Background: MED1 is a key coactivator for androgen receptor (AR)-dependent transcription in prostate cancer cells.
Results: The mechanisms and binding motifs through which the MED1-Mediator complex functionally interact with AR were determined.
Conclusion: MED1 functionally interacts with the Tau-1 domain of AR via a newly discovered noncanonical binding motif.
Significance: Our findings provide new insights into the molecular mechanisms of AR-mediated transcriptional activation in prostate cancer cells.
Keywords: Androgen receptor, Coactivator transcription, Gene expression, Nuclear receptors, Prostate cancer, MED1, Mediator
Abstract
Nuclear receptor (NR) activation by cognate ligand generally involves allosteric realignment of C-terminal α-helices thus generating a binding surface for coactivators containing canonical LXXLL α-helical motifs. The androgen receptor (AR) is uncommon among NRs in that ligand triggers an intramolecular interaction between its N- and C-terminal domains (termed the N/C interaction) and that coactivators can alternatively bind to surfaces in the AR N-terminal or hinge regions. The evolutionary conserved Mediator complex plays a key coregulatory role in steroid hormone-dependent transcription and is chiefly targeted to NRs via the LXXLL-containing MED1 subunit. Whereas MED1 has been demonstrated to serve as a key transcriptional coactivator for AR, the mechanisms by which AR recruits MED1 have remained unclear. Here we show that MED1 binds to a distinct AR N-terminal region termed transactivation unit-1 (Tau-1) via two newly discovered noncanonical α-helical motifs located between MED1 residues 505 and 537. Neither of the two MED1 LXXLL motifs is required for AR binding, whereas loss of the intramolecular AR N/C interaction decreases MED1 binding. We further demonstrate that mitogen-activated protein kinase phosphorylation of MED1 enhances the AR-MED1 interaction in prostate cancer cells. In sum, our findings reveal a novel AR-coactivator binding mechanism that may have clinical implications for AR activity in prostate cancer.
Introduction
The male sex steroids testosterone and dihydrotestosterone (DHT)2 affect the expression of genes essential for the development and maintenance of male reproductive and accessory sex tissues. The physiological actions of these androgenic hormones are mediated primarily through the androgen receptor (AR), a 110-kDa member of the nuclear receptor (NR) family of ligand-activated transcription factors (1, 2). Similar to other members of the NR family, the AR has a modular structure, including a large, poorly conserved N-terminal domain (NTD), a highly conserved DNA binding domain, a C-terminal ligand binding domain (LBD), and a hinge region connecting the DNA binding domain with the LBD. For most NRs, the predominant transcriptional activation domain (termed activation function 2 or AF2) resides in the LBD and facilitates ligand-dependent transcriptional activation (3, 4). Yet for the AR, the major transcriptional activation domain resides almost entirely in the NTD and can be subdivided into two distinct regions: transactivation unit-1 (Tau-1) (residues 100–360) and transactivation unit-5 (Tau-5) (residues 360–528) (1).
Upon binding ligand, most NRs undergo a significant repositioning of a conserved α-helix 12 located near the extreme C terminus of the AF2 domain. The realigned AF2 domain creates a hydrophobic groove on the surface of the LBD that serves as a specific binding site for transcriptional coactivators containing signature LXXLL motifs (4–8). While AR also contains a C-terminal AF2 domain capable of binding LXXLL-bearing polypeptides in the presence of ligand (9), the AR-AF2 domain differs from that of other NRs in that it more strongly binds to signature FXXLF motifs such as the 23FQNLF27 motif found at the AR-NTD (10–13). The resulting intramolecular N/C interaction is thought to generate alternative coactivator binding sites at the AR NTD and hinge region (1, 2). For example, members of the p160/SRC family of coactivators directly bind to the Tau-5 domain of the AR NTD via conserved Gln-rich regions in a manner that is independent of their intrinsic LXXLL motifs (14–16). Coactivator complexes shown to interact with the AR hinge region include the BAF57-containing SWI/SNF complex and the p300/PCAF complex (17, 18). Interestingly, mutational analyses of the AR NTD have also implicated the Tau-1 domain as a potential coactivator binding surface (19), yet the identity of the corresponding interacting coregulatory factors remain unclear.
Mediator is an evolutionarily conserved multisubunit complex that plays an essential coregulatory role in eukaryotic transcription of protein-encoding genes (reviewed in Ref. 20). The complex can facilitate multiple functions in transcription, including recruitment of RNA polymerase II, activation of the pre-initiation complex, regulation of distinct chromatin modification events, and promotion of transcriptional elongation (21). Mediator is composed of over 30 subunits, several of which interact with different signal-activated gene-specific transcription factors (22). In human cells, Mediator was initially isolated as a coactivator activity bound to NRs in the presence of ligand (23). A single subunit of Mediator, termed MED1 (also known as TRAP220, PBP, and DRIP205), can directly target the complex to the AF2 domain of DNA-bound NRs via two signature LXXLL motifs (24). Despite its functional importance as the binding target for most NRs, MED1 is only variably associated with Mediator existing in a small subpopulation (∼20%) of steady-state Mediator complexes (25, 26). Notably in this regard, the extracellular signal-regulated kinase 1/2 (ERK1/2) regulates MED1 activity via phosphorylation by promoting its association with Mediator and enhancing its NR-dependent transcriptional coactivator activity (27, 28).
The MED1-Mediator complex plays a key coregulatory role for AR-dependent transcription in prostate cells (29–32) as well as from in vitro chromatinized templates (33). Moreover, MED1 expression is enriched in the prostate epithelium (34), a tissue whose growth and development are dependent on androgen signaling (2). In a comparative study of the requirement of different AR coactivators, MED1 was found to be indispensible for androgen-dependent, prostate-specific transcription of the well characterized AR target gene prostate-specific antigen (PSA) (31). Loss of MED1 expression in prostate cancer cells was accompanied by a significant reduction in RNA polymerase II and basal transcription factor recruitment at the PSA gene promoter (31). More recent studies utilizing androgen-dependent and -independent prostate cancer cells have revealed that MED1 is required for robust transcription of other AR target genes and further suggest that amplification or hyper-activation of MED1 promotes prostate oncogenesis (34–36). Although these studies clearly implicate functional interactions between AR and MED1, their respective binding domains have surprisingly remained poorly defined. Previous reports suggested that the two signature MED1 LXXLL motifs facilitate a very weak interaction with an isolated GST-AR AF2 fusion (9, 32), yet the physiological relevance of these data is unclear in light of the much stronger N/C interaction that takes place between the AR N-terminal 23FQNLF27 motif and its core AF2 domain.
In an effort to better understand how MED1 coactivates AR activity, we utilized a broad array of deletion and point mutants to investigate how the two proteins bind to one another in the presence of ligand. We report here that MED1 binds to the AR N-terminal Tau-1 domain via two tandem α-helices located between MED1 residues 505 and 537. Surprisingly, neither of MED1's two LXXLL motifs is required for this association, whereas loss of the AR N/C interaction inhibits AR binding with MED1. Our results also reveal that ERK1/2 phosphorylation of MED1 promotes its binding to AR in prostate cancer cells by directly stabilizing the AR-MED1 interaction and by stabilizing MED1 cellular expression, which in turn drives its interaction with AR. Together our data reveal a novel AR-MED1 binding mechanism that may have clinical implications for AR-mediated gene expression in prostate cancer.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Antibodies against AR, MED1, MED17, MED24, MED7, MED6, and α-tubulin were from Santa Cruz Biotechnology. Antibody against phosphothreonine was from Cell Signaling. Mouse monoclonal antibody against FLAG epitope was from Sigma. Mouse monoclonal antibody against the hemagglutinin (HA) epitope was obtained from Roche Applied Science.
Plasmids
The pSG5-HA-MED1-wt, -CΔ454, -CΔ690, -CΔ918, -CΔ1215, -NΔ1233, -ERK mutant (T1032A and T1457A), -LXXAA(A), and -LXXAA(B) mammalian expression vectors were described previously (24, 28). The pGEX-2TK-RBD, -RBD-LXXAA, -RBD1, and -RBD2 bacterial expression vectors were described previously (24). pSG5-FLAG-AR-wt, -ΔNTD, -ΔTau5, -ΔTau1, -ΔcTau1, -ΔF, and -G21E were described previously (16, 19, 37). Transfer vectors baculovirus-expressing for HA-MED1-wt, -CΔ454, -CΔ690, -CΔ918, -CΔ1215, -ERK mutant (T1032A and T1457A), and full-length FLAG-AR were generated by subcloning the corresponding epitope-tagged open reading frames into plasmids pAcSG2 or pVL1392 (BD Biosciences). The androgen-responsive MMTV-Luc reporter gene was reported earlier (32). The pMCL-HA-MKK1–8E and pMCL-HA-MKK1-NΔ4 expression vectors (38) and pCMV-ERK2-L73P/S151D expression vector (39) were provided by Natalie Ahn (University of Colorado). pGEX-2TK-RBD-1Δα1,α2 and pGEX-2TK-RBD-1Δα3–5 were generated by PCR-amplifying MED1 amino acids 542–635, and MED1 amino acids 501–575, respectively. The PCR fragments were then ligated into pGEX-2TK (Pharmacia Biotech, Piscataway, NJ). pSG5-HA-MED1Δα1,α2 was created via PCR-generated mutagenesis by deletion of amino acid residues from 501 to 541 from the parental pSG5-HA-MED1wt template. pSG5-HA-MED1-LXXAA(A/B) was generated by using the parental pSG5-HA-MED1-LXXAA(A) template together with a specific mutant oligonucleotide (LL residues 648 and 649 to AA). pGEX-2TK-RBD1-Δα1 and pGEX-2TK-RBD1-Δα2 were generated by using mutagenic oligonucleotides deleting residues 506–517 for Δα1 and residues 523–533 for Δα2, respectively.
Cell Culture
LNCaP, DU145, 1532T, and COS cells were obtained from American Type Culture Collection. LNCaP and 1532T cells were routinely maintained in RPMI 1640 (Invitrogen) with 10% fetal bovine serum (FBS, Gemini Bioproducts) along with penicillin and streptomycin (Invitrogen). COS and DU145 cells were grown in DMEM with 10% FBS and penicillin/streptomycin. In the androgen starvation experiments, cells were grown in phenol red-free medium containing 10% charcoal/dextran-stripped FBS (CDS-FBS, Gemini Bioproducts). All cells were maintained in a humidified incubator at 37 °C and 5% CO2. DHT and R1881 were from Sigma. EGF was from Invitrogen. U0126 was from Alexis Biochemicals.
Immunoprecipitation of Endogenous AR-Mediator
Smart pool small interfering RNA (siRNA) specific for MED1 was from Dharmacon Research as previously described (28). A scrambled siRNA smart pool (Dharmacon) was used as a control. LNCaP cells were grown to 60% confluency in 10% CDS-FBS containing RPMI 1640 and transfected with MED1-specific or control siRNA at the final concentration of 100 nm using the Lipofectamine and Plus reagents (Invitrogen). Three hours post-transfection, medium was replaced with fresh medium containing or lacking DHT (10 nm final) for an additional 48 h. The cells were then lysed in lysis buffer (50 mm Tris-Cl (pH 7.4), 150 mm NaCl, 5 mm EDTA, and 0.5% Nonidet P-40, 0.2 mm phenylmethylsulfonyl fluoride) for 1 h at 4 °C. One milligram of protein from each sample was combined with 1 μg of anti-AR antibodies and 10 μl (packed) of protein A-agarose beads (Roche Applied Science) overnight at 4 °C. The beads were then washed three times in lysis buffer, and the precipitated immune complexes were resolved by SDS-PAGE. The gel was then processed for Western blotting analyses.
Luciferase Assays
LNCaP cells were cultured in CDS-FBS containing media for 3 days and then seeded (1 × 105 cells) in 12-well plates and cotransfected with 100 ng of MMTV-Luc and pSG5-HA-MED1 (0.5 μg) or pSG5-HA-MED1Δα1,α2 (0.5 μg) using Lipofectamine and Plus reagents (Invitrogen). For competition assays, pSG5-HA-MED1Δ454 (50 ng), pSG5-HA-MED1Δ918 (0.2 μg), or pSG5-empty vector were cotransfected with 100 ng of MMTV-Luc and 0.5 μg of pSG5-HA-MED1. Three hours post-transfection, the medium was replaced with fresh media containing or lacking R1881 (10 nm final) for an additional 24 h. Cells were harvested, and equivalent amounts of protein were assayed for luciferase activity using an assay kit (Promega) and a luminometer. Luciferase values were normalized by using a β-galactosidase (pSV-β-gal, Promega) expression vector as internal control.
Recombinant Baculovirus Protein Expression in Sf9 Cells
Generation of recombinant baculovirus and purification of recombinant proteins from infected Sf9 cells were carried out as previously described (27, 32) with the exception that the FLAG-AR-infected Sf9 cells were additionally cultured and purified in the presence or absence of 10 nm DHT.
GST Pulldown Assay
Glutathione S-transferase (GST) and GST fusion proteins were expressed and purified as described previously (24). AR was [35S]methionine-labeled in vitro in the presence of 10 nm DHT using the TnT in vitro translation kit (Promega). GST pulldown assays with 35S-labeled AR were carried out as previously described (24) in the presence of 10 nm DHT.
In Vitro Phosphorylation Assay
50 ng of purified baculovirus-expressed MED1 (bv-MED1-FL) or MED1-Erk mut (bv-MED1-Erk mut) protein was incubated with ATP (0.1 mm) plus 10 ng of ERK1 (Upstate) in kinase buffer exactly as described previously (28).
In Vitro Biotinylated-ARE Pulldown Assay
A double-stranded biotinylated androgen response element (ARE) corresponding to the androgen-responsive unit in the first intron of the C3(1) gene (40) was generated by annealing 5′-biotinylated oligonucleotides: Forward, 5′Bio-GAT CAT AGT ACG TGA TGT TCT AGG CCT AGT ACG TGA TGT TCT CAA GAT C-3′, and Reverse, 5′Bio-GAT CTT GAG AAC ATC ACG TAC TAG GCC TAG AAC ATC ACG TAC TAT GAT C-3′. 10 pmol of the biotin-conjugated ARE was incubated at 4 °C with 50 ng of baculovirus-AR (bv-AR) in 250 μl (total volume) of binding buffer (20 mm Tris-HCl (pH 7.3), 20% glycerol, 0.2 mm EDTA, 0.5 mm phenylmethylsulfonyl fluoride, 3 mm β-mercaptoethanol, 300 mm KCl, 0.05% Nonidet P-40) containing 10 nm DHT and 10 μl (packed volume) of streptavidin-Sepharose beads (Sigma). After 90 min, 10–50 ng of bv-MED1 or bv-MED1 mutants was added to the binding reactions, and the mixture was incubated for another 2 h at 4 °C. Each sample was washed 5× with binding buffer, resolved on an 8% SDS-PAGE, and then detected by immunoblotting using antibodies against HA, MED1, and AR.
AR-MED1 Coimmunoprecipitation
Transient transfections were carried out in 6-well plates using Lipofectamine 2000 (Invitrogen). COS-7 cells were first cultured in 10% CDS-FBS medium for 48 h to starve cells of androgen. For AR deletion mutant analyses, 5 × 105 COS cells were transfected with pSG5-FLAG-ARΔNTD (1.25 μg), pSG5-FLAG-ARΔTau5 (1 μg), pSG5-FLAG-ARΔTau1 (1.25 μg), pSG5-FLAG-ARΔcTau1 (1.5 μg), pSG5-FLAG-AR (2 μg), pSG5-FLAG-AR-ΔF (2 μg), or pSG5-FLAG-AR-G21E (2 μg) along with 2 μg of pSG5-HA-MED1. For the analyses of MED1 mutants, pSG5-HA-MED1 (2 μg), pSG5-HA-MED1Δ454 (0.25 μg), pSG5-HA-MED1Δ690 (0.5 μg), pSG5-HA-MED1Δ918 (1 μg), pSG5-HA-MED1Δ1215 (1.5 μg), pSG5-HA-MED1Δ1233 (0.25 μg), pSG5-HA-MED1-LXXAA(A) (2 μg), pSG5-HA-MED1-LXXAA(B) (2 μg), pSG5-HA-MED1-LXXAA(A/B) (2 μg), pSG5-HA-MED1-ERKmut (2 μg), or pSG5-HA-MED1Δα1,α2 (2 μg) were transfected, along with pSG5-FLAG-AR (2 μg). Three hours post-transfection, cells were then treated with or without 10 nm R1881 for another 24 h. Cell lysates were prepared in lysis buffer (50 mm Tris HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.2 mm phenylmethylsulfonyl fluoride). Protein concentrations were determined by Bradford assay. Equal amounts of protein lysate were then incubated with 5 μl of packed anti-HA-agarose beads (Sigma) at 4 °C overnight and then washed three times with lysis buffer. The immunoprecipitated protein was fractioned by 6% SDS-PAGE and analyzed by immunoblotting as indicated in the text. For MED1 phosphorylation studies, 1 μg of pCMV-ERK2-L73P/S151D, pMCL-HA-MKK1–8E, or pMCL-HA-MKK1-NΔ4 was transfected as indicated in the text. Cell lysate was prepared 24 h post-transfection in lysis buffer (50 mm Tris HCl (pH 7.4), 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.2 mm phenylmethylsulfonyl fluoride). Equal amounts of protein lysate were resolved on a 6% SDS-PAGE and analyzed by Western blotting using anti-MED1 antibody.
ChIP Analyses
LNCaP cells were cultured for 3 days in CDS-FBS-containing medium and then treated with or without DHT (10 nm) for 4 h and with or without EGF (100 ng/ml) or U0126 (50 μm) for 30 min. Antibodies specific for AR and Mediator subunits were used to immunoprecipitate formaldehyde cross-linked chromatin-protein complexes as outlined in detail previously (30). The immunoprecipitated DNA was analyzed via semi-quantitative PCR using primers spanning the enhancer region of the PSA gene or promoter region of Cdc6 gene as described previously (30, 32). All ChIP experiments were carried out at least three times. Image processing and quantitation of the semi-quantitative PCR data were performed using Quantity One software (Bio-Rad).
RNA Extraction, Reverse Transcription, and Real-time PCR
LNCaP cells were androgen-starved in phenol red-free RPMI 1640/10% CDS-FBS for 2 days and then treated with or without DHT (10 nm) for 4 h and with or without EGF (100 ng/ml) or U0126 (50 μm) for additional 30 min. Total RNA was then isolated using an RNeasy Mini Kit (Qiagen). First strand cDNA was generated using 1 μg of total RNA via MuMLV-reverse transcriptase (Invitrogen) in a final volume of 20 μl. Real-time PCR was performed using an Opticon Continuous Fluorescence Detection System (MJ Research) with a power SYBR Green PCR Mix (Applied Biosystems). The primers specific for either PSA or β-actin were described previously (34).
RESULTS
AR Recruits Mediator via Direct Interaction with MED1
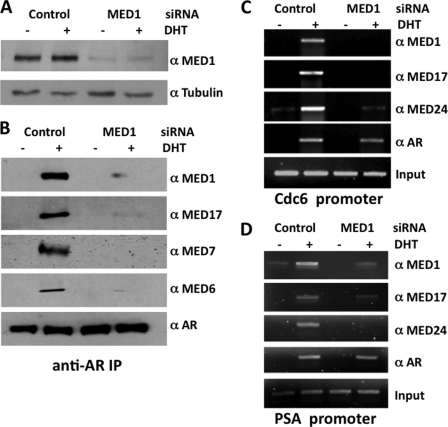
Previous coimmunoprecipitation assays showed that AR is associated with the MED1-Mediator complex in androgen-stimulated cell lines expressing AR (32), whereas several independent chromatin immunoprecipitation (ChIP) analyses clearly demonstrate that AR, MED1, and other Mediator subunits are concomitantly recruited to androgen-responsive gene promoters and enhancers (29–33, 36). Although the earlier findings implicate MED1 in directly tethering the core Mediator complex to AR, the possibility exists that AR might recruit Mediator via interactions with other Mediator subunits as was reported for the glucocorticoid and estrogen receptors (41, 42). To address this possibility, we utilized MED1 siRNA to knock down MED1 expression in AR-positive prostate cancer LNCaP cells and then precipitated endogenously expressed AR with specific antibodies. As shown in Fig. 1B, MED1 along with several other subunits of the core Mediator complex (MED6, MED7, and MED17) were associated with AR in androgen-treated LNCaP cells, but when MED1 expression was silenced, the association of these proteins with AR was significantly decreased. We next performed AR and Mediator ChIP assays in LNCaP cells at two distinct AREs: the distal enhancer region of the PSA gene (43) and the distal promoter region of the Cdc6 gene (30). As demonstrated earlier by our laboratory and others (30–32, 36), we found that AR recruits MED1 and other components of the core Mediator complex (MED17 and MED24) to both ARE regions in the presence of DHT (Fig. 1, C and D). However, when MED1 expression was silenced, the recruitment of MED1 and associated Mediator subunits to the AR target genes was significantly decreased (Fig. 1, C and D). Collectively, these findings are consistent with the notion that, in the presence of ligand, AR directly associates with MED1, which in turn tethers the core Mediator complex to AR.
FIGURE 1.
AR recruits Mediator via interaction with MED1. A, LNCaP cells were transfected with MED1 siRNA or a nonspecific scrambled control siRNA and then probed by immunoblot with antibodies against MED1 and α-tubulin. B, androgen-starved LNCaP cells were transfected with siRNAs for 48 h and then treated with or without 10 nm DHT for 24 h. Whole cell extract was prepared and incubated with antibodies against AR conjugated with protein-A beads. Immunoprecipitated protein was then analyzed by immunoblot with antibodies indicated on the right of each panel. C and D, chromatin prepared from LNCaP cells (treated as described in B) was used for ChIP analyses using the indicated antibodies. Semi-quantitative PCR was performed using primer sets spanning AREs located at the Cdc6 promoter (C) or PSA enhancer (D).
MED1 Interacts with the AR Tau-1 Domain
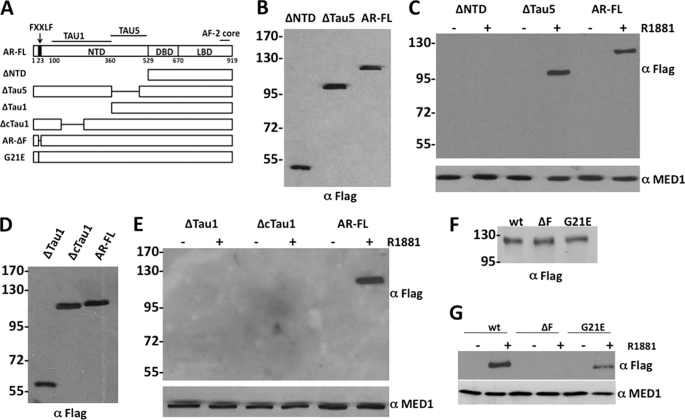
To identify the specific AR domain that binds to MED1, we transiently overexpressed HA-tagged full-length MED1 together with various FLAG-tagged AR deletion mutants in COS cells in the presence or absence of androgen and then performed anti-HA immunoprecipitation assays. We initially found that MED1 only bound to full-length AR and to an AR deletion mutant lacking the Tau-5 domain (ΔTau5) in an androgen-dependent manner, but failed to bind an AR deletion mutant lacking both the Tau-1 and Tau-5 domains (ΔNTD) (Fig. 2C). Consistent with the notion that MED1 specifically binds to the AR Tau-1 domain, we observed that MED1 failed to bind an AR deletion mutant lacking Tau-1 (ΔTau1) (Fig. 2E). Earlier mutational analysis of Tau-1 revealed two putative α-helices between amino acids 173 and 203 defining a presumptive Tau-1 core motif containing strong autonomous activation function independent of p160/SRC coactivators (19). Interestingly, we found that MED1 failed to bind to an AR deletion mutant lacking the Tau-1 core domain (ΔcTau1) thus suggesting that this sub-domain is critical for MED1 interaction (Fig. 2E).
FIGURE 2.
MED1 interacts with the AR Tau-1 domain. A, schematic representation of AR deletion and point mutants. Internal deletions are indicated by horizontal lines. B, D, and F, COS cells transfected with FLAG-tagged AR or AR mutant expression vectors were probed with an anti-FLAG antibody. Molecular mass markers (in kilodaltons) are indicated on the left. C, E, and G, androgen-starved COS cells were transiently transfected with FLAG-tagged AR or AR mutant expression vectors (indicated at the top of panels) along with a full-length HA-MED1 expression vector and then cultured with or without 10 nm R1881 for 24 h. Whole cell lysate was then prepared and incubated with anti-HA agarose beads. Immunoprecipitated protein was then analyzed by anti-FLAG and anti-MED1 immunoblotting.
The AR undergoes a ligand-induced intramolecular N/C interaction that positively affects its ability to activate transcription (1, 2). Deletion of AR's N-terminal 23FQNLF27 motif or mutation of glycine 21 to glutamic acid (G21E) has been shown to blunt the N/C interaction (37). To directly address whether the N/C interaction influences AR's interaction with MED1, we tested MED1 for interaction with AR containing either of these mutations. Interestingly, we found that, although the G21E point mutation reduced MED1 binding to AR, deletion of the entire 23FQNLF27 motif completely abolished MED1 binding (Fig. 2G). Taken together, the results of Fig. 2 indicate that MED1 binds to the Tau-1 domain of AR in the presence of ligand and that AR's N/C interaction apparently serves to promote or stabilize this interaction.
AR Interacts with the MED1 N Terminus Independently of LXXLL Motifs
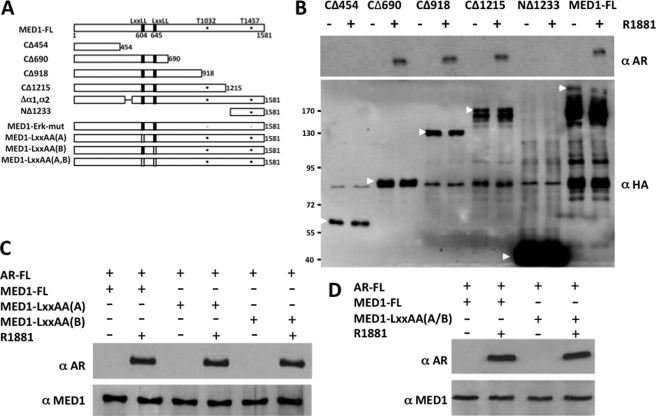
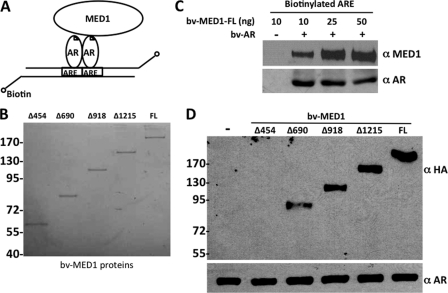
Having identified the domain of AR that binds to MED1, we next set out to define the corresponding region of MED1 that contacts AR. To this end, we overexpressed full-length AR together with various N- and C-terminal HA-tagged MED1 deletion mutants in COS cells and again performed anti-HA immunoprecipitation assays. We observed that AR only bound MED1 deletion mutants minimally containing amino acids 455–690 (Fig. 3B) on the N-terminal half of MED1. To validate these findings in vitro in the context of DNA-bound AR, we re-expressed AR and the HA-tagged MED1 deletion mutants in insect Sf9 cells and then incubated the purified proteins with a biotinylated ARE (Fig. 4). The DNA-bound AR-MED1 complexes were then precipitated using streptavidin-conjugated beads and probed by anti-HA immunoblot. As a negative control for this assay, addition of MED1 alone to the biotinylated-ARE failed to form a complex with the DNA (Fig. 4C). Similar to our coimmunoprecipitation results, we found that AR specifically bound and recruited MED1 deletion mutants minimally containing amino acids 455–690 (Fig. 4D). We have previously termed this region of MED1 the “receptor binding domain” (RBD), because it contains two signature LXXLL motifs (at amino acids 604 and 645) that are necessary for MED1 binding to the AF2 domains of other nonsteroid NRs (24). To determine whether either MED1 LXXLL motif is required for AR binding, we mutated each motif (LXXAA) within the full-length MED1 protein and then tested the mutants for AR binding in COS cells via coimmunoprecipitation. Surprisingly, we found that both LXXLL motifs were dispensable for ligand-dependent AR binding (Fig. 3, C and D). Our findings are thus reminiscent of the situation observed earlier with the p160/SRC coactivators that can bind to AR independently of their intrinsic LXXLL motifs (14–16) and suggest that alternative binding motifs within the MED1 RBD region can facilitate interactions with AR.
FIGURE 3.
AR interacts with the MED1 N terminus independently of LXXLL motifs. A, schematic representation of MED1 deletion and point mutants. LXXLL motifs are indicated by black bars; LXXLL to LXXAA are indicated by open bars. ERK1/2 phosphorylation sites (threonines 1032 and 1457) are indicated by filled circles; ERK1/2 phosphorylation site mutations (Thr to Ala) are indicated by open circles. B–D, androgen-starved COS cells were transiently transfected with HA-tagged full-length MED1 (FL) or MED1 deletion mutant expression vectors (indicated above the panels) together with a full-length FLAG-AR expression vector and subsequently cultured with or without 10 nm R1881 for another 24 h. Whole cell lysate was then incubated with anti-HA agarose beads, and the precipitated proteins were probed with anti-AR, anti-HA, and anti-MED1 immunoblotting. Arrows indicate specific ectopically expressed MED1 full-length or truncated polypeptides.
FIGURE 4.
DNA-bound AR binds to the MED1 N terminus in vitro. A, schematic representation of biotinylated-ARE pull-down assay. B, purified baculovirus-expressed full-length MED1 and MED1 deletion mutants fractionated by SDS-PAGE and stained with Coomassie blue. C, MED1 and AR form a complex on DNA. Purified baculovirus-expressed full-length MED1 (bv-MED1-FL) and AR (bv-AR) were incubated with a biotinylated ARE corresponding to the androgen-responsive unit in the first intron of the C3(1) gene (40). DNA-bound protein complexes were precipitated with streptavidin beads, washed, fractionated by SDS-PAGE, and then immunoblotted with anti-MED1 and anti-AR antibodies. D, purified baculovirus-expressed full-length MED1 or MED1 deletion mutants were incubated together with bv-AR and a biotinylated ARE and processed as described in C.
Two Novel α-Helical Motifs in the MED1 RBD Are Required for Functional Interactions with AR
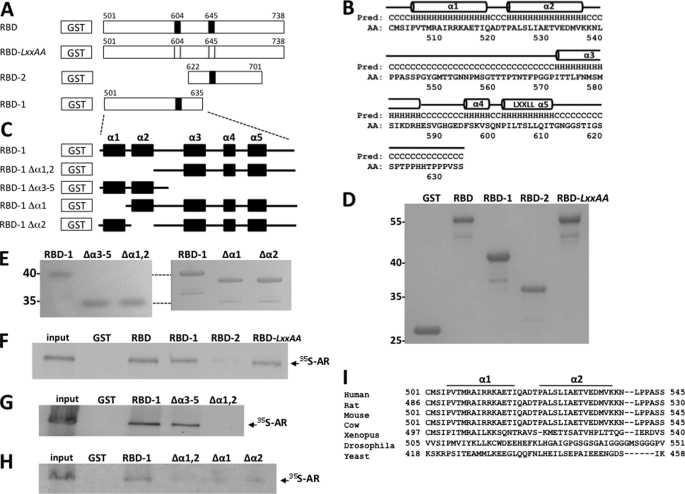
To more precisely delineate the domain of MED1 responsible for binding to AR, we utilized two GST-MED1 fusion constructs that essentially separate the RBD region into two parts: RBD-1 (amino acids 501–635) and RBD-2 (amino acids 622–701) (Fig. 5A). By performing GST pulldown assays with full-length radiolabeled AR in the presence of ligand, we found that AR clearly bound with RBD-1 but not with RBD-2, whereas mutation of either LXXLL motif within the context of the entire RBD region (RBD-LXXAA) had no effect on AR binding (Fig. 5F). Interestingly, an examination of the predicted amino acid secondary structure of the RBD-1 region revealed the presence of five putative α-helices (hereafter denoted α1, α2, α3, α4, and α5) (Fig. 5B). To assess the relative importance of the five α-helices in facilitating an interaction with AR, we deleted the first two, or last three, helices within the context of the GST-RBD-1 construct (i.e. RBD-1Δα1,α2, and RBD-1Δα3–α5, respectively) and tested their ability to bind to AR. We observed that deletion of both α1 and α2 abolished AR binding, whereas deletion of α3–α5 had no significant effect on AR binding (Fig. 5G). Moreover, a separate deletion of either α1 or α2 alone inhibited RBD-1 interaction with AR to the same extent as deletion of both α-helices together (Fig. 5H).
FIGURE 5.
AR binds to a novel noncanonical α-helical array in the MED1 RBD. A, schematic representation of the GST-MED1-RBD (amino acids 501–738) and mutant derivative fusion proteins. LXXLL motifs are indicated by black bars; LXXLL to LXXAA are indicated by open bars. B, predicted secondary structure of MED1 amino acid residues 501–635. Five α-helical motifs (α1, α2, α3, α4, and α5) are indicated by horizontal cylinders. C, schematic representation of the GST-MED1-RBD1 (amino acids 501–635) and mutant derivative fusion proteins. The α-helices α1, α2, α3, α4, and α5 are indicated by black boxes. D and E, purified GST-MED1 fusion proteins were fractionated by SDS-PAGE and stained with Coomassie blue. F–H, GST pulldown assays were carried out by incubating [35S]methionine-labeled AR (labeled in the presence of 10 nm DHT) together with GST-MED1-RBD and mutant derivative fusion proteins (see A and C). The bound proteins were detected by autoradiography. I, sequence alignment of the identified AR-binding noncanonical α-helical array in MED1 of different species.
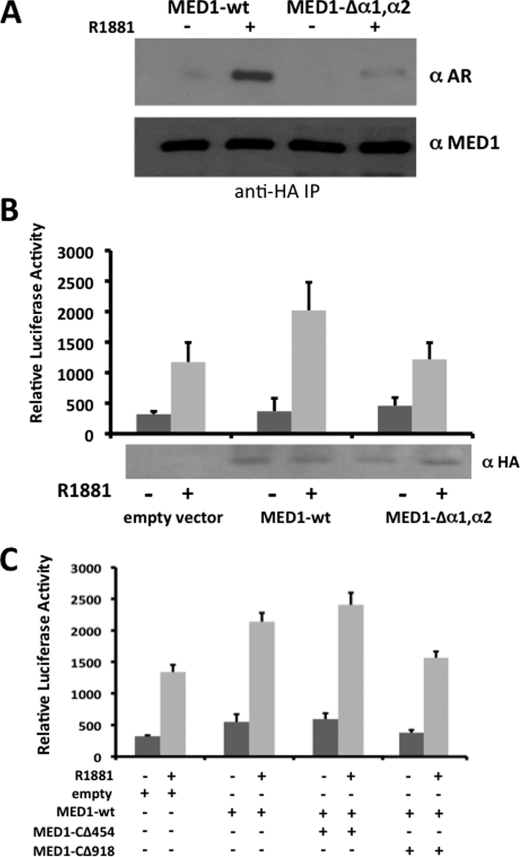
To assess the importance of α1 and α2 within the context of the entire MED1 polypeptide, we internally deleted both α-helices within a full-length MED1 expression construct (MED1-Δα1,α2) and tested the mutant for AR binding in COS cells via coimmunoprecipitation. Consistent with the GST pulldown assays, we found that deletion of the two α-helices markedly decreased androgen-dependent binding between AR and MED1 (Fig. 6A). Considering that MED1 serves as a key transcriptional coactivator for AR, we next examined the functional relevance of the α1 and α2 motifs in terms of MED1-mediated transcriptional coactivation of androgen-dependent gene expression. Accordingly, we transiently overexpressed wild-type MED1 or MED1-Δα1,α2 in LNCaP cells and then measured androgen-dependent transcription from a cotransfected MMTV-luciferase reporter gene (Fig. 6B). In agreement with previous studies from our laboratory (27, 32), overexpression of wild-type MED1 enhanced AR-dependent transcription from the androgen-responsive reporter gene by almost 2-fold. By contrast, overexpression of MED1-Δα1,α2 had no significant costimulatory effect on androgen-dependent reporter gene transcription. We also observed that when deletion mutant MED1-CΔ918, which contains both the α1 and α2 motifs, was cotransfected along with wild-type MED1, androgen-dependent transcription from the MMTV reporter was notably decreased thus suggesting that the MED1-CΔ918 polypeptide can compete with wild-type MED1 for binding with AR in a dominant negative manner (Fig. 6C). By contrast, cotransfection of deletion mutant MED1-CΔ454, lacking both the α1 and α2 motifs, had no significant inhibitory effect on wild-type MED1 coactivation. Collectively, our data here suggest that MED1 helices α1 (amino acids 505–519) and α2 (amino acids 523–537) comprise a composite binding surface that facilitates physiologically important interactions with AR.
FIGURE 6.
The tandem noncanonical α-helical array in the MED1 RBD is required for transcriptional coactivation of AR. A, COS cells were androgen-starved and transfected with expression vectors for HA-MED1 or HA-MED1Δα1,α2 (see Fig. 3A for schematic representation) along with full-length FLAG-AR and subsequently cultured with or without 10 nm R1881 for another 24 h. Whole cell lysate was then incubated with anti-HA agarose beads, and the precipitated proteins were probed with anti-AR and anti-MED1 immunoblotting. B, androgen-starved LNCaP cells were transfected with expression vectors for wild-type MED1, MED1Δα1,α2 or an empty vector control together with a MMTV-Luc reporter template. 3 h post-transfection, cells were treated with or without 10 nm R1881 for another 24 h. Whole cell lysate was then prepared and assayed for luciferase reporter activity and for MED1 expression by immunoblotting (shown at the bottom). C, androgen-starved LNCaP cells were transfected with expression vectors for wild-type MED1, MED1-CΔ454, MED1-CΔ918, or an empty vector control along with the MMTV-Luc reporter. Cells were treated with or without R1881 and assayed for luciferase reporter activity as outlined in A. Luciferase values were normalized by using a β-galactosidase expression vector as internal control and are presented as the mean ± S.E. of triplicate transfections.
Phosphorylation of MED1 by ERK1/2 Enhances Its Association with AR
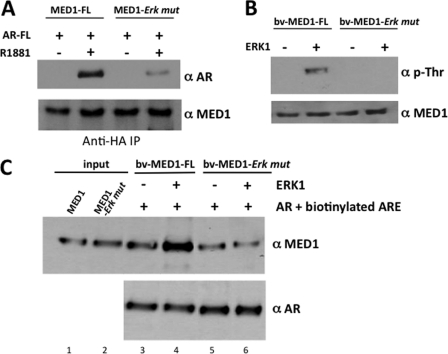
We previously discovered that mitogen-activated protein kinase ERK1/2 phosphorylates human MED1 (28). We subsequently found that ERK1/2 phosphorylation promotes MED1 association with the core Mediator complex and further enhances its ability to coactivate ligand-dependent transcription by the thyroid hormone receptor (27). Given that ERK1/2 covalently modifies the MED1 protein and regulates its functional activity, we were interested in examining whether MED1 phosphorylation influences its association with AR. To first address this question, we compared the AR binding affinity of wild-type MED1 to that of a mutant MED1 protein in which the two ERK phosphorylation sites are mutated (MED1-ERK-mut). The binding assays were carried out as before in transiently transfected COS cells treated with or without R1881, a synthetic androgen that can also specifically stimulate the MAPK-ERK1/2 signaling pathway in cultured cell lines (44, 45). Interestingly, we found that mutation of the two ERK phosphorylation sites in MED1 significantly decreased its binding affinity with AR (Fig. 7A). To investigate this more precisely, we utilized purified recombinant ERK1 enzyme to phosphorylate either wild-type MED1 or MED1-ERK-mut in vitro and then tested their relative binding affinity with recombinant AR bound to a biotinylated ARE. As shown in Fig. 7C, phosphorylation of MED1 increased its binding affinity with the DNA-bound AR >2-fold (compare lanes 3 and 4), whereas incubation of the MED1-ERK-mut protein with ERK1 had no significant effect on its association with AR (lanes 5 and 6). These results suggest that, in addition to promoting MED1 association with the core Mediator complex, ERK phosphorylation of MED1 serves to stabilize and enhance its association with AR.
FIGURE 7.
ERK1 phosphorylation of MED1 enhances its association with AR. A, COS cells were androgen-starved and then transiently transfected with expression vectors for HA-MED1 or MED1-ERK mutant together with full-length FLAG-AR and subsequently cultured with or without 10 nm R1881 for another 24 h. Whole cell lysate was then incubated with anti-HA agarose beads, and the precipitated proteins were probed with anti-AR and anti-MED1 immunoblotting. B, purified baculovirus-expressed full-length MED1 (bv-MED1-FL) or MED1-ERK mutant (bv-MED1-Erk mut) were incubated in kinase buffer containing (+) or lacking (−) purified ERK1 and then probed by immunoblot using anti-phospho-threonine (α-p-Thr) and anti-MED1 antibodies. C, purified bv-MED1-FL and bv-MED1-Erk mut were preincubated in kinase buffer containing (+) or lacking (−) purified ERK1. The entire kinase reactions were then incubated with bv-AR and a biotinylated ARE. DNA-bound protein complexes were precipitated with streptavidin beads, washed, fractionated by SDS-PAGE, and then immunoblotted with anti-MED1 and anti-AR antibodies. 30% of the bv-MED1-FL and bv-MED1-Erk mut protein input were loaded in lanes 1 and 2.
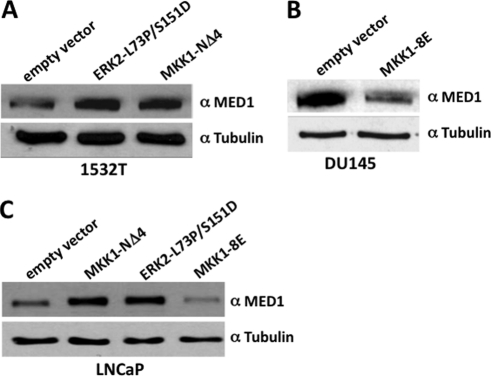
ERK1/2 phosphorylation significantly increases MED1 protein stability and half-life in cultured human HeLa cells (28). We have thus hypothesized that ERK1/2-induced stabilization of MED1 protein expression in mammalian cells presumably increases its availability as a coactivator for gene-specific transcriptional activators like AR. In view of the fact that AR is highly expressed in the prostate and plays an integral role in prostate cancer growth (2), we were curious to determine whether ERK phosphorylation of MED1 likewise stabilizes its expression in prostate cancer cells, which in turn, might have clinical implications for AR-mediated gene expression and prostate oncogenesis. To begin to explore this issue, we utilized prostate cancer 1532T cells that are derived from a primary adenocarcinoma (46) and that express relatively low levels of MED1 (34). Interestingly, we found that transfection of 1532T cells with a constitutively active ERK2 mutant construct (L73P/S151D) (39), or a constitutively active MAPK-kinase 1 mutant construct (MKK1-NΔ4) (38), markedly amplified MED1 expression levels (Fig. 8A). Contrarily, when prostate cancer DU145 cells, a metastatic cell line expressing relatively high levels of MED1 (34), were transfected with a dominant negative kinase-dead MKK1 mutant construct (MKK1–8E) (38), MED1 levels significantly decreased (Fig. 8B). Furthermore, we also found that we could modulate MED1 levels in prostate cancer LNCaP cells either up or down via the transient overexpression of the constitutively active MKK1-NΔ4 or ERK2-L73P/S151D mutant constructs or the dominant negative MKK1–8E mutant construct, respectively (Fig. 8C). These data confirm that MED1 phosphorylation via activated ERK1/2 can stabilize MED1 protein expression in prostate cancer cells.
FIGURE 8.
ERK phosphorylation of MED1 stabilizes protein expression in prostate cancer cells. A–C, prostate cancer cell lines 1532T (A), DU145 (B), and LNCaP (C) were transiently transfected with expression vectors for ERK2-L73P/S151D, MKK1-NΔ4, MKK1–8E, or empty vector controls as indicated. Whole cell extract was prepared, and equivalent amounts of protein were probed by immunoblot using anti-MED1 and anti-tubulin antibodies.
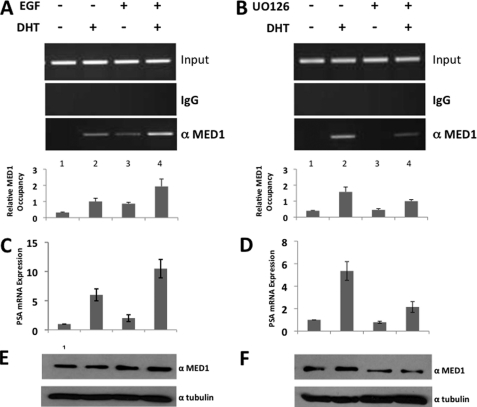
To further investigate whether ERK1/2 phosphorylation influences MED1 association with endogenous AR-target genes in vivo, we performed ChIP assays at the distal enhancer region of the PSA gene using LNCaP cells. In these studies, LNCaP cells were treated with or without DHT and epidermal growth factor (EGF), a potent activator of MAPK-ERK signaling pathways (47). In agreement with the MED1-AR binding assays (above), we observed higher occupancy of MED1 at the PSA enhancer in the presence of both EGF and DHT as compared with that observed with DHT treatment alone (Fig. 9A, lanes 2 and 4). We also observed a modest DHT-independent occupancy of MED1 at the PSA enhancer in the presence of EGF alone, possibly suggesting that MED1 phosphorylation by ERK1/2 may facilitate its recruitment to the PSA promoter via a mechanism independent of AR. Importantly, and consistent with the increased MED1 occupancy at the enhancer, DHT-dependent PSA gene expression was higher in the presence of EGF than in its absence (Fig. 9C).
FIGURE 9.
ERK phosphorylation of MED1 promotes its recruitment to the PSA enhancer. A–F, LNCaP cells were androgen-starved for 48 h and then treated with or without DHT and EGF (A, C, and E) or DHT and U0126 (B, D, and F) as indicated. A and B, chromatin was prepared and immunoprecipitated with anti-MED1 antibodies or nonspecific IgG. Semi-quantitative PCR was then performed using primer sets spanning the PSA distal enhancer. Image processing and quantification of the semi-quantitative PCR data were performed using Quantity One software (Bio-Rad). The results are presented as relative -fold induction of the enrichment over nonspecific IgG and normalized to input. C and D, PSA mRNA expression was determined by RT-PCR in real-time. Values were normalized to β-actin expression. E and F, equal amounts of whole cell extract were probed by immunoblot with antibodies against MED1 and α-tubulin. Error bars in A–D represent the S.D. calculated from three independent experiments.
When LNCaP cells were treated with both DHT and U0126, a specific chemical inhibitor of ERK1/2 activation, we observed a marked decrease in MED1 occupancy at the PSA enhancer as compared with that observed with DHT treatment alone (Fig. 9B, lanes 2 and 4). This result was accompanied by a significant decrease in DHT-dependent PSA gene expression (Fig. 9D). When taken into consideration with the phospho-MED1 binding experiments and expression assays presented in Figs. 7 and 8, our data thus suggest that ERK1/2 phosphorylation of MED1 can promote its association with AR in two ways: first, by directly stabilizing the MED1-AR protein-protein interaction, and second, by stabilizing MED1 cellular protein expression in prostate cells, which in turn increases its availability for interaction with AR.
DISCUSSION
The fundamental mechanism for ligand-dependent activation of NRs involves an allosteric realignment of conserved α-helix 12 in the C-terminal LBD thus generating a binding surface for transcriptional coactivators that contain signature LXXLL motifs. AR is uncommon among most NRs in that ligand triggers and intramolecular N/C interaction and that its major transcriptional activation domain resides at the NTD rather than at the C-terminal LBD (1, 2). The AR NTD can be subdivided into two distinct transactivation units termed Tau-1 and Tau-5 (48), but the mechanisms by which these regions recruit coregulatory factors and facilitate transcriptional activation are not completely understood. The evolutionary conserved Mediator complex plays a key coactivator role for nearly all ligand-activated NRs, including AR (23). Mediator is generally recruited to NRs via the LXXLL-containing MED1 subunit whose functional activity can be regulated by cellular MAPK transduction pathways. In this report, we investigated the mechanism by which AR interacts with MED1. We show for the first time that MED1 binds to the AR Tau-1 domain via two novel α-helical motifs located between MED1 amino acid residues 505 and 537. Neither of MED1's two signature LXXLL motifs appear to be required for this interaction, whereas loss of the intramolecular AR N/C interaction inhibits MED1 binding. We further show that MAPK ERK1/2 phosphorylation of MED1 enhances the MED1-AR interaction in vitro and within prostate cancer cells.
The AR Tau-5 domain is reported to be the primary recruitment surface for p160/SRC coactivators (14–16, 19). Tau-1 has also been strongly implicated as a potential coactivator binding site independent of p160/SRC proteins, but the identities of the corresponding interacting coregulatory factors have remained unclear (19). The data presented here suggest that MED1 is a direct interacting coactivating partner for Tau-1. Although Tau-5 retains transactivation potential in the absence of the AR LBD, Tau-1 was originally defined as requiring the AR LBD for its full transactivation function (48). In agreement with the functional requirement of Tau-1 for the LBD, and its functional role as an interaction site for MED1, we found that AR mutations that blunt the N/C interaction (the G21E point mutation or deletion of 23FQNLF27) significantly reduced or completely abolished MED1 binding (Fig. 2G). One interpretation of these findings is that the ligand-induced AR N/C interaction results in a conformational change that renders the Tau-1 domain accessible for MED1 binding. Notably, the N/C interaction was also shown to be required for AR binding with the SWI/SNF chromatin remodeling complex (49), although in this case, the primary AR recruitment surface appears to be located within the hinge region (18). Another possible explanation for the N/C interaction requirement for MED1-AR binding may be that MED1 contacts additional surfaces on the C terminus of AR that in turn may serve to stabilize the MED1-Tau-1 interaction.
An important finding of this study is the delineation of two tandem α-helices in the primary MED1 polypeptide sequence (α1 and α2, amino acids 505–519 and 523–537, respectively) that apparently serve as a binding surface for AR in the presence of ligand. Apart from the sequence LXLI found in α2 (amino acids 525–528), both α-helices appear to be structurally distinct from the signature LXXLL motifs found in other NR coactivators (4, 7). Moreover, both α-helices appear to be equally important as deletion of either motif abolishes AR binding. Collectively, our data suggest that the MED1 region containing this tandem α-helical array (amino acids 505–537) comprises a composite binding surface that facilitates functional interactions with AR. Indeed, transient overexpression of an MED1 mutant protein lacking this tandem α-helical array had no significant costimulatory effect on androgen-dependent reporter gene transcription as compared with the wild-type MED1 protein (Fig. 6A). Notably, the noncanonical α-helical array is highly conserved in mammals, yet poorly conserved in metazoans lacking androgen signaling pathways (Fig. 5I). Although this newly discovered α-helical array is located proximal to the two signature LXXLL motifs of MED1 (located at amino acids 604 and 645), we surprisingly found that both LXXLL motifs were dispensable for ligand-dependent AR binding (Fig. 3, C and D). Our results are thus reminiscent of the situation observed earlier with the p160/SRC coactivators that are also able to bind AR independently of their intrinsic LXXLL motifs and retain their ability to potentiate AR activity (14–16). Nevertheless, previous in vitro studies showed that the two MED1 LXXLL motifs are capable of facilitating a weak interaction with an isolated GST-AR-LBD fusion in the absence of the AR-NTD (9, 32). Thus it remains conceivable that MED1 may be capable of establishing two distinct contacts with AR, one at the Tau-1 region in the NTD and another weaker interaction at the AF-2 region in the LBD, possibly even serving as a molecular N/C bridge.
Interestingly, mutational analyses of the AR Tau-1 domain has revealed a presumptive Tau-1 core motif that, similar to MED1's AR binding motif, is composed of two putative tandem α-helices located between AR amino acids 173 and 203 (19). The first α-helix in this region has been proposed to serve as a binding surface for the Tab2 component of the NCoR corepressor complex when AR is bound to hormone antagonists and facilitates transcriptional silencing (50). In the presence of agonist, we found that MED1 failed to bind to an AR deletion mutant lacking the Tau-1 core motif (ΔcTau1) thus suggesting that this α-helical sub-domain is critical for MED1 interaction (Fig. 2D). Along these same lines, deletion of the core Tau-1 motif within the AR NTD enhanced p160/SRC recruitment at Tau-5 (19), possibly indicating that AR binding to MED1 versus p160/SRC may be mutually exclusive. Taken together, these data suggest that the association of MED1 with AR is primarily based on an intermolecular interaction involving two distinct α-helical arrays on each respective polypeptide. Biochemical and mutagenesis studies should reveal whether this interaction additionally involves key hydrophobic side chains and/or charged amino acid residues found within or flanking each respective α-helical array. In this regard, it is intriguing to speculate that the AR Tau-1 domain may serve as a binding surface for other coregulatory factors containing conserved α-helical motifs with structural similarity to that found in MED1.
Our findings also show that ERK1/2 phosphorylation of MED1 enhances its binding with AR. These data suggest that MED1 phosphorylation by ERK1/2 promotes its association with AR at two levels: first, it directly stabilizes the MED1-AR protein-protein interaction, and second, it stabilizes MED1 cellular protein expression in prostate cancer cells, which, in turn, stabilizes the amount of nuclear MED1 available for interaction with AR. The mechanism by which MED1 phosphorylation increases the one-to-one MED1-AR protein interaction is presently unclear. Given that the ERK1/2 phosphorylation sites in the MED1 polypeptide are not particularly close to the novel α-helical array region, MED1 phosphorylation might effect a conformational change that stabilizes or enhances the MED1-AR interaction between Tau-1 and the α-helical array. It is also possible that conformational changes triggered by MED1 phosphorylation might generate additional AR binding surfaces distinct from the α-helical array region. In view of the fact that testosterone, DHT, and other androgenic steroids can activate MAPK-ERK1/2 signaling in an extranuclear manner (27, 44, 45), we have proposed that MED1 phosphorylation may be a part of a feed-forward signaling mechanism by androgenic steroids in which AR coactivators, as well as AR itself, become activated and a maximal transcriptional response is achieved and sustained (27). In the case of MED1, androgen-stimulated phosphorylation not only enhances MED1 association with the core Mediator complex (27) but also with AR (Fig. 7).
It has been suggested that amplification of AR coactivators via hyperactivated MAPK-ERK signaling pathways may play a role in promoting prostate tumorigenesis to the androgen-independent stage (51). Interestingly, we have observed that MED1 expression levels are amplified in a number of prostate cancer cell lines and clinically localized human prostate cancer specimens (34). Because MAPK-ERK signaling pathways are commonly constitutively activated in prostate cancer cells (52–56), especially the androgen-independent disease (53–55), it is plausible that hyperactivated MAPKs might promote MED1 overexpression during prostate oncogenesis. Indeed, our findings showing that activated ERK1/2 stabilizes MED1 protein expression in cultured prostate cancer cells is consistent with this hypothesis (Fig. 8). A recent study suggests that MED1 may also be a phosphorylation target for other signal transduction pathways in androgen-independent prostate cancer cells and that phosphorylated protein can facilitate other key oncogenic events such as chromosomal looping (35). Future studies should more precisely reveal the structural determinants that underlie the different phosphorylation-dependent functions of MED1.
This work was supported by a United States Department of Defense Prostate Cancer Research Program Award W81XWH-10-1-0227 (to J. D. F.).
- DHT
- dihydrotestosterone
- AR
- androgen receptor
- NR
- nuclear receptor
- NTD
- N-terminal domain
- LBD
- ligand binding domain
- Tau-1
- transactivation unit-1
- RBD
- receptor binding domain
- PSA
- prostate-specific antigen
- CDS
- charcoal/dextran-stripped
- ARE
- androgen response element
- bv
- baculovirus-expressed.
REFERENCES
- 1. Claessens F., Denayer S., Van Tilborgh N., Kerkhofs S., Helsen C., Haelens A. (2008) Nucl. Recept Signal 6, e008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dehm S. M., Tindall D. J. (2007) Mol. Endocrinol. 21, 2855–2863 [DOI] [PubMed] [Google Scholar]
- 3. Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 4. Moras D., Gronemeyer H. (1998) Curr. Opin. Cell Biol. 10, 384–391 [DOI] [PubMed] [Google Scholar]
- 5. Danielian P. S., White R., Lees J. A., Parker M. G. (1992) EMBO J. 11, 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., Yamamoto K. R. (1998) Genes Dev. 12, 3343–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Heery D. M., Kalkhoven E., Hoare S., Parker M. G. (1997) Nature 387, 733–736 [DOI] [PubMed] [Google Scholar]
- 8. Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. (1998) Nature 395, 137–143 [DOI] [PubMed] [Google Scholar]
- 9. Zhou X. E., Suino-Powell K. M., Li J., He Y., Mackeigan J. P., Melcher K., Yong E. L., Xu H. E. (2010) J. Biol. Chem. 285, 9161–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dubbink H. J., Hersmus R., Verma C. S., van der Korput H. A., Berrevoets C. A., van Tol J., Ziel-van der Made A. C., Brinkmann A. O., Pike A. C., Trapman J. (2004) Mol. Endocrinol. 18, 2132–2150 [DOI] [PubMed] [Google Scholar]
- 11. He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. (2004) Mol. Cell 16, 425–438 [DOI] [PubMed] [Google Scholar]
- 12. He B., Kemppainen J. A., Wilson E. M. (2000) J. Biol. Chem. 275, 22986–22994 [DOI] [PubMed] [Google Scholar]
- 13. Hur E., Pfaff S. J., Payne E. S., Grøn H., Buehrer B. M., Fletterick R. J. (2004) PLoS Biol. 2, E274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alen P., Claessens F., Verhoeven G., Rombauts W., Peeters B. (1999) Mol. Cell. Biol. 19, 6085–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bevan C. L., Hoare S., Claessens F., Heery D. M., Parker M. G. (1999) Mol. Cell. Biol. 19, 8383–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Christiaens V., Bevan C. L., Callewaert L., Haelens A., Verrijdt G., Rombauts W., Claessens F. (2002) J. Biol. Chem. 277, 49230–49237 [DOI] [PubMed] [Google Scholar]
- 17. García-Pedrero J. M., Kiskinis E., Parker M. G., Belandia B. (2006) J. Biol. Chem. 281, 22656–22664 [DOI] [PubMed] [Google Scholar]
- 18. Link K. A., Balasubramaniam S., Sharma A., Comstock C. E., Godoy-Tundidor S., Powers N., Cao K. H., Haelens A., Claessens F., Revelo M. P., Knudsen K. E. (2008) Cancer Res. 68, 4551–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Callewaert L., Van Tilborgh N., Claessens F. (2006) Cancer Res. 66, 543–553 [DOI] [PubMed] [Google Scholar]
- 20. Kornberg R. D. (2005) Trends Biochem. Sci. 30, 235–239 [DOI] [PubMed] [Google Scholar]
- 21. Malik S., Roeder R. G. (2010) Nat. Rev. Genet. 11, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knuesel M. T., Taatjes D. J. (2011) Transcription 2, 28–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Belakavadi M., Fondell J. D. (2006) Rev. Physiol. Biochem. Pharmacol. 156, 23–43 [DOI] [PubMed] [Google Scholar]
- 24. Ren Y., Behre E., Ren Z., Zhang J., Wang Q., Fondell J. D. (2000) Mol. Cell. Biol. 20, 5433–5446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Taatjes D. J., Tjian R. (2004) Mol. Cell 14, 675–683 [DOI] [PubMed] [Google Scholar]
- 26. Zhang X., Krutchinsky A., Fukuda A., Chen W., Yamamura S., Chait B. T., Roeder R. G. (2005) Mol. Cell 19, 89–100 [DOI] [PubMed] [Google Scholar]
- 27. Belakavadi M., Pandey P. K., Vijayvargia R., Fondell J. D. (2008) Mol. Cell. Biol. 28, 3932–3942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pandey P. K., Udayakumar T. S., Lin X., Sharma D., Shapiro P. S., Fondell J. D. (2005) Mol. Cell. Biol. 25, 10695–10710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Baek S. H., Ohgi K. A., Nelson C. A., Welsbie D., Chen C., Sawyers C. L., Rose D. W., Rosenfeld M. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 3100–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jin F., Fondell J. D. (2009) Nucleic Acids Res. 37, 4826–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Q., Carroll J. S., Brown M. (2005) Mol. Cell 19, 631–642 [DOI] [PubMed] [Google Scholar]
- 32. Wang Q., Sharma D., Ren Y., Fondell J. D. (2002) J. Biol. Chem. 277, 42852–42858 [DOI] [PubMed] [Google Scholar]
- 33. Huang Z. Q., Li J., Sachs L. M., Cole P. A., Wong J. (2003) EMBO J. 22, 2146–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vijayvargia R., May M. S., Fondell J. D. (2007) Cancer Res. 67, 4034–4041 [DOI] [PubMed] [Google Scholar]
- 35. Chen Z., Zhang C., Wu D., Chen H., Rorick A., Zhang X., Wang Q. (2011) EMBO J. 30, 2405–2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Q., Li W., Zhang Y., Yuan X., Xu K., Yu J., Chen Z., Beroukhim R., Wang H., Lupien M., Wu T., Regan M. M., Meyer C. A., Carroll J. S., Manrai A. K., Jänne O. A., Balk S. P., Mehra R., Han B., Chinnaiyan A. M., Rubin M. A., True L., Fiorentino M., Fiore C., Loda M., Kantoff P. W., Liu X. S., Brown M. (2009) Cell 138, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Callewaert L., Verrijdt G., Christiaens V., Haelens A., Claessens F. (2003) J. Biol. Chem. 278, 8212–8218 [DOI] [PubMed] [Google Scholar]
- 38. Mansour S. J., Matten W. T., Hermann A. S., Candia J. M., Rong S., Fukasawa K., Vande Woude G. F., Ahn N. G. (1994) Science 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 39. Emrick M. A., Hoofnagle A. N., Miller A. S., Ten Eyck L. F., Ahn N. G. (2001) J. Biol. Chem. 276, 46469–46479 [DOI] [PubMed] [Google Scholar]
- 40. Celis L., Claessens F., Peeters B., Heyns W., Verhoeven G., Rombauts W. (1993) Mol. Cell. Endocrinol. 94, 165–172 [DOI] [PubMed] [Google Scholar]
- 41. Hittelman A. B., Burakov D., Iñiguez-Lluhí J. A., Freedman L. P., Garabedian M. J. (1999) EMBO J. 18, 5380–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee J. E., Kim K., Sacchettini J. C., Smith C. V., Safe S. (2005) J. Biol. Chem. 280, 8819–8830 [DOI] [PubMed] [Google Scholar]
- 43. Cleutjens K. B., van der Korput H. A., van Eekelen C. C., van Rooij H. C., Faber P. W., Trapman J. (1997) Mol. Endocrinol. 11, 148–161 [DOI] [PubMed] [Google Scholar]
- 44. Migliaccio A., Castoria G., Di Domenico M., de Falco A., Bilancio A., Lombardi M., Barone M. V., Ametrano D., Zannini M. S., Abbondanza C., Auricchio F. (2000) EMBO J. 19, 5406–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu X., Li H., Liu J. P., Funder J. W. (1999) Mol. Cell. Endocrinol. 152, 199–206 [DOI] [PubMed] [Google Scholar]
- 46. Bright R. K., Vocke C. D., Emmert-Buck M. R., Duray P. H., Solomon D., Fetsch P., Rhim J. S., Linehan W. M., Topalian S. L. (1997) Cancer Res. 57, 995–1002 [PubMed] [Google Scholar]
- 47. Pearson G., Robinson F., Beers Gibson T., Xu B. E., Karandikar M., Berman K., Cobb M. H. (2001) Endocr. Rev. 22, 153–183 [DOI] [PubMed] [Google Scholar]
- 48. Jenster G., van der Korput H. A., Trapman J., Brinkmann A. O. (1995) J. Biol. Chem. 270, 7341–7346 [DOI] [PubMed] [Google Scholar]
- 49. Li J., Fu J., Toumazou C., Yoon H. G., Wong J. (2006) Mol. Endocrinol. 20, 776–785 [DOI] [PubMed] [Google Scholar]
- 50. Zhu P., Baek S. H., Bourk E. M., Ohgi K. A., Garcia-Bassets I., Sanjo H., Akira S., Kotol P. F., Glass C. K., Rosenfeld M. G., Rose D. W. (2006) Cell 124, 615–629 [DOI] [PubMed] [Google Scholar]
- 51. Gregory C. W., Fei X., Ponguta L. A., He B., Bill H. M., French F. S., Wilson E. M. (2004) J. Biol. Chem. 279, 7119–7130 [DOI] [PubMed] [Google Scholar]
- 52. Bakin R. E., Gioeli D., Sikes R. A., Bissonette E. A., Weber M. J. (2003) Cancer Res. 63, 1981–1989 [PubMed] [Google Scholar]
- 53. Gao H., Ouyang X., Banach-Petrosky W. A., Gerald W. L., Shen M. M., Abate-Shen C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 14477–14482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gioeli D., Mandell J. W., Petroni G. R., Frierson H. F., Jr., Weber M. J. (1999) Cancer Res. 59, 279–284 [PubMed] [Google Scholar]
- 55. Unni E., Sun S., Nan B., McPhaul M. J., Cheskis B., Mancini M. A., Marcelli M. (2004) Cancer Res. 64, 7156–7168 [DOI] [PubMed] [Google Scholar]
- 56. Weber M. J., Gioeli D. (2004) J. Cell. Biochem. 91, 13–25 [DOI] [PubMed] [Google Scholar]