain these transcripts at a low cellular abundance in resting T cells in order to maintain the quiescent phenotype. Some of these short lived transcripts along with their respective half-lives and biological function can be found in Table 1.
Although a subset of CUGBP1 target transcripts exhibited rapid decay, most CUGBP1 target transcripts were relatively stable in resting T cells. The average half-life of CUGBP1 target transcripts in resting T cells was 1405 min, whereas the average half-life of all transcripts expressed in resting T cells was 1588 min. This finding shows that CUGBP1 targets showed a statistically significant skewing toward shorter half-lives (p = 0.001, Welch's t test) consistent with the observation that CUGBP1 promotes transcript decay. Interestingly, when comparing CUGBP1 target transcripts that do and do not contain a consensus GRE in their 3′-UTR, it was observed that GRE-containing transcripts had shorter half-lives in resting T cells than those that did not contain a GRE (p = 0.029, Welch's t test). No difference was found in the average half-life of those transcripts containing the UGUUUGUUUGU version of the GRE versus the GU repeat version of the GRE.
Altered CUGBP1 Association with Target Transcripts upon T Cell Activation
We next sought to compare the targets of CUGBP1 in resting T cells with T cells that were stimulated for 6 h with anti-CD3 and anti-CD28 antibodies in order to determine if CUGBP1 played a role in the dynamic changes in gene expression that occur following T cell activation. Using the same RNA-IP approach followed by microarray analysis, as described above, we discovered that CUGBP1 associated with only 168 unique transcripts, corresponding to 153 probe sets in activated T cells. Comparing the CUGBP1 target transcripts in resting T cells with activated T cells, we found that there was an overlap of these populations of only 53 probe sets, suggesting that most of the 1309 probe sets that associated with CUGBP1 in resting T cells were no longer associated with CUGBP1 after T cell activation. Of the 1256 probe sets bound by CUGBP1 only in resting T cells, 1219 (97%) were present in the transcriptomes of both resting and stimulated T cells, suggesting that the lack of association with CUGBP1 in activated cells was not due to the disappearance of these transcripts. Using traditional RT-PCR, we verified that a subset of these transcripts showed decreased association with CUGBP1 in immunoprecipitates from activated T cells (Fig. 1, compare lanes 5 and 6), despite the fact that the association of these transcripts with PABP increased (compare lanes 1 and 2). To more quantitatively examine the effect of T cell activation on the relationship between the change in the abundance of a transcript in the cell relative to its abundance in the CUGBP1 RNA-IP, we defined a -fold change in enrichment (FCE) for each transcript on the microarray as follows,
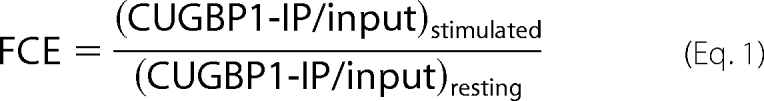 |
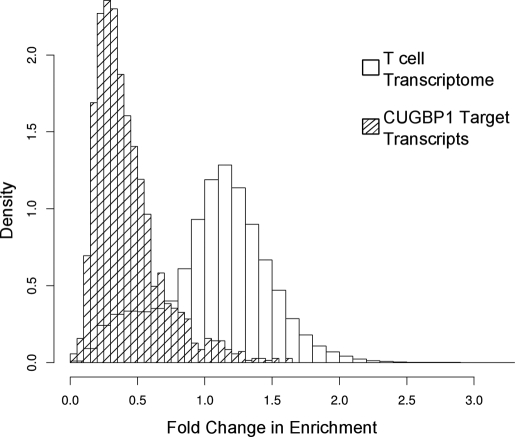
where the ratio of the microarray signal from the CUGBP1 RNA-IP (CUGB1-IP) to the microarray signal from input RNA for stimulated T cells is divided by the same ratio for resting T cells. The average FCE for all microarray probe sets was 1.084, suggesting that the association of most transcripts with CUGBP1 did not change following T cell activation. In contrast, the average FCE for the CUGBP1 target transcripts was 0.422, suggesting that changes in the abundance of these transcripts in the cell could not explain their lack of association with CUGBP1. The distribution of FCE for both the T cell transcripts and the CUGBP1 target transcripts is shown in Fig. 3.
FIGURE 3.
Decreased association of CUGBP1 with target transcripts following T cell activation. The FCE was calculated for all T cell transcripts (blank bars) and CUGBP1 target transcripts (dashed bars), and a histogram of these values is depicted. On the y axis, density is defined as the normalized number of transcripts falling in a given FCE bin, such that the total area of the histogram is 1. The CUGBP1 target transcript population exhibits a reduced FCE compared with all T cell transcripts, suggesting decreased association of these transcripts with CUGBP1 following T cell activation.
Interestingly, although most CUGBP1 target transcripts bound to CUGBP1 only in resting T cells, we identified 100 probe sets that were bound by CUGBP1 only in stimulated T cells and not in resting T cells. Of these, 40 probe sets were induced by T cell activation and were not present in the resting condition. The remaining 60 probe sets were expressed in both the resting and stimulated conditions but were only bound in the stimulated condition. A list of a subset of CUGBP1 target transcripts in stimulated cells can be found in Table 3. We investigated the prevalence of the GRE, allowing for zero mismatches or one mismatch, in the 3′-UTRs of the CUGBP1 target transcripts in stimulated T cells as described above. As can be seen in Fig. 2B, we found enrichment of transcripts containing a GRE in the CUGBP1 targets compared with the activated T cell transcriptome or the total human transcriptome (p < 0.001 for both zero mismatches and one mismatch, χ2 test). Among transcripts bound in both resting and activated T cells, 46 (82%) contained a GRE, whereas among transcripts bound in only stimulated cells but expressed in both resting and stimulated conditions, 38 (68%) contained a GRE. Finally, in those transcripts bound only in stimulated cells and newly expressed in the stimulated condition, 20 (54%) contained a GRE. Ingenuity pathway analysis showed that the biological pathways of RNA metabolism, apoptosis, and cell cycle control were enriched in the CUGBP1 targets in stimulated T cells.
TABLE 3.
CUGBP1 target transcripts in stimulated T cells
* -Fold change.
** -Fold change in enrichment as defined under “Results.”
CUGBP1 Target Transcripts Exhibit Changes in Gene Expression upon T Cell Activation
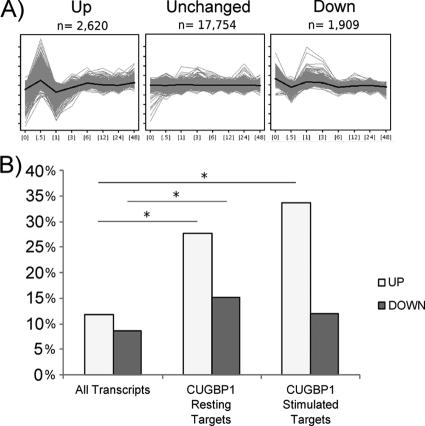
We used previously generated data that assessed gene expression using Affymetrix U133A microarrays in primary human T cells stimulated with anti-CD3/28 antibodies for 0 min, 30 min, 1 h, 3 h, 6 h, 12 h, 24 h, and 48 h (24) to determine the patterns of expression of CUGBP1 targets following T cell activation. As can be seen in Fig. 4A, we found that T cell transcripts exhibited three predominant patterns of expression, which we have termed “unchanged”, “up,” and “down.” Our findings showed that most transcripts in the T cell transcriptome were unchanged, consistent with recent studies suggesting that ∼80% of the transcriptome is ubiquitously expressed (28) and thus would not be expected to change in abundance upon stimulation. As can be seen in Fig. 4B, we found that CUGBP1 target transcripts expressed in resting cells were statistically enriched for transcripts that showed significant activation-induced regulation, exhibiting both the up (p < 2.2 × 10−16, χ2 test) and down (p = 2.2 × 10−9, χ2 test) patterns of expression, with a strong preference for the up pattern. Similarly, CUGBP1 target transcripts in stimulated cells were enriched for transcripts following the up pattern of expression (p = 1.1 × 10−6, χ2 test). Of those transcripts that followed the up pattern of expression, there was a strong enrichment of transcripts coding for proteins that participate in pre-mRNA processing and alternative splicing, including several heterogeneous ribonucleoproteins. We validated the expression profile of three of the transcripts encoding RNA-binding proteins following the up pattern and one following the down pattern using qRT-PCR. As can be seen in supplemental Table 3, the -fold change in expression after 6 h of stimulation determined by qRT-PCR correlated closely to that observed in the microarray data set. A list of a subset of CUGBP1 target transcripts involved in mRNA processing and following the up pattern of expression can be found in Table 1. Overall, our data suggest that CUGBP1 target transcripts were highly enriched for transcripts that show dynamic changes in expression over the course of T cell activation.
FIGURE 4.
Expression patterns of CUGBP1 target transcripts following T cell activation. A, gene expression was previously assessed using microarrays over the first 48 h of T cell stimulation with anti-CD3 and anti-CD28 antibodies (24). Using these data, the T cell transcriptome was segregated into three common patterns of expression using a K-means clustering approach. The “up” pattern of expression is shown on the left with a sharp increase in abundance at 30 min followed by a return to baseline at 1 h and a subsequent slow increase over the following 47 h. The “down” pattern of expression is shown on the right and exhibits an inverse pattern compared with the up pattern. Finally the majority of transcripts follow the “unchanged” pattern, which shows little change in abundance over T cell stimulation. On the y axis of each panel are normalized, log2 microarray expression values, and the x axis depicts hours of T cell stimulation. For each pattern of gene expression, the average expression pattern of each cluster is depicted by the black line with the range of expression values of each cluster depicted by the gray lines. B, we compared the percentage of transcripts expressing the up or down patterns for all transcripts expressed in primary T cells at one or more time points throughout the first 48 h of T cell activation (All Transcripts), CUGBP1 target transcripts in resting T cells (CUGBP1 Resting Targets), and CUGBP1 target transcripts in stimulated T cells (CUGBP1 Stimulated Targets). *, p < 0.05, χ2 test.
Decreased CUGBP1 Binding to RNA Is Due to Activation-induced Phosphorylation
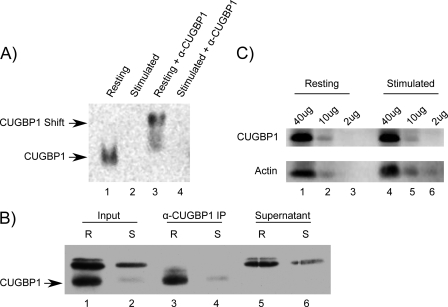
The finding that the majority of CUGBP1 target transcripts had an FCE of <1 at 6 h following T cell activation suggested that an activation-dependent change in CUGBP1 binding occurred. To test this hypothesis, we performed electromobility shift assays (EMSAs) using a GRE-containing RNA probe and cytoplasmic lysates prepared from resting T cells and T cells that were stimulated for 6 h with anti-CD3 and anti-CD28 antibodies. As can be seen in Fig. 5A, we observed CUGBP1 binding to the GRE probe in resting T cell lysates (lane 1). This binding activity was due to CUGBP1 because it was specifically supershifted with an anti-CUGBP1 antibody (lane 3) but was not supershifted with a control anti-HA antibody (data not shown). In stimulated T cell lysates, however, we observed minimal to no binding to the GRE (lane 2). Additionally, we performed a UV cross-linking assay using cytoplasmic lysates from resting and activated T cells and confirmed that CUGBP1 bound to the GRE probe (Fig. 5B). The identity of the CUGBP1 band was determined by immunoprecipitation with an anti-CUGBP1 antibody (lanes 3 and 4) but not a control antibody (data not shown). The lack of a CUGBP1 band in the supernatant from the immunoprecipitation reaction (lanes 5 and 6) indicated that the immunoprecipitation depleted CUGBP1 from the lysates. The intensity of the CUGBP1 band decreased significantly in the lysates from stimulated T cells, confirming the results of the EMSA showing that binding by CUGBP1 to the GRE probe decreased following T cell activation. The lack of binding by CUGBP1 in extracts from stimulated T cells was not due to a decrease in CUGBP1 protein abundance, because Western blot performed on the same extracts showed that similar CUGBP1 protein levels were present in extracts from resting and activated T cells (Fig. 5C). These findings suggest that the decrease in CUGBP1 binding to RNA upon T cell activation was due to a posttranslational effect.
FIGURE 5.
Binding by CUGBP1 to the GRE decreased following T cell activation. A, EMSAs were performed by incubating a biotinylated GRE-containing oligonucleotide with cytoplasmic extracts from resting T cells (Resting) or T cells that had been activated with anti-CD3 and anti-CD28 antibodies for 6 h (Stimulated). An antibody against CUGBP1 was added to the indicated reactions (α-CUGBP1). The CUGBP1-containing band and the supershifted band are shown with arrows. B, cytoplasmic lysates from resting (R) and stimulated (S) T cells were mixed with a biotinylated GRE probe and treated with UV light. Reactions were mixed with an anti-CUGBP1 antibody, and the non-immunoprecipitated reactions (Input), the anti-CUGBP1 immunoprecipitated fractions (α-CUGBP1 IP), and the immunoprecipitation supernatants (Supernatant) were separated by SDS-PAGE. The CUGBP1 band is indicated with an arrow. C, Western blot analysis was performed on titrated amounts (2–40 μg of protein) of cytoplasmic lysates from resting and activated T cells using anti-CUGBP1 and anti-actin antibodies.
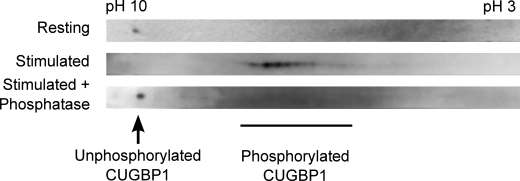
CUGBP1 is a phosphoprotein, and the binding specificity of CUGBP1 has been shown to be altered, depending on the phosphorylation state (19, 22). We hypothesized that posttranslational phosphorylation of CUGBP1 may occur in human T cells and may explain the reduced RNA-binding activity of CUGBP1 upon T cell activation. In order to investigate this hypothesis, two–dimensional Western blot analysis of CUGBP1 was performed in resting and activated T lymphocytes (Fig. 6). Compared with the CUGBP1 signal in extracts prepared from resting T cells, the CUGBP1 signal in extracts prepared from activated T cells exhibited an acidic shift (Fig. 6, top two panels). This shift was a result of phosphorylation, because treatment of lysates with λ-phosphatase resulted in the reversion of the CUGBP1 signal back to its position in resting cells. These results suggest that CUGBP1 was not phosphorylated in resting T cells, but upon T cell stimulation, CUGBP1 became phosphorylated, and this phosphorylation correlated with reduced binding to GRE-containing RNA.
FIGURE 6.
CUGBP1 was phosphorylated upon T cell activation. Cytoplasmic lysates were prepared from resting T cells and T cells that were stimulated for 6 h with anti-CD3 and anti-CD28 antibodies. The lysates were immunoprecipitated using an anti-CUGBP1 antibody, and the immunoprecipitated material was treated with λ-phosphatase or mock-treated. Samples were then separated by two-dimensional electrophoresis, and CUGBP1 was identified by Western blot analysis.
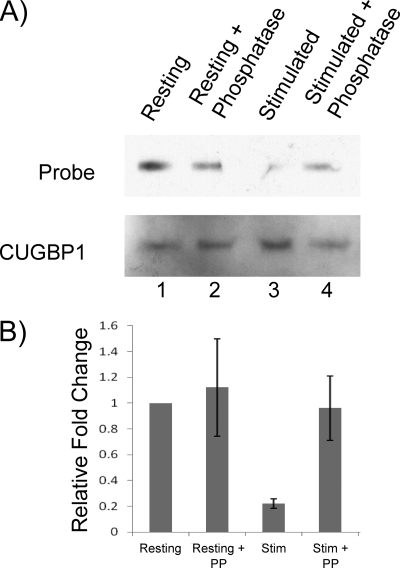
We next sought to determine directly if the reduction in CUGBP1 affinity for the GRE was a result of CUGBP1 phosphorylation. We immunoprecipitated CUGBP1 from resting and stimulated T cell lysates and then subjected it to λ-phosphatase or mock treatment. The immunopurified CUGBP1 was subsequently incubated with either biotinylated GRE sequences or mutated GRE sequences. These reactions were then UV-cross-linked, and the amount of RNA that cross-linked to CUGBP1 was determined after separating the mixtures by electrophoresis and probing for biotinylated RNA (Fig. 7A, top). Subsequently, the same membranes were probed with an anti-CUGBP1 antibody to determine the amount of CUGBP1 protein immunopurified from resting and activated T cells (Fig. 7A, bottom). Using this assay, we observed that CUGBP1 immunopurified from resting T cells bound to the GRE probe (lane 1) but not the mutated GRE probe (data not shown). Phosphatase treatment had no effect on binding by CUGBP1 from resting T cells to the GRE probe (compare lanes 1 and 2). In contrast, CUGBP1 immunopurified from stimulated T cells bound poorly to the GRE RNA probe (Fig. 7A, lane 3), but we observed an increase in CUGBP1 binding to the GRE probe upon phosphatase treatment (Fig. 7A, compare lanes 3 and 4). The amount of GRE RNA bound, normalized to the amount of CUGBP1 precipitated, was quantified in triplicate experiments for resting or activated T cells (Fig. 7B). The results showed a significant decrease in CUGBP1 binding to the GRE in the stimulated condition (p = 0.001). Upon phosphatase treatment, binding by immunopurified CUGBP1 to GRE increased significantly (p = 0.03) and was similar to the binding observed for the CUGBP1 immunopurified form resting T cells. Overall, these results indicate that phosphorylation of CUGBP1 caused a decrease in CUGBP1 binding to GRE RNA in stimulated T cells, and this effect was reversed by phosphatase treatment.
FIGURE 7.
CUGBP1 phosphorylation caused reduced affinity of CUGBP1 binding to GRE RNA. A, CUGBP1 was immunopurified from resting (Resting) and stimulated (Stim) T cell lysates and subsequently treated with either λ-phosphatase (PP) or underwent mock treatment. The immunopurified CUGBP1 was then mixed with a biotinylated GRE probe and treated with UV light. The CUGBP1-RNA complexes were separated by SDS-PAGE, and the biotinylated GRE probe (Probe) and CUGBP1 (CUGBP1) levels were visualized. B, the experiment in A was performed three times, and the results were quantified. For each reaction, the RNA/CUGBP1 ratio was determined, and the ratios were normalized to the resting, mock-treated condition for each experiment. Error bars, S.D. of three experiments.
DISCUSSION
CUGBP1 regulates posttransciptional gene expression at a number of levels, including alternative splicing, translation, and mRNA degradation, and functions by binding directly to RNA (14). A network of GRE-containing genes is coordinately regulated at posttranscriptional levels through the interaction of the CUGBP1 with its target sequence, the GRE (12). In the present study, we sought to understand the dynamics of the CUGBP1-GRE interaction throughout the process of T cell activation and performed RNA-IP followed by microarray analysis to identify CUGBP1 target transcripts. We found ∼1300 CUGBP1 target transcripts in resting T cells and ∼150 target transcripts in activated T cells. CUGBP1 targets in both resting and activated T cells displayed significant enrichment for GRE-containing transcripts, confirming that the GRE is indeed a CUGBP1-binding site within cells. We also showed that CUGBP1 phosphorylation following T cell activation correlated with the decrease in the number of CUGBP1 target transcripts that we observed as well as a decreased ability of CUGBP1 to bind to GRE sequences in vitro. We further showed that phosphatase treatment of CUGBP1 immunopurified from stimulated T cells restored its ability to bind to the GRE by increasing the affinity of CUGBP1 for the GRE.
Several results suggest that CUGBP1 function is regulated by phosphorylation, but a unifying theme has not emerged. In some situations, CUGBP1 phosphorylation is associated with increased binding to RNA. For example, phosphorylation of CUGBP1 at Ser-28 by Akt kinase led to increased affinity of CUGBP1 for cyclin D1 mRNA (22), and phosphorylation at Ser-302 by cyclin D3-Cdk4/6 led to increased CUGBP1 binding to p21 and C/EBPβ mRNA (18). In a myotonic dystrophy model, CUGBP1 was hyperphosphorylated by PKCα and βII, causing increased protein half-life and altered splicing patterns (19, 30). Expression of a (CUG)960-expanded myotonic dystrophy protein kinase gene caused CUGBP1 phosphorylation and inhibition of CUGBP1-mediated decay of the TNF-α transcript (21). It is thought that CUGBP1 binds to a CG-rich motif in the 5′-UTR of certain transcripts and promotes their translation (17, 31). It has been found in a liver model that following partial hepatectomy, there is phosphorylation of CUGBP1. It was further shown that this phosphorylation event led to an increase in the association and translational efficiency of specific CUGBP1 target transcripts. This effect was shown to be mediated by an increased association with the translational initiation factor eIF2 (17). Here, we found that CUGBP1 is phosphorylated upon T cell activation and that this caused a decrease in binding by CUGBP1 to cytoplasmic mRNA targets. Multiple kinase cascades are triggered upon T cell activation, leading to several potential candidates for CUGBP1 phosphorylation. Further efforts will focus on identifying the phosphorylation site and kinase(s) involved in activation-dependent phosphorylation of CUGBP1. Our data show that the affinity of CUGBP1 for the GRE is regulated by phosphorylation and suggest a powerful mechanism of coordinate posttranscriptional regulation whereby a cell could quickly alter the stability of a whole network of transcripts. For transcripts exhibiting the up pattern of expression, a possible scenario is that unphosphorylated CUGBP1 promotes the coordinate decay of transcripts in resting T cells, but upon stimulation, CUGBP1 becomes phosphorylated, leading to stabilization of these transcripts and allowing for transient increased expression. It is also possible that CUGBP1 phosphorylation may play a role in regulating the translation of these transcripts. For transcripts exhibiting the down pattern of expression, CUGBP1-mediated mRNA decay may play a role in mediating the down-regulation of these transcripts, and CUGBP1 phosphorylation may regulate their subsequent return to base-line levels.
The targets of CUGBP1 have been identified in two previous studies. The first study was performed on the HeLa human cervical carcinoma cell line and identified ∼600 CUGBP1 target transcripts, many of which were involved in RNA processing, cellular proliferation, and apoptosis (13). The second study was performed on the mouse C2C12 myoblast cell line and identified 881 CUGBP1 target transcripts, many of which were also involved in RNA processing, cellular proliferation, and apoptosis (25). These other studies were both performed using transformed cell lines that have been shown to display widespread alterations in the 3′-UTR isoform (32). We found relatively little overlap between the actual targets of CUGBP1 between our study in primary human T cells and these previous studies performed in transformed cell lines. This disparity could be a result of differential regulation of other trans-acting factors as well as differences in the transcriptomes of the different cell types. CUGBP1 targets showed significant enrichment of GRE-containing transcripts in all three cell types, and although there was little overlap in specific target transcripts, the biological processes of proliferation, apoptosis, and RNA processing were enriched among target transcripts in all three cell types.
Interestingly, CUGBP1 target transcripts were enriched for transcripts encoding RNA-binding proteins, including its own transcript. Thus, CUGBP1 may be similar to other RNA-binding proteins in exhibiting autoregulatory behavior and functions as a “regulator of regulators” (33, 34). Here we also confirmed the previous reports that CUGBP1 seems to be targeting transcripts that are involved in cellular proliferation and cell death (13, 35). One pathway we found to be particularly enriched in CUGBP1 targets in resting cells was that of mRNA formation. T cell activation induces changes in alternative polyadenylation and alternative splicing (36, 37), and it would be interesting to determine if CUGBP1 plays a regulatory role in these processes.
Overall, our data are consistent with a model whereby CUGBP1 is actively involved in coordinately repressing transcripts involved in mRNA maturation, proliferation, and cell death to maintain the cell in a quiescent state. Upon stimulation, phosphorylation of CUGBP1 leads to its inability to bind to certain transcripts, allowing for accumulation of target transcripts necessary for acquisition of the characteristics of activated T cells, such as proliferation and increased sensitivity to apoptotic stimuli. Inactivation of CUGBP1 plays a role in the development of leukemia through dysregulation of the transcription factor C/EBPβ (38). Additionally, a forward genetic screen in mice identified CUGBP1 loss of function as a potential driving mutation in the development of colorectal cancer (29). This suggests that CUGBP1 may be regulating cell survival and proliferation pathways. CUGBP1 could regulate these pathways both via direct binding to transcripts coding for proteins involved in these pathways and through regulation of splicing factors and other RNA-binding proteins, which in turn regulate mRNA isoform usage, mRNA decay, and/or translation patterns that promote survival and proliferation.
In conclusion, we have shown that CUGBP1 coordinates networks of transcripts crucial for primary human T cell activation. Upon T cell activation, we observed dramatic reduction in the CUGBP1 target transcript pool and showed that this change in CUGBP1 binding to RNA was due to phosphorylation of CUGBP1. Further work will focus on delineating the signaling pathways involved in CUGBP1 phosphorylation in primary human T cells and the mechanism of CUGBP1-mediated regulation of the activated T cell phenotype.
Supplementary Material
Acknowledgments
We thank the Biomedical Genomics Center in the Academic Health Center at the University of Minnesota and the Minnesota Supercomputing Institute for expertise and services. We also thank Dr. Cavan S. Reilly for helpful discussions regarding the bioinformatic analysis.
This work was supported, in whole or in part, by National Institutes of Health Grants AIO57484 and AIO72068 (to P. R. B.).

This article contains supplemental Tables 1–3.
- ARE
- AU-rich element
- GRE
- GU-rich element
- RNA-IP
- RNA immunoprecipitation
- PABP
- poly(A)-binding protein
- IPG
- immobilized pH gradient
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- FCE
- -fold change in enrichment.
REFERENCES
- 1. Dustin M. L. (2009) The cellular context of T cell signaling. Immunity 30, 482–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crispín J. C., Tsokos G. C. (2009) Transcriptional regulation of IL-2 in health and autoimmunity. Autoimmun. Rev. 8, 190–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Merup M. (1998) Genetic abnormalities in non-Hodgkin's lymphomas and chronic lymphocytic leukaemia. Med. Oncol. 15, 79–88 [DOI] [PubMed] [Google Scholar]
- 4. Pearson P. L., Van der Luijt R. B. (1998) The genetic analysis of cancer. J. Intern. Med. 243, 413–417 [DOI] [PubMed] [Google Scholar]
- 5. Sanchez-Lockhart M., Marin E., Graf B., Abe R., Harada Y., Sedwick C. E., Miller J. (2004) Cutting edge. CD28-mediated transcriptional and posttranscriptional regulation of IL-2 expression are controlled through different signaling pathways. J. Immunol. 173, 7120–7124 [DOI] [PubMed] [Google Scholar]
- 6. Stellato C., Gubin M. M., Magee J. D., Fang X., Fan J., Tartar D. M., Chen J., Dahm G. M., Calaluce R., Mori F., Jackson G. A., Casolaro V., Franklin C. L., Atasoy U. (2011) Coordinate regulation of GATA-3 and Th2 cytokine gene expression by the RNA-binding protein HuR. J. Immunol. 187, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheadle C., Fan J., Cho-Chung Y. S., Werner T., Ray J., Do L., Gorospe M., Becker K. G. (2005) Control of gene expression during T cell activation. Alternate regulation of mRNA transcription and mRNA stability. BMC Genomics 6, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vlasova I. A., McNabb J., Raghavan A., Reilly C., Williams D. A., Bohjanen K. A., Bohjanen P. R. (2005) Coordinate stabilization of growth-regulatory transcripts in T cell malignancies. Genomics 86, 159–171 [DOI] [PubMed] [Google Scholar]
- 9. Keene J. D. (2007) RNA regulons. Coordination of post-transcriptional events. Nat. Rev. Genet. 8, 533–543 [DOI] [PubMed] [Google Scholar]
- 10. Ogilvie R. L., Abelson M., Hau H. H., Vlasova I., Blackshear P. J., Bohjanen P. R. (2005) Tristetraprolin down-regulates IL-2 gene expression through AU-rich element-mediated mRNA decay. J. Immunol. 174, 953–961 [DOI] [PubMed] [Google Scholar]
- 11. Ogilvie R. L., Sternjohn J. R., Rattenbacher B., Vlasova I. A., Williams D. A., Hau H. H., Blackshear P. J., Bohjanen P. R. (2009) Tristetraprolin mediates interferon-γ mRNA decay. J. Biol. Chem. 284, 11216–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vlasova I. A., Tahoe N. M., Fan D., Larsson O., Rattenbacher B., Sternjohn J. R., Vasdewani J., Karypis G., Reilly C. S., Bitterman P. B., Bohjanen P. R. (2008) Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell 29, 263–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rattenbacher B., Beisang D., Wiesner D. L., Jeschke J. C., von Hohenberg M., St Louis-Vlasova I. A., Bohjanen P. R. (2010) Analysis of CUGBP1 targets identifies GU-repeat sequences that mediate rapid mRNA decay. Mol. Cell. Biol. 30, 3970–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vlasova-St Louis I., Bohjanen P. R. (2011) Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1. Curr. Opin. Genet. Dev. 21, 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han J., Cooper T. A. (2005) Identification of CELF splicing activation and repression domains in vivo. Nucleic Acids Res. 33, 2769–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kalsotra A., Xiao X., Ward A. J., Castle J. C., Johnson J. M., Burge C. B., Cooper T. A. (2008) A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 105, 20333–20338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Timchenko N. A., Wang G. L., Timchenko L. T. (2005) RNA CUG-binding protein 1 increases translation of 20-kDa isoform of CCAAT/enhancer-binding protein β by interacting with the α and β subunits of eukaryotic initiation translation factor 2. J. Biol. Chem. 280, 20549–20557 [DOI] [PubMed] [Google Scholar]
- 18. Timchenko L. T., Salisbury E., Wang G. L., Nguyen H., Albrecht J. H., Hershey J. W., Timchenko N. A. (2006) Age-specific CUGBP1-eIF2 complex increases translation of CCAAT/enhancer-binding protein β in old liver. J. Biol. Chem. 281, 32806–32819 [DOI] [PubMed] [Google Scholar]
- 19. Kuyumcu-Martinez N. M., Wang G. S., Cooper T. A. (2007) Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell 28, 68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang G. S., Kuyumcu-Martinez M. N., Sarma S., Mathur N., Wehrens X. H., Cooper T. A. (2009) PKC inhibition ameliorates the cardiac phenotype in a mouse model of myotonic dystrophy type 1. J. Clin. Invest. 119, 3797–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang L., Lee J. E., Wilusz J., Wilusz C. J. (2008) The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells. Implications for myotonic dystrophy. J. Biol. Chem. 283, 22457–22463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salisbury E., Sakai K., Schoser B., Huichalaf C., Schneider-Gold C., Nguyen H., Wang G. L., Albrecht J. H., Timchenko L. T. (2008) Ectopic expression of cyclin D3 corrects differentiation of DM1 myoblasts through activation of RNA CUG-binding protein, CUGBP1. Exp. Cell Res. 314, 2266–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tenenbaum S. A., Carson C. C., Lager P. J., Keene J. D. (2000) Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. U.S.A. 97, 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raghavan A., Dhalla M., Bakheet T., Ogilvie R. L., Vlasova I. A., Khabar K. S., Williams B. R., Bohjanen P. R. (2004) Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics 84, 1002–1013 [DOI] [PubMed] [Google Scholar]
- 25. Lee J. E., Lee J. Y., Wilusz J., Tian B., Wilusz C. J. (2010) Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PLoS One 5, e11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu X., Brutlag D. L., Liu J. S. (2001) BioProspector. Discovering Conserved DNA Motifs in Upstream Regulatory Regions of Co-Expressed Genes. Proceedings of the Pacific Symposium on Biocomputing, 127–138 [PubMed] [Google Scholar]
- 27. Raghavan A., Ogilvie R. L., Reilly C., Abelson M. L., Raghavan S., Vasdewani J., Krathwohl M., Bohjanen P. R. (2002) Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 30, 5529–5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramsköld D., Wang E. T., Burge C. B., Sandberg R. (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 5, e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Starr T. K., Allaei R., Silverstein K. A., Staggs R. A., Sarver A. L., Bergemann T. L., Gupta M., O'Sullivan M. G., Matise I., Dupuy A. J., Collier L. S., Powers S., Oberg A. L., Asmann Y. W., Thibodeau S. N., Tessarollo L., Copeland N. G., Jenkins N. A., Cormier R. T., Largaespada D. A. (2009) A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science 323, 1747–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koshelev M., Sarma S., Price R. E., Wehrens X. H., Cooper T. A. (2010) Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol. Genet. 19, 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Iakova P., Wang G. L., Timchenko L., Michalak M., Pereira-Smith O. M., Smith J. R., Timchenko N. A. (2004) Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 23, 406–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang E. T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S. F., Schroth G. P., Burge C. B. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Al-Ahmadi W., Al-Ghamdi M., Al-Haj L., Al-Saif M., Khabar K. S. (2009) Alternative polyadenylation variants of the RNA binding protein, HuR. Abundance, role of AU-rich elements and auto-regulation. Nucleic Acids Res. 37, 3612–3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mukherjee N., Lager P. J., Friedersdorf M. B., Thompson M. A., Keene J. D. (2009) Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol. Syst. Biol. 5, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kress C., Gautier-Courteille C., Osborne H. B., Babinet C., Paillard L. (2007) Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol. Cell. Biol. 27, 1146–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sandberg R., Neilson J. R., Sarma A., Sharp P. A., Burge C. B. (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320, 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Whistler T., Chiang C. F., Lonergan W., Hollier M., Unger E. R. (2010) Implementation of exon arrays. Alternative splicing during T-cell proliferation as determined by whole genome analysis. BMC Genomics 11, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi W. T., Folsom M. R., Azim M. F., Meyer C., Kowarz E., Marschalek R., Timchenko N. A., Naeem R. C., Lee D. A. (2007) C/EBPβ suppression by interruption of CUGBP1 resulting from a complex rearrangement of MLL. Cancer Genet. Cytogenet. 177, 108–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.