Background: MOV10 inhibits HIV-1 replication after being packaged.
Results: A Gag binding plus all but one of seven helicase domains are required for MOV10 packaging. Nearly all residues are required for anti-HIV-1 activity.
Conclusion: Gag binding is not sufficient for MOV10 packaging, and multiple discontinuous domains regulate MOV10 activity.
Significance: These findings uncover a new packaging mechanism and provide new insights into MOV10 antiviral activity.
Keywords: Human Immunodeficiency Virus, P body, Retrovirus, RNA Helicase, Stress Granule, Virus-Host Interaction
Abstract
Discovery of novel antiretroviral mechanism is essential for the design of innovative antiretroviral therapy. Recently, we and others reported that ectopic expression of Moloney leukemia virus 10 (MOV10) protein strongly inhibits retrovirus replication. MOV10, a putative RNA helicase, can be packaged into HIV-1 virions by binding to the nucleocapsid (NC) region of Gag and inhibit viral replication at a postentry step. Here, we report critical determinants for MOV10 virion packaging and antiviral activity. MOV10 has 1,003 amino acids and seven helicase motifs. We found that MOV10 packaging requires the NC basic linker, and Gag binds to the N-terminal amino acids 261–305 region of MOV10. Our predicted MOV10 three-dimensional structure model indicates that the Gag binding region is located in a structurally exposed domain, which spans amino acids 93–305 and is Cys-His-rich. Simultaneous mutation of residues Cys-188, Cys-195, His-199, His-201, and His-202 in this domain significantly compromised MOV10 anti-HIV-1 activity. Notably, although MOV10-Gag interaction is required, it is not sufficient for MOV10 packaging, which also requires its C-terminal all but one of seven helicase motifs. Moreover, we have mapped the minimal MOV10 antiviral region to amino acids 99–949, indicating that nearly all MOV10 residues are required for its antiviral activity. Mutations of residues Cys-947, Pro-948, and Phe-949 at the C terminus of this region completely disrupted MOV10 anti-HIV-1 activity. Taken together, we have identified two critical MOV10 packaging determinants and eight other critical residues for anti-HIV-1 activity. These results provide a molecular basis for further understanding the MOV10 antiretroviral mechanism.
Introduction
Moloney leukemia virus 10 homolog (MOV10)2 was originally discovered from mice as a GTP-binding protein (1). Later, it was found that MOV10 has seven highly conserved helicase motifs in the C-terminal region, and MOV10 was then proposed as a putative superfamily-1 (SF-1) RNA helicase (2). Helicases separate duplex DNA and/or RNA into single strands in an ATP-dependent reaction. They may have up to seven helicase motifs (I, Ia, II, III, IV, V, and IV) and are classified into three superfamilies (SF-1, SF-2, SF-3) and two small families (F-4, F-5) (3). The physiological function of MOV10 is still unclear, but MOV10 was found in the RNA-induced silencing complex (4) as well as in the mRNA processing (P) bodies and stress granules (5), indicating that MOV10 may play an important role in RNA biology and metabolism. Recently, we and others reported that ectopic expression of MOV10 inhibits a wide range of retrovirus replication, including HIV-1, simian immunodeficiency virus, murine leukemia virus, and equine infectious anemia virus (6–8). Notably, MOV10 has multiple antiviral activities. It not only inhibits viral infectivity (6–8), but also reduces viral production, blocks Gag proteolytic processing, and lowers Gag protein stability (6). To inhibit viral infectivity, MOV10 is packaged into HIV-1 virions from producer cells via an interaction with the nucleocapsid (NC) region of Gag in an RNA-dependent manner (8). After that, it blocks viral reverse transcription directly or indirectly in the target cells, resulting in ∼20–100-fold reduction of viral infectivity. Thus, MOV10 has very broad and potent antiretroviral activities, and understanding this novel antiretroviral mechanism will provide new strategies for developing more effective antiretroviral therapies.
Here, we studied the mechanism of MOV10 packaging into HIV-1 virions, which is an essential step for inhibition of viral infectivity. Using a combination of deletion, mutation, and homology modeling techniques, we have identified two critical domains that determine its packaging. In addition, we also identified several other residues that are critical for antiviral activity. These results provide new insights into the as yet poorly defined MOV10 antiretroviral mechanism.
EXPERIMENTAL PROCEDURES
Plasmids
pNL-LucΔenv, pcDNA3.1-MOV10-V5-His6, and pcDNA3.1-A3G-V5-His6 were described before (8, 9). The Gag-CFP fusion constructs expressing MA-CFP, Gag377-CFP, Gag384-CFP, Gag391-CFP, Gag405-CFP, Gag411-CFP, Gag426-CFP, Gag432-CFP, and full-length Gag-CFP were obtained from the Spearman laboratory (10, 11). These Gag genes were derived from a codon-optimized HXB2 Gag sequence (14). Previously, we also cloned this Gag gene into the pcDNA3.1 vector to express a C-terminally GST-V5-tagged Gag protein (8). The pcDNA3.1-Gag-HA-FLAG expression vector was created from this pcDNA3.1-Gag-GST-V5-His6 vector by replacing the GST tag with a HA-FLAG linker after NotI/XbaI digestion. To express MOV10 deletion mutants, partially deleted MOV10 genes were amplified by PCR and then cloned into the pcDNA3.1-V5-His6 vector using the pcDNA3.1 Directional TOPO® Expression kit (Invitrogen). Other MOV10 mutations were created directly in the pcDNA3.1-MOV10-V5-His6 vector using the QuikChange XL site-directed mutagenesis kit (Stratagene). All of these constructs were verified by DNA sequencing.
MOV10 Virion Incorporation Assay
HIV-1 particles and virus-like particles (VLPs) containing MOV10 were produced similarly from 293T cells cultured in 6-well plates by transfecting them with a 3-μg HIV-1 proviral construct pNL-LucΔenv and 3-μg MOV10 expression vector, or 3-μg Gag-CFP fusion construct and 3-μg MOV10 expression vector, respectively. Supernatants were harvested 48 h after transfection and clarified by 12,000 rpm centrifugation for 2 min at 4 °C. Virions were further purified by spinning the clarified supernatants through a 20% sucrose cushion at 296,000 × g for 30 min at 4 °C using the S100AT6 rotor (Sorvall). Viral pellets were resuspended in phosphate-buffered saline (PBS) and analyzed by Western blotting.
Determination of MOV10 and Gag Interaction by Anti-HA Immunoprecipitation
293T cells cultured in 6-well plates were transfected with a 3-μg MOV10 expression vector and 3-μg codon-optimized Gag-HA-FLAG expression vector. Cells were then lysed with a buffer containing 50 mm Tris-HCl, pH 7.6, 100 mm NaCl, 1% Triton X-100, and 5 mm EDTA. The cytosolic fractions were collected and incubated with anti-HA-agarose beads (Sigma) for 3 h at 4 °C. Beads were then loaded on Micro Bio-Spin chromatography columns (catalogue 732-6204; Bio-Rad) and washed extensively with Tris-buffered saline containing 25 mm Tris-HCl, pH 7.2, and 150 mm NaCl. After that, beads were boiled in sample loading buffer containing 60 mm Tris-HCl, pH 6.8, 7.8% glycerol, 2% SDS, 1.55% dithiothreitol, and 0.001% bromphenol blue, and bead-associated proteins were analyzed by Western blotting.
MOV10 Anti-HIV-1 Activity Assay
Viruses were produced from 293T cells cultured in 6-well plate by transfecting them with a 3-μg MOV10 expression vector, 3-μg pNL-LucΔenv, and 0.3-μg VSV-G expression vector. Viral production was measured by the p24Gag capture enzyme-linked immunosorbent assay (ELISA). Virions equivalent to ∼5 ng of p24Gag were used to infect 3 × 105 CEM-SS cells in a 48-well plate by spinoculation at 1,200 × g, 25 °C, for 2 h. Cells were cultured for 36 h and were lysed in a buffer (25 mm Tris-HCl, pH 7.8, 2 mm dithiothreitol, 2 mm 1,2-diaminocyclohexane-N,N,N',N'-tetraacetic acid, 10% glycerol, 1% Triton X-100). After removing the nuclei, the cytosolic fraction was used to determine the luciferase activity with a luciferase assay kit (Promega).
Western Blotting
Horseradish peroxidase (HRP)-conjugated anti-HA antibody (Roche Applied Science) or HRP-conjugated anti-V5 antibody (Invitrogen) was used to directly detect Gag, APOBEC3G (A3G), and MOV10 proteins that express HA or V5 tag. The rabbit anti-human MOV10 polyclonal antibody was from Proteintech Group. The mouse anti-GFP monoclonal antibody and goat anti-actin polyclonal antibody (C-11) were from Santa Cruz Biotechnology. The mouse anti-HIV-1 Gag monoclonal antibody (183-H12-5C) was from the National Institutes of Health AIDS Research and Reference Reagent Program. HRP-conjugated anti-mouse, rabbit, and goat immunoglobulin G secondary antibodies were from Pierce. Detection of the HRP-conjugated antibody was performed using an enhanced chemiluminescence detection kit (Amersham Biosciences).
Homology Modeling of MOV10 Protein Structure
The MOV10 protein three-dimensional structure was modeled using Discovery Studio Client 2.5 (Accelrys Software, San Diego, CA). The template for sequence-based homology modeling is from human nonsense-mediated decay factor up frameshift 1 (UPF1) (Protein Data Bank code 2WJY) (12). Five models were generated and evaluated. The final model was selected after energy minimization. The energy calculations were all based on CHARMm (Chemistry at Harvard Macromolecular Mechanics). The parameters for sequence alignment, build homology, and energy minimization were basically setup as default.
RESULTS
Packaging of MOV10 into HIV-1 Virions from Human CD4+ T Cells
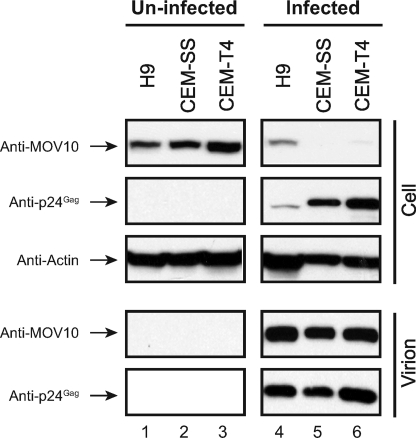
Virion packaging is an essential and critical step for MOV10 to inhibit HIV-1 infectivity. Our previous demonstration of MOV10 virion packaging was done in an overexpressed condition (8). It is important to know whether MOV10 is incorporated into HIV-1 virions during normal replication in CD4+ T cells. To address this issue, we infected human CD4+ T cell lines H9, CEM-SS, and CEM-T4 with HIV-1. After detection of productive HIV-1 replication, HIV-1 virions were purified from the culture medium by ultracentrifugation, and proteins were analyzed by Western blotting. It was found that HIV-1 virions were specifically detected from the supernatant of infected cells, and importantly, MOV10 was detected from these virions (Fig. 1, lanes 4-6, lower panels). This result is consistent with the previous finding by Chertova et al. that MOV10 is packaged into HIV-1 virions from monocyte-derived macrophages (13). It was also found that MOV10 expression was significantly reduced in HIV-infected cells (Fig. 1, upper panels), which supports our previous conclusion that HIV-1 can suppress MOV10 expression (8). Thus, MOV10 is packaged into HIV-1 virions during natural HIV-1 infection.
FIGURE 1.
MOV10 is packaged into HIV-1 virions from human CD4+ T cells. 3 × 105 H9, CEM-SS, or CEM-T4 cells were infected with NL HIV-1 equivalent to 20 ng of p24Gag, respectively. Six days later, virions were purified from the culture supernatants by ultracentrifugation through a 20% sucrose cushion. Cells and virions were lysed, and proteins were analyzed by Western blotting. MOV10 and A3G were detected by anti-V5 antibodies, and Gag-CFP fusions were detected by anti-GFP antibodies.
Requirement of HIV-1 NC Basic Linker for MOV10 Packaging
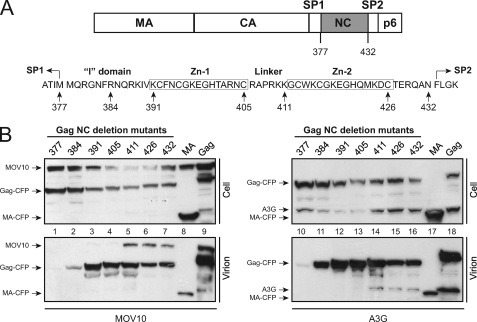
Although we demonstrated that MOV10 interacts with NC for packaging (8), the precise binding domain is still unclear. To map the minimal NC region, we utilized a panel of previously described Gag-CFP fusion constructs that express matrix (MA), capsid (CA), spacer peptide 1 (SP1), and different NC regions of Gag (10, 11). The Gag gene in these constructs was derived from the codon-optimized version of HXB2 Gag sequence (14), so their expression is Rev-independent.
HIV-1 NC consists of a N-terminal interaction (I) domain, followed by two zinc fingers, which are separated by a basic linker (Fig. 2A). Seven NC deletion constructs that span the NC region were selected for mapping: the Gag377-CFP expresses MA, CA, and SP1; the Gag384-, Gag391-, Gag405-, Gag411-, and Gag426-CFP additionally express the N-terminal NC region up to the minimal I domain, the entire I domain, the first zinc finger, the basic linker, or the second zinc finger, respectively; and the Gag432-CFP expresses the full-length NC (Fig. 2A). Two other constructs, MA-CFP, which only expresses MA, and Gag-CFP, which expresses the full-length Gag, were used as controls. All these constructs except for MA-CFP and Gag377-CFP were shown to produce VLPs with normal density (10, 11), and they were used successfully to demonstrate that A3G packaging requires the NC basic linker (10).
FIGURE 2.
MOV10 packaging into HIV-1 requires the NC basic linker. A, schematic illustrates p55Gag and NC for creating Gag-CFP fusion constructs. The number indicates the C-terminal AA residue expressed, with the first Gag residue considered 1. B, MOV10 or A3G was expressed with indicated Gag-CFP fusion proteins in 293T cells. Virions were purified through a 20% sucrose cushion via ultracentrifugation. Cellular and virion-associated proteins were determined by Western blotting with the indicated antibodies.
We then transfected each of these constructs with a MOV10 or A3G expression vector into 293T cells and purified VLPs from the supernatants by ultracentrifugation. It was found that A3G, MOV10, and all CFP fusion proteins were expressed at similar levels in cells. In addition, all these CFP fusion proteins except for the Gag377-CFP and MA-CFP were able to produce VLPs (Fig. 2B), which is consistent with previous reports (10, 11). Nonetheless, A3G and MOV10 proteins were only detected in VLPs produced from the Gag411-, Gag426-, Gag432-, and full-length Gag-CFP fusion constructs (Fig. 2B, lanes 5–7, 9, 14–16, and 18). These results indicate that like the A3G protein, MOV10 also requires the NC basic linker for virion packaging.
N-terminal AA 261–305 Region of MOV10 Interacts with HIV-1 Gag
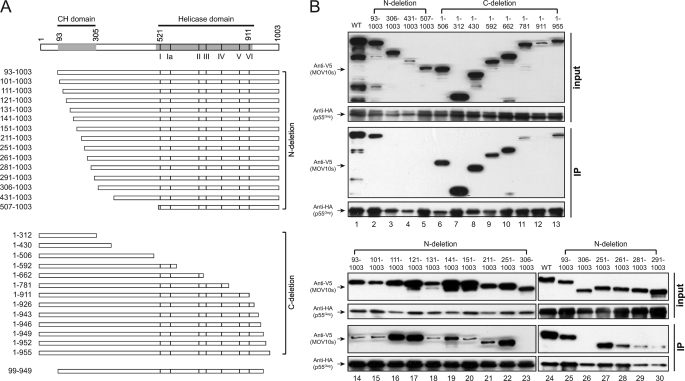
Next, we studied which domain of MOV10 interacts with HIV-1 Gag. MOV10 has 1,003 AA and seven C-terminal helicase motifs (I, Ia, II, III, IV, V, and VI) (Fig. 3A). Initially, we created 12 MOV10 N-terminal and C-terminal deletion mutants (Fig. 3B, lanes 2–13). After that, the wild-type (WT) or mutant MOV10 proteins were expressed with codon-optimized HIV-1 HXB2 Gag that has a C-terminal HA-FLAG tag in 293T cells, and MOV10-Gag interaction was determined by immunoprecipitation with anti-HA antibodies.
FIGURE 3.
HIV-1 Gag binds to the N-terminal AA 261–305 region of MOV10. A, schematic illustrates MOV10 N-terminal or C-terminal deletion mutations. Numbers indicate AA positions. The locations of the putative CH domain (see Fig. 6) and seven helicase motifs are indicated. The border of the putative CH domain, the first residue of the helicase motif I, and the last residue of the helicase motif VI are numbered. B, Gag binding domain of MOV10 was mapped by immunoprecipitation (IP). The indicated MOV10 deletion mutants were expressed with HA-tagged codon-optimized HIV-1 HXB2 Gag in 293T cells. Proteins were pulled down by anti-HA antibody and analyzed by Western blotting.
It was found that all these MOV10 proteins except for two mutants 431–1003 and 1–911 were well expressed (Fig. 3B, lanes 2–13). Among these well expressed MOV10 proteins, only two mutants 306–1003 and 507–1003 were not pulled down by Gag (Fig. 3B, lanes 3 and 5). Because Gag could pull down mutant 93–1003 but not mutant 306–1003, and all the other MOV10 mutants that were pulled down by Gag express the AA 93–305 region, we concluded that the AA 93–305 region is required for MOV10-Gag interaction. Next, we created another 12 MOV10 mutants bearing a series of N-terminal deletion in the AA 93–305 region to map the exact Gag binding domain (Fig. 3B, lanes 15–22 and 27-30). It was found that mutants bearing N-terminal deletions up to the AA 251 position were all efficiently pulled down by Gag (Fig. 3B, lanes 15–22). However, mutant 261–1003 was pulled down at relatively lower efficiency, and this efficiency for mutants 281–1003 and 291–1003 was further reduced (Fig. 3B, lanes 28–30). We then created another mutant 271–1003, but this mutant was not expressed (data not shown). Thus, we concluded that the AA 261–305 region is required for MOV10 interaction with Gag.
Gag Binding Domain of MOV10 Is Required but Not Sufficient for Virion Packaging
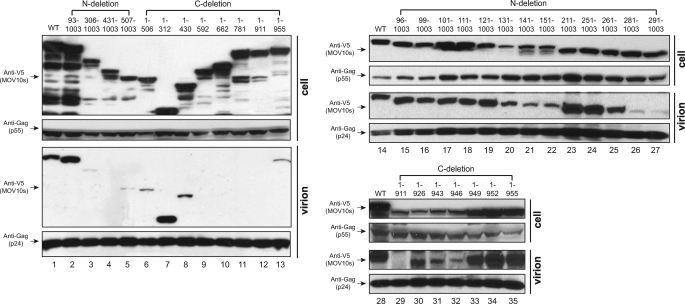
Having mapped the Gag binding domain, we determined how it contributes to virion packaging. MOV10 mutants were expressed with a HIV-1 proviral construct in 293T cells, and virions were purified from culture supernatant by ultracentrifugation. After that, virion-associated proteins were detected by Western blotting.
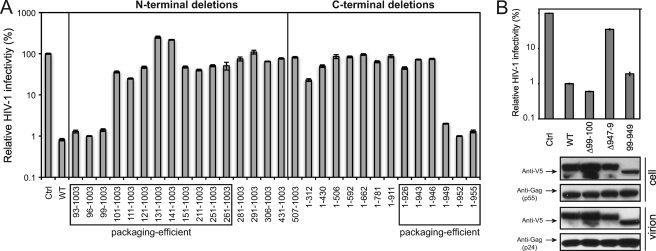
When those MOV10 N-terminal deletion mutants were compared (Fig. 4, lanes 2–5 and 15–27), we found that those mutants interacting with Gag were all packaged into virions (Fig. 4, lanes 2 and 15–25), and mutants 281–1003, 291–1003, 306–1003, 431–1003, and 507–1003, which did not interact with Gag, were all poorly packaged (Fig. 4, lanes 3–5, 26, and 27). These results indicated that Gag binding is required for MOV10 virion packaging.
FIGURE 4.
Gag binding domain of MOV10 is not sufficient for virion packaging. The indicated MOV10 mutants were expressed with HIV-1 proviral construct in 293T cells. Virions were purified, and virion incorporation of these mutants was determined by Western blotting.
Next, we compared MOV10 C-terminal deletion mutants (Fig. 4, lanes 6–13 and 29–35). It was found that mutants 1–312, 1–430, 1–506, 1–949, 1–952, and 1–955 were detected in the virions (Fig. 4, lanes 6–8, 13, and 33–35). Mutants 1–926, 1–943, and 1–946 were also detected at relatively lower efficiency, but this was due to their lower expression levels in cells (Fig. 4, lanes 30–32). Thus, all of these mutants are packaged into the virions, which is consistent with that they all have an intact Gag binding domain. Nonetheless, we found that four mutants 1–592, 1–662, 1–781, and 1–911 were not packaged (Fig. 4, lanes 9–12), although they still bound to Gag (Fig. 3B). This result indicated that Gag binding might not be sufficient, and there may be additional determinant(s) in the C-terminal region for MOV10 packaging.
Helicase Domain Is Required for MOV10 Virion Packaging
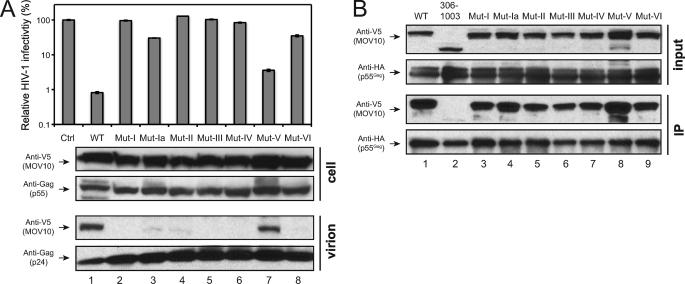
To search for additional packaging determinant(s), we compared those four C-terminal deletion mutants that were not packaged. We noticed that they lost at least one helicase motif, as indicated in Fig. 3A. We wondered whether these helicase motifs could play a role in virion incorporation. Previously, we created seven helicase motif mutants: Mut I, Mut Ia, Mut II, Mut III, Mut IV, Mut V, and Mut VI, which have alanine substitution(s) of the critical residues in the corresponding helicase motifs (8).
When their anti-HIV-1 activity was determined, it was found that all of these mutants except for Mut V significantly lost anti-HIV-1 activity (Fig. 5A, upper panel), which is consistent with our previous observation (8). We then determined their ability for virion incorporation as well as interactions with Gag. It was found that only Mut V was detected in HIV-1 virions, whereas Mut I, Mut Ia, Mut II, Mut III, Mut IV, and Mut VI were not, although they were expressed at similar levels in cells (Fig. 5A, lower panels). In addition, unlike the mutant 306–1003 that lacks the Gag binding domain, all of these mutants were still able to interact with HIV-1 Gag (Fig. 5B). These results indicated that although the seven helicase motifs are not required for MOV10-Gag interaction, all but one of them are required for MOV10 packaging.
FIGURE 5.
All but one of seven helicase motifs are also required for MOV10 virion packaging. A, anti-HIV-1 activity and virion incorporation of MOV10 helicase mutants. The indicated MOV10 mutants were expressed with VSV-G pseudotyped pNL-Luc reporter viruses in 293T cells, and viral infectivity was analyzed in CEM-SS cells. Viral infectivity is presented as relative value, with the infectivity of viruses produced in the presence of a control vector as 100%. The S.E. were calculated from three independent experiments. In addition, virions were purified by ultracentrifugation, and cellular and virion proteins were analyzed by Western blotting. B, interaction of MOV10 helicase mutants with HIV-1 Gag, as determined by the same as in Fig. 3B.
MOV10 Three-dimensional Structure Model
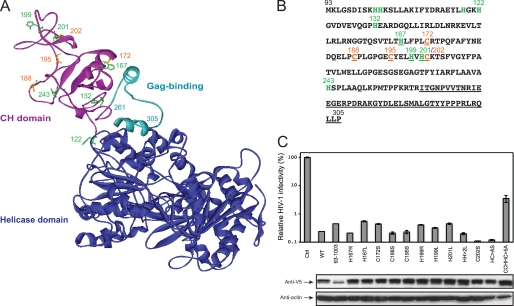
To understand why the MOV10 N-terminal region is so critical for Gag interaction and virion packaging, we predicted the MOV10 three-dimensional structure by homology modeling. MOV10 shares ∼40% AA sequence homology with another SF-1 RNA helicase, the UPF1 protein (supplemental Fig. S1). Based on the known UPF1 three-dimensional structure (12), we created a MOV10 structure model. In this model, MOV10 shows a RNA helicase core domain possessing seven conserved helicase motifs (Fig. 6A). Similar to UPF1, MOV10 also shows a structurally exposed N-terminal region. Interestingly, UPF1 has a Cys-His-rich (CH) domain in this region, functioning as a zinc finger that binds to three zinc atoms. Similarly, MOV10 has four cysteines and nine histidines in the AA 93–260 region (Fig. 6, A and B), which is immediately followed by the Gag binding domain from AA 261 to 305. Our model predicts that the AA 261–305 region has two α-helices separated by a coiled coil (Fig. 6A). It is possible that the entire AA 93–305 region consists of a putative CH domain, which explains why it is so important for Gag binding and virion packaging.
FIGURE 6.
MOV10 three-dimensional structure model. A, ribbon structure model of human MOV10 protein from AA 122 to 968. It was generated from the x-ray structure of human UPF1 protein from AA 116 to 914 (Protein Data Bank code 2WJY). The C-terminal core structure of RNA helicase domain from residue 306 to 968 is shown in blue; the Gag binding domain from residue 261 to 305 is shown in light blue; and the N-terminal part of a potential CH-domain from residue 122 to 260 is shown in magenta. In this CH domain, histidines are shown in green, cysteines are shown in orange, and their position is indicated by the corresponding number. B, protein sequence of MOV10 from AA 93 to 305. Histidines are shown in green, cysteines are shown in orange, and the Gag binding sequence from AA 261 to 305 is underlined. Residues that were targeted for mutagenesis are also underlined. C, anti-HIV-1 activity of MOV10 CH domain mutants, as determined in Fig. 5A. Their cellular expression was determined by Western blotting.
To dissect the function of this domain further, we mutated His-167, Cys-172, Cys-188, Cys-195, His-199, His-201, and Cys-202 by introducing single-point and multiple-point mutations (Fig. 6C). After that, their anti-HIV-1 activity was measured as before. It was found that the H167R, H167L, C172S, C188S, C195S, H199R, H199L, H201L, and C202S single-point mutations as well as the H199L/H201L (HH>2L) and H201A/C202S (HC>AS) double-point mutations could not disrupt MOV10 anti-HIV activity; however, the C188A/C195A/H199A/H201A/C202A (CCHHC>5A) quintuple-point mutation significantly disrupted MOV10 anti-HIV-1 activity (Fig. 6C). This result indicated that residues Cys-188, Cys-195, His-199, His-201, and Cys-202 are involved in MOV10 antiviral activity, and they may be able to compensate each other to support this MOV10 activity.
Identification of Other MOV10 Antiviral Regions
Having determined Gag binding and virion packaging, we next directly measured the anti-HIV-1 activity of these N- or C-terminal deletion mutants of MOV10. It was found that the WT protein, three N-terminal deletion mutants 93–1003, 96–1003, and 99–1003, and three C-terminal deletion mutants 1–949, 1–952, and 1–955 had similar levels of anti-HIV-1 activities, which reduced viral infectivity by ∼100-fold; and all the other mutants almost completely failed to reduce viral infectivity (Fig. 7A). Thus, MOV10 mutants that were not packaged into virions all lost antiviral activity, and several mutants that were packaged into virions also lost antiviral activity. These results indicate that after being packaged into virions, MOV10 needs additional domains to inhibit viral replication. Because the 99–1003 and 1–949 mutants were active whereas the 101–1003 and 1–946 mutants were inactive, the AA 99–949 region might be essential for MOV10 anti-HIV-1 activity.
FIGURE 7.
A and B, anti-HIV-1 activity of MOV10 deletion mutants, as determined in Fig. 5A. Protein expression in cells and virions in B was determined as in Fig. 4.
To test this possibility, we created another MOV10 mutant that only expresses the AA 99–949 region and measured its activity. It was found that mutant 99–949 had almost the same levels of anti-HIV activity as the WT proteins (Fig. 7B, top panel). Next, we created two other mutants: the Δ99–100 mutant, which has a deletion of residues 99 and 100, and the Δ947–949 mutant, which has a deletion of residues 947, 948, and 949. All of these residues are located in the border of the AA 99–949 region. It was found that the Δ99–100 mutant was still active, whereas the Δ947–949 mutant completely lost antiviral activity (Fig. 7B, top panel). We then determined the virion packaging of mutants Δ99–100, Δ947–949, and 99–949, and it was found that all of these mutants were efficiently packaged into virions (Fig. 7B, lower panels). Thus, residues 99–949 compose the minimal MOV10 antiviral region, whereas residues 947, 948, and 949 are also critical for MOV10 antiviral activity.
DISCUSSION
In this report, we studied MOV10 antiretroviral activity by mapping its critical antiviral domains and residues. We first studied the MOV10 packaging mechanism because virion packaging is a critical step for its antiviral activity. Previously, we reported that MOV10 is packaged into HIV-1 virions via an interaction with NC in an RNA-dependent manner (8). It has been reported that A3G also interacts with HIV-1 NC in a similar manner and gets packaged into HIV-1 virions (8, 15–19) and that the basic linker of NC is required for A3G packaging (10). Using the same system, we found that the NC basic linker is also required for MOV10 packaging (Fig. 2B). Thus, A3G and MOV10 share a very similar packaging mechanism. This similarity raises a question of whether they compete with each other for virion packaging during natural HIV-1 infection, which may compromise their anti-HIV-1 activity. However, it should be noted that Vif potently inhibits the encapsidation of A3G by HIV-1 (20–24), which should completely disrupt this competition. On the other hand, it has been reported that A3G binds to MOV10 (25–27). Thus, even when the neutralization of A3G by Vif is incomplete, remnant A3G proteins may enhance MOV10 packaging as long as the domain on A3G that binds to MOV10 does not overlap with the domain that binds to NC.
We have mapped the Gag binding domain of MOV10 to the N-terminal AA 261–305 region (Fig. 3B). Interestingly, this region is located in a putative CH domain, which spans the AA 93–305 region (Fig. 6A). The CH domain was recently recognized as a novel class of protein-protein interaction module (28). Indeed, the UPF1 CH domain binds to UPF2 (29). Thus, it is not coincidental that Gag binds to this domain of MOV10. Nonetheless, we found that Gag binding is not sufficient for MOV10 packaging. A similar observation was made in the study of A3G virion packaging by Huthoff and Malim (30). They found that when the packaging motif 124YYFW127 was mutated, A3G became resistant to virion packaging, but it still interacted with HIV-1 Gag in an RNA-dependent manner. In addition, TSG101 also interact with HIV-1 Gag (31), but it is poorly packaged into HIV-1 virions. Thus, Gag binding is not the sole requirement for virion packaging. Indeed, we found that in addition to the Gag binding domain, MOV10 helicase motifs play an important role in its virion packaging. Our analysis shows that all helicase mutants except for Mut V had a defect in virion packaging. Consistently, we found that only Mut V did not lose anti-HIV-1 activity (Fig. 5A). Motif I has a GXXXXGKT/S consensus and binds to phosphates; and motif II has a DEXX consensus and binds to magnesium. These two motifs catalyze the hydrolysis of purine nucleoside triphosphate, providing energy for helicase activity. The other five motifs are more diverse, and they could contribute to RNA or DNA binding (32). It is unclear why motif V is not required for MOV10 virion packaging. It is possible that we did not completely inactivate this helicase motif when this mutant was created. Nonetheless, we conclude that the helicase domain serve as another packaging signal for MOV10. It is known that helicase motifs have high affinity to RNAs. We speculate that RNA binding may change protein conformation and facilitate MOV10 encapsidation by the virus.
In addition to these packaging signals, we have identified several other critical residues that are required for MOV10 anti-HIV-1 activity. We have found that nearly all MOV10 residues (AA 99–949) are required for its antiviral activity. In this region, we found that in addition to those packaging signals, both the N-terminal region and the C-terminal region are critical for MOV10 anti-HIV-1 activity. Although individually mutating histidine and cysteine residues in the CH domain did not disrupt MOV10 anti-HIV-1 activity, simultaneously mutating residues Cys-188, Cys-195, His-199, His-201, and His-202 seriously compromised MOV10 activity (Fig. 6C). We also found that the C-terminal residues Cys-947, Pro-948, and Phe-949 are also critical for MOV10 antiviral activity (Fig. 7B). These results indicate that in addition to Gag binding and virion packaging, there is another critical step(s) that controls MOV10 antiviral activity. Further identification of this step(s) will lead to a more comprehensive understanding of MOV10 antiviral activity.
In summary, we have identified two determinants for MOV10 packaging and eight other residues for MOV10 anti-HIV-1 activity. These results provide a molecular basis for understanding MOV10 antiretroviral mechanism.
Supplementary Material
Acknowledgments
We thank P. Spearman and the National Institutes of Health AIDS Research and Reference Reagent Program for critical reagents.
This work was supported, in whole or in part, by National Institutes of Health Grants AI063944 and AI080225 (to Y.-H. Z).

This article contains supplemental Fig. S1.
- MOV10
- Moloney leukemia virus 10 homolog
- AA
- amino acid(s)
- A3G
- APOBEC3G
- CA
- capsid
- CFP
- cyan fluorescent protein
- CH
- Cys-His-rich
- I domain
- interaction domain
- MA
- matrix
- NC
- nucleocapsid
- SF
- superfamily
- SP1
- spacer peptide 1
- UPF1
- up frame-shift 1
- VLP
- virus-like particle.
REFERENCES
- 1. Mooslehner K., Müller U., Karls U., Hamann L., Harbers K. (1991) Structure and expression of a gene encoding a putative GTP-binding protein identified by provirus integration in a transgenic mouse strain. Mol. Cell. Biol. 11, 886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koonin E. V. (1992) Trends Biochem. Sci. 17, 495–497 [DOI] [PubMed] [Google Scholar]
- 3. Jeang K. T., Yedavalli V. (2006) Role of RNA helicases in HIV-1 replication. Nucleic Acids Res. 34, 4198–4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chendrimada T. P., Finn K. J., Ji X., Baillat D., Gregory R. I., Liebhaber S. A., Pasquinelli A. E., Shiekhattar R. (2007) MicroRNA silencing through RISC recruitment of eIF6. Nature 447, 823–828 [DOI] [PubMed] [Google Scholar]
- 5. Meister G., Landthaler M., Peters L., Chen P. Y., Urlaub H., Lührmann R., Tuschl T. (2005) Identification of novel argonaute-associated proteins. Curr. Biol. 15, 2149–2155 [DOI] [PubMed] [Google Scholar]
- 6. Burdick R., Smith J. L., Chaipan C., Friew Y., Chen J., Venkatachari N. J., Delviks-Frankenberry K. A., Hu W. S., Pathak V. K. (2010) P body-associated protein Mov10 inhibits HIV-1 replication at multiple stages. J. Virol. 84, 10241–10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Furtak V., Mulky A., Rawlings S. A., Kozhaya L., Lee K., Kewalramani V. N., Unutmaz D. (2010) Perturbation of the P body component Mov10 inhibits HIV-1 infectivity. PLoS One 5, e9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang X., Han Y., Dang Y., Fu W., Zhou T., Ptak R. G., Zheng Y. H. (2010) Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J. Biol. Chem. 285, 14346–14355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng Y. H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B. M. (2004) Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 78, 6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burnett A., Spearman P. (2007) APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 81, 5000–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Derdowski A., Ding L., Spearman P. (2004) A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78, 1230–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clerici M., Mourão A., Gutsche I., Gehring N. H., Hentze M. W., Kulozik A., Kadlec J., Sattler M., Cusack S. (2009) Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 28, 2293–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chertova E., Chertov O., Coren L. V., Roser J. D., Trubey C. M., Bess J. W., Jr., Sowder R. C., 2nd, Barsov E., Hood B. L., Fisher R. J., Nagashima K., Conrads T. P., Veenstra T. D., Lifson J. D., Ott D. E. (2006) Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80, 9039–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang Y., Kong W. P., Nabel G. J. (2001) Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75, 4947–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cen S., Guo F., Niu M., Saadatmand J., Deflassieux J., Kleiman L. (2004) The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279, 33177–33184 [DOI] [PubMed] [Google Scholar]
- 16. Luo K., Liu B., Xiao Z., Yu Y., Yu X., Gorelick R., Yu X. F. (2004) Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 78, 11841–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schäfer A., Bogerd H. P., Cullen B. R. (2004) Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the Gag polyprotein precursor. Virology 328, 163–168 [DOI] [PubMed] [Google Scholar]
- 18. Svarovskaia E. S., Xu H., Mbisa J. L., Barr R., Gorelick R. J., Ono A., Freed E. O., Hu W. S., Pathak V. K. (2004) Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279, 35822–35828 [DOI] [PubMed] [Google Scholar]
- 19. Zennou V., Perez-Caballero D., Göttlinger H., Bieniasz P. D. (2004) APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78, 12058–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mariani R., Chen D., Schröfelbauer B., Navarro F., König R., Bollman B., Münk C., Nymark-McMahon H., Landau N. R. (2003) Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114, 21–31 [DOI] [PubMed] [Google Scholar]
- 21. Marin M., Rose K. M., Kozak S. L., Kabat D. (2003) HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9, 1398–1403 [DOI] [PubMed] [Google Scholar]
- 22. Sheehy A. M., Gaddis N. C., Malim M. H. (2003) The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 9, 1404–1407 [DOI] [PubMed] [Google Scholar]
- 23. Stopak K., de Noronha C., Yonemoto W., Greene W. C. (2003) HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell 12, 591–601 [DOI] [PubMed] [Google Scholar]
- 24. Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X. F. (2003) Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302, 1056–1060 [DOI] [PubMed] [Google Scholar]
- 25. Chiu Y. L., Witkowska H. E., Hall S. C., Santiago M., Soros V. B., Esnault C., Heidmann T., Greene W. C. (2006) High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. U.S.A. 103, 15588–15593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gallois-Montbrun S., Kramer B., Swanson C. M., Byers H., Lynham S., Ward M., Malim M. H. (2007) Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81, 2165–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kozak S. L., Marin M., Rose K. M., Bystrom C., Kabat D. (2006) The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281, 29105–29119 [DOI] [PubMed] [Google Scholar]
- 28. Heise C. T., Le Duff C. S., Boter M., Casais C., Airey J. E., Leech A. P., Amigues B., Guerois R., Moore G. R., Shirasu K., Kleanthous C. (2007) Biochemical characterization of RAR1 cysteine- and histidine-rich domains (CHORDs): a novel class of zinc-dependent protein-protein interaction modules. Biochemistry 46, 1612–1623 [DOI] [PubMed] [Google Scholar]
- 29. Kadlec J., Guilligay D., Ravelli R. B., Cusack S. (2006) Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA 12, 1817–1824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Huthoff H., Malim M. H. (2007) Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and virion encapsidation. J. Virol. 81, 3807–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garrus J. E., von Schwedler U. K., Pornillos O. W., Morham S. G., Zavitz K. H., Wang H. E., Wettstein D. A., Stray K. M., Côté M., Rich R. L., Myszka D. G., Sundquist W. I. (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107, 55–65 [DOI] [PubMed] [Google Scholar]
- 32. Caruthers J. M., McKay D. B. (2002) Helicase structure and mechanism. Curr. Opin. Struct. Biol. 12, 123–133 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.