Background: The existence of gap junctions in differentiated skeletal muscles has been recently appreciated in vertebrates.
Results: Connexin 39.9-mediated gap junctions in slow-twitch muscles are necessary for robust activation of muscle in zebrafish.
Conclusion: Gap junction-mediated electrical coupling in skeletal muscle plays an essential role in coordinated behavior.
Significance: Gap junctions ensure robust muscle activation despite unreliable neural outputs during early motor development.
Keywords: Connexin, Electrophysiology, Gap Junctions, Skeletal Muscle, Zebrafish
Abstract
In many tissues and organs, connexin proteins assemble between neighboring cells to form gap junctions. These gap junctions facilitate direct intercellular communication between adjoining cells, allowing for the transmission of both chemical and electrical signals. In rodents, gap junctions are found in differentiating myoblasts and are important for myogenesis. Although gap junctions were once believed to be absent from differentiated skeletal muscle in mammals, recent studies in teleosts revealed that differentiated muscle does express connexins and is electrically coupled, at least at the larval stage. These findings raised questions regarding the functional significance of gap junctions in differentiated muscle. Our analysis of gap junctions in muscle began with the isolation of a zebrafish motor mutant that displayed weak coiling at day 1 of development, a behavior known to be driven by slow-twitch muscle (slow muscle). We identified a missense mutation in the gene encoding Connexin 39.9. In situ hybridization found connexin 39.9 to be expressed by slow muscle. Paired muscle recordings uncovered that wild-type slow muscles are electrically coupled, whereas mutant slow muscles are not. The further examination of cellular activity revealed aberrant, arrhythmic touch-evoked Ca2+ transients in mutant slow muscle and a reduction in the number of muscle fibers contracting in response to touch in mutants. These results indicate that Connexin 39.9 facilitates the spreading of neuronal inputs, which is irregular during motor development, beyond the muscle cells and that gap junctions play an essential role in the efficient recruitment of slow muscle fibers.
Introduction
Gap junctions are channels that directly connect the cytoplasms of adjacent cells (1–3). They allow intercellular passage of ions, second messengers, metabolites, and other small molecules up to 1000 Da and, thus, play important roles in many aspects of tissue/organ homeostasis. A gap junction channel is formed by the conjunction of two hemichannels, each composed of six connexin proteins.
More than 20 connexin genes have been identified so far in mammals and 37 in teleosts (4, 5). Measurement of connexin (Cx)5 gene expression levels and ultrastructural observation of gap junctions have revealed that gap junctions are present in many animal tissues (6, 7). In rodents, Cx39, Cx43, and Cx45 are expressed in differentiating myoblasts, and Cx43 promotes myogenesis (8–12). After the differentiation of skeletal muscle, however, the expression of these connexins attenuates in vivo. On the other hand, Northern blot analysis of human RNA revealed that transcripts of Cx40.1 and Cx43, which are orthologs of rodent Cx39 and Cx43, respectively, as well as transcripts of Cx59 and Cx62, are found in adult skeletal muscle, although expression of these connexin protein was not reported and their function in differentiated muscle is unclear (13). In zebrafish, intriguingly, orthologs of rodent Cx43 are not expressed in either developing myoblasts or differentiated skeletal muscle (5, 14, 15). Nevertheless, gap junctions are present in differentiated skeletal muscle of zebrafish (16–19). The connexin molecules present in zebrafish skeletal muscle have not yet been identified, and their functional importance in differentiated skeletal muscle remains unclear. In fact, physiological function of gap junctions in differentiated skeletal muscles is not well understood in any vertebrate species.
Forward genetics can be applied to zebrafish to identify genes that are essential for the development and function of muscle (20–28). Additionally, zebrafish embryos are transparent, a characteristic that facilitates the visualization of dynamic events such as cell migration and calcium transients in live fish (29); can be examined electrophysiologically in vivo (16–19, 26, 30); and exhibit readily assayable and stereotyped behaviors. By 22 h post-fertilization (hpf), zebrafish embryos respond to tactile stimulation with alternating coilings of the trunk (31–33). In zebrafish, as in other vertebrates, skeletal muscles are subdivided into two types on the basis of their physiology: slow-twitch muscle (slow muscle) and fast-twitch muscle (fast muscle) (34, 35). Touch-evoked coiling in embryos at 24 hpf is mediated primarily by slow muscle because differentiation of fast muscle is not yet complete (36, 37). As fast muscles differentiate, touch-evoked coiling is replaced at 48 hpf by touch-evoked burst swimming that requires fast muscles (32, 33, 38–40). This burst swimming is an urgent escape behavior being different from routine slow swimming in adult fish, the latter being mediated by slow muscles (41–43).
In this paper, we characterized zebrafish motility mutant cx39.9mi264 that carried a missense mutation in the cx39.9 gene, a member of the vertebrate GJA3 connexins (Cx46 subfamily). cx39.9 was expressed in differentiated slow muscles where it played an essential role in electrical coupling between slow muscle fibers. At 48 hpf, cx39.9 mutants responded to touch with normal burst swimming. However, at 26 hpf, mutants exhibited weak touch-evoked coiling because of a coupling defect between slow muscle cells.
EXPERIMENTAL PROCEDURES
Animal Care and Use
Zebrafish were bred and raised using established protocols (44, 45) that meet the guidelines set forth by the National Institute of Genetics. The cx39.9mi264 mutation was identified from an N-ethyl-N-nitrosourea mutagenesis performed at the University of Michigan (22).
Video Recording of Zebrafish Behavior
Embryonic behaviors were observed and video-recorded using a dissection microscope (Leica MZ16, Wetzlar, Germany) as described previously (46). Touch responses elicited by tactile stimulation delivered to the tail with forceps were captured by a charge-coupled device camera at 200 Hz (Fastcam-Ultima1024, Photron, Tokyo, Japan). For quantitative analysis of behaviors, we measured the trunk angle of embryos from above using a modified version of an established protocol (47). Embryos were visualized from above, and a line was drawn along the midline of the head and rostral trunk of embryos. A second line connecting the most anterior portion of the tail, with the point where the trunk becomes detached from the yolk sac, was drawn to represent the bend of the trunk. The angle between these two lines was defined as the trunk angle.
Muscle Recording and Dye Coupling Assay
The dissection protocols for in vivo patch recordings have been described elsewhere (17). Briefly, zebrafish embryos were anesthetized in 0.02% tricaine (ethyl 3-aminobenzoate methanesulfonate, Sigma-Aldrich, St. Louis, MO), pinned with tungsten wires (30-μm diameter, Nilaco, Tokyo, Japan) through the notochord onto a silicone-coated dish (SILPOT 184, Dow Corning, Midland, MI), and immersed in Evans solution (134 mm NaCl, 2.9 mm KCl, 2.1 mm CaCl2, 1.2 mm MgCl2, 10 mm glucose, and 10 mm HEPES at 290 mOsm (pH to 7.8) with NaOH). The skin was peeled off with forceps to allow access to the underlying muscles. During electrophysiological recordings, embryos were partially curarized in Evans solution containing 3 μm D-tubocurarine (Sigma-Aldrich) without tricaine. The patch pipettes were pulled from borosilicate glass (GD1.5, Narishige, Tokyo, Japan) to yield electrodes with a resistance of 5–10 MΩ. Electrodes were visually guided to patch muscle cells using Hoffman modulation optics (×40 water immersion objective, BX51WI, Olympus, Tokyo, Japan). The intracellular electrode solution consisted of 0.1% sulforhodamine B for cell type identification and 105 mm potassium gluconate, 16 mm KCl, 2 mm MgCl2, 10 mm HEPES, 10 mm EGTA and 4 mm Na3ATP at 273 mOsm (pH to 7.2) with KOH. The morphology of each patched cell was examined to determine whether the fiber was slow or fast muscle. Recordings were amplified with a Multiclamp 700A amplifier (Axon Instruments, Sunnyvale, CA) and sampled at 10 kHz. Data were collected with Clampex 10 software (Axon Instruments) and analyzed with Clampfit 10 software (Axon Instruments). Tactile stimuli were delivered by ejecting bath solution (30 psi, 20 ms) from a glass pipette with a 20-μm tip to the head of the pinned embryo using a Picospritzer III (Parker Hannifin Corp., Cleveland, OH). Paired recordings were performed as described previously (18). Pairs of neighboring muscle fibers within the same segment were selected for recording. The intracellular electrode solution contained 0.1% sulforhodamine B and 105 mm potassium gluconate, 15 mm KCl, 10 mm HEPES and 5 mm 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid at 290 mOsm (pH to 7.2) with KOH. Dye coupling was assayed 30 min after each single muscle cell was patch-clamped. Images of intercellular diffusion of sulforhodamine B were captured with a fluorescent microscope. To block gap junctions pharmacologically, heptanol (2 mm, Sigma-Aldrich) was applied to the bath solution.
Genetic Mapping
cx39.9 carrier fish were outcrossed with wild-type wild-type in Kalkutta fish to generate mapping carriers which were then crossed to identify mutants for meiotic mapping using microsatellites markers as described previously (48–50). The following PCR primers were designed to narrow down the mutant locus. mtmr2, 5′-GGAAGGCCAATTGAAAAGGT-3′ (forward) and 5′-TGGGTCTTTGTACATCTGCG-3′ (reverse) and zgc:66417, 5′-GTCACAAGAACAAGAGGTAAGC-3′ (forward) and 5′-TGGAAACGTTTACTGCCCTG-3′ (reverse).
Cloning, mRNA Rescue, and Antisense Knockdown
The following primers were used for the cloning of zebrafish cx39.9 cDNA. cx39.9, 5′-CTAGCAAGGTGGATGCACTGCAGCG-3′ (forward) and 5′-CATTCAAACTCCCTGATTCTGATGTCACTC-3′ (reverse).
For mutant rescue experiments, in vitro transcription of RNAs encoding wild-type and mutant cx39.9 was performed using an mMESSAGE mMACHINE T7 kit (Ambion, Austin, TX). 100 ng of capped wild-type cx39.9 RNA or mutant cx39.9 RNA was injected into cx39.9 heterozygous incross embryos at the one- to two-cell stage. Following further development, RNA-injected embryos were assayed for touch-evoked responses.
For morpholino (MO) knockdown, the following translational blocking cx39.9 antisense MO was raised: 5′-TAAAGTCTCCCATCGCTGCAGTGC-3′. The CAT sequence in bold corresponds to the start ATG codon. Antisense MOs for smyhc1, smyhc2 and ryr1b have been described previously (37). Standard control MO (randomized sequence available from Gene Tools, Philomath, OR) was used as a control. Wild-type embryos were injected with 5–15 ng of MO at the one- to two-cell stage as described previously (44, 51). At these doses, control MO produced no discernible change in phenotype.
In Situ Hybridization
In situ hybridizations of whole-mounted embryos were performed as described previously (52). For frozen sectioning after color development, embryos were equilibrated in mounting media (15% sucrose and 7.5% gelatin in PBS) at 37 °C and then stored at −80 °C until sectioning. 10-μm sections were cut with a cryostat (Leica, CM1850). Probes were synthesized using full-length cx39.9 as a template. Antisense staining was considered specific when absent from sense control-treated embryos.
Immunostaining
Zebrafish embryos were anesthetized in 0.02% tricaine and pinned on a silicone-coated dish with tungsten wires (30 μm in diameter). Following removal of the skin atop the trunk region, embryos were fixed in 4% paraformaldehyde at room temperature for 1 h and subjected to immunostaining as described previously (53). The following antibodies and labeling reagents were used: anti-slow myosin (F59, mouse IgG1, 1/50 dilution, Developmental Studies Hybridoma Bank, Iowa City, IA), anti-fast myosin (MF20, mouse IgG2b, 1/50 dilution, Developmental Studies Hybridoma Bank), anti-laminin (rabbit polyclonal, 1/500, Sigma-Aldrich), Alexa Fluor 568 phalloidin (1/1000 dilution, Molecular Probes, Carlsbad, CA), Alexa Fluor 488-conjugated anti-mouse IgG (1/1000 dilution, Molecular Probes), and Alexa Fluor 488-conjugated anti-rabbit IgG (1/1000 dilution, Molecular Probes). Fluorescent images were captured through a confocal microscope (FV-300, Olympus).
RESULTS
mi264 Mutants Exhibit Weak Coiling Behavior because of Reduced Contractility
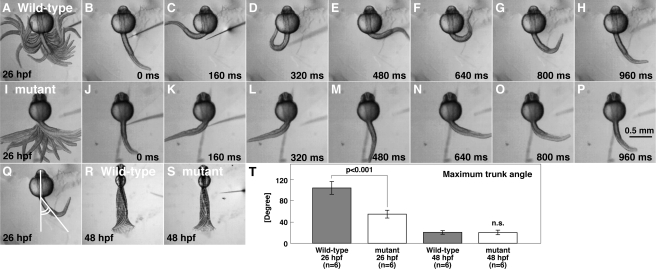
Early during development, zebrafish embryos display two highly stereotyped touch-evoked behaviors: coiling and burst swimming. Touch-evoked coiling at 26 hpf consists of one to three alternating contractions of the trunk and tail. At 48 hpf, tactile stimuli evoke burst swimming typically lasting a few seconds. In a screen for mutations, which affected zebrafish touch-evoked motor behaviors, one family, mi264, was found to yield a fraction of offspring that displayed reduced touch-evoked coiling at 26 hpf but seemingly normal touch-evoked burst swimming at 48 hpf. The reduction in touch-evoked coiling amplitude in mi264 mutants was observed between 21–34 hpf, with the greatest difference occurring at 26 hpf. We therefore quantified touch-evoked coiling at 26 hpf by measuring the displacement of the trunk of embryos with their heads and yolks restrained in agarose (supplemental movies 1 and 2). When analyzed, the frequency of coiling (wild-type, 1.92 ± 0.32 Hz, n = 6, Fig. 1A--H; mutant, 1.71 ± 0.29 Hz, n = 6, Fig. 1, I–P) and initial coil duration (wild-type, 169 ± 11 ms, n = 6; mutant, 178 ± 14 ms, n = 6) were comparable between wild-type and mutants, suggesting that the timing of motor output of muscles was not perturbed in mutants. However, the amplitude of coiling, measured as the maximum angle of the trunk from the longitudinal axis of the head (Fig. 1Q, see “Experimental Procedures” for details), was significantly decreased in mutants. A closer examination revealed that wild-type embryos bent their trunks 103 ± 12° from rest (n = 6, Fig. 1T), whereas mutant embryos bent only 54 ± 7° (n = 6, Student's t test, p < 0.001). As the duration of the initial coil was unaffected in mutants, the angular velocity was also reduced in mutants (wild-type, 611 ± 71°/s, n = 6; mutants, 304 ± 41°/s, n = 6; Student's t test, p < 0.001). Later in development (> 36 hpf), touch-evoked responses became seemingly normal in mutants, a time point when the majority of zebrafish embryos display sustained bouts of burst swimming (Fig. 1, R and S). A comparison of burst swimming attributes between wild-type and mutants at 48 hpf found that the contraction frequencies (wild-type, 34.3 ± 3.0 Hz, n = 6; mutant, 32.2 ± 2.5 Hz, n = 6), the maximum amplitude of trunk contractions (wild-type, 19.2 ± 3.5°, n = 6; mutant, 19.5 ± 4.5°, n = 6), and swimming speed (wild-type, 6.48 ± 1.04 cm/s, n = 6; mutant, 6.15 ± 0.78 cm/s, n = 6) did not differ significantly. Thus, the mi264 mutation selectively affects touch-evoked coiling behaviors in zebrafish.
FIGURE 1.
mi264 embryos exhibit weak touch-evoked coiling. A–H, tactile stimulation induced vigorous coiling in a wild-type embryo (26 hpf). The head and yolk of the embryo was embedded in an agarose gel, leaving the trunk and tail free to move. Superimposed (A) and individual movie frames (B–H) show vigorous coiling behavior in the wild-type embryo. I–P, tactile stimulation induced weak coiling in a mi264 embryo. Superimposed (I) and single frames (J–P) reveal weak coiling in the mutant embryo. Q, schematic of quantitative analysis of the amplitude of the coils as measured by the caudal trunk angle. R and S, superimposition of movie frames showing that the displacement of the trunk and tail in a wild-type embryo (R) and a mi264 mutant (S) during burst swimming are comparable at 48 hpf. T, histograms showing that the maximum amplitude of touch-induce coiling, but not burst swimming, is reduced in mutants. n.s., not significant.
mi264 Encodes for Cx39.9
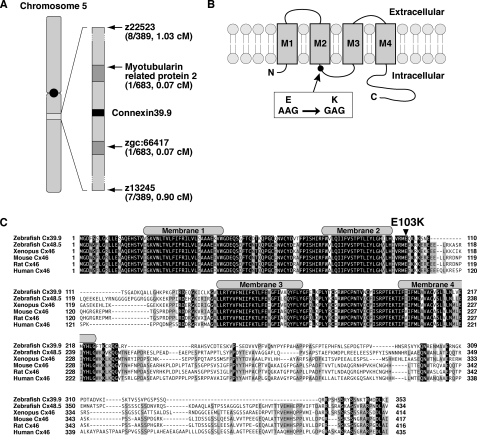
To identify the gene responsible for reduced touch-evoked coiling in mutants, we performed meiotic mapping from 683 mutants, which identified a region of chromosome 5 defined by two flanking microsatellites, z22523 (1.03 cM, 8 recombinants in 778 meioses) and z13245 (0.90 cM, 7/778, Fig. 2A). Subsequent higher-resolution mapping identified a critical region containing three genes. The cloning and sequencing of these three candidate genes uncovered a missense mutation in connexin 39.9 (GenBankTM accession no. AB678409), converting a conserved glutamate residue in an intracellular loop between membrane domains 2 and 3 to a lysine residue (E103K, Fig. 2B). Zebrafish Cx39.9 (353 amino acids, 39.9 kDa) belongs to the GJA3 subgroup of connexin proteins, which includes mammalian and Xenopus Cx46 and the zebrafish Cx48.5 (5). The existence of two orthologs for a mammalian gene is common within zebrafish because of a partial genome duplication in the bony fish lineage (54). Protein sequence alignment revealed that zebrafish Cx48.5 is more closely related to mammalian Cx46 than zebrafish Cx39.9 (Fig. 2C), suggesting that Cx48.5 is the ortholog of Cx46, whereas Cx39.9 is a derivative (see “Discussion”). The N terminus membrane domains of Cx39.9 were highly conserved with other GJA3 connexins, whereas the C terminus intracellular tail was divergent.
FIGURE 2.
mi264 embryos have a mutation in Connexin 39.9. A, meiotic mapping placed the mi264 locus between myotubularin related protein 2 and zgc:66417 in chromosome 5. B, in mutants, a missense mutation (E103K) was found in an intracellular loop between membrane domains 2 and 3. C, protein alignment of the GJA3 group of connexins in vertebrates. Shaded residues indicate conserved amino acids. The gray boxes represent the membrane domains. Note that the Glu-103 residue, which is mutated in cx39.9 mutants, is completely conserved among vertebrates.
To confirm that cx39.9 is the causative gene in mi264 mutants, we attempted to restore touch-evoked coiling in mutants by injection of RNA encoding wild-type Cx39.9 into embryos obtained from incrosses of mi264 carriers. We found that only 5% (1 of 20) of carrier incross progeny injected with wild-type cx39.9 RNA exhibited reduced touch-evoked coiling. In contrast, 30% (6 of 20) of carrier incross progeny injected with mutant (E103K) RNA exhibited diminished touch-evoked coiling, indicating a failure to rescue. In addition to RNA rescue, we attempted to recapitulate the mutant phenotype by antisense MO-mediated knockdown of Cx39.9. To this end, an antisense MO was raised against the translational start site of cx39.9. Injection of the antisense cx39.9 MO into wild-type embryos resulted in embryos with weak touch-evoked coils at 26 hpf (85%, 17 of 20, maximum trunk angle < 70°), whereas no control MO-injected embryos with reduced responses were observed (0%, 0 of 20). Thus, cx39.9 is the causative gene responsible for the weak touch-evoked coils seen in mi264 mutants.
cx39.9 Is Expressed by Muscle
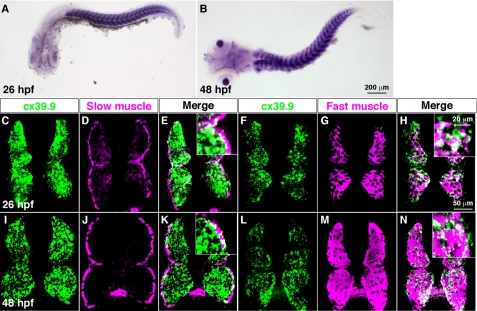
Whole-mount in situ hybridization revealed that cx39.9 was expressed by axial skeletal muscles and lens at 26 and 48 hpf (Fig. 3, A and B). In zebrafish, axial skeletal muscle is segregated into two groups: a single superficial layer of slow muscle fibers, and multiple medial layers of fast muscle fibers (34). To determine whether cx39.9 was selectively expressed by one skeletal muscle type, we performed fluorescent in situ hybridization coupled with immunohistochemistry. We found that cx39.9 was expressed by both slow and fast muscles at 26 and 48 hpf (Fig. 3C-N). Furthermore, cx39.9 expression was not detected in the spinal cord, indicating that the mutant phenotype arises from a defect in skeletal muscle.
FIGURE 3.
cx39.9 is expressed in slow and fast skeletal muscles. A and B, in situ hybridization with cx39.9 riboprobe showed that cx39.9 was expressed in axial muscles at 26 hpf (A) and 48 hpf (B). C–N, double labeling of cross sections of the trunk with a cx39.9 riboprobe and anti-slow muscle myosin showed that cx39.9 is expressed in slow muscle at 26 hpf (C–E) and 48 hpf (I–K). Double labeling with a cx39.9 riboprobe and anti-fast muscle myosin showed that cx39.9 is expressed in fast muscle at 26 hpf (F–H) and 48 hpf (L--N).
Muscle Morphology Is Unaffected in cx39.9 Mutants
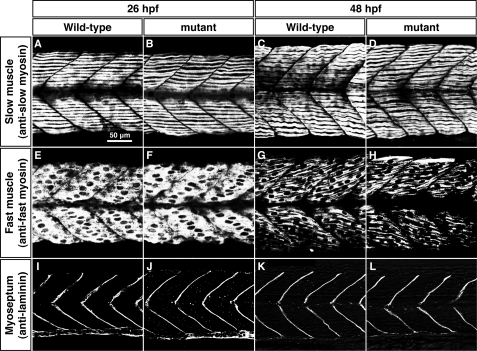
In rodents, Cx43, which is expressed in differentiating myoblasts, plays an important role in muscle differentiation (10, 12). To address whether the cx39.9 mutation affects myogenesis, we examined morphology of muscle. Differential interference contrast imaging of muscles revealed no obvious defects, with mutant fibers exhibiting normal striations. In fact, labeling with anti-slow and anti-fast myosin showed that myosin fibers in both fiber types are well organized in mutants just like in wild-type embryos at both 26 and 48 hpf (Fig. 4, A–H). We also labeled the myosepta, the structures that form somite boundaries, with anti-laminin and found that they were unaffected in mutants (Fig. 4, I–L). These results indicate that differentiation of muscle is unperturbed in cx39.9 mutants, suggesting that the mutant phenotype arises from a functional defect in muscle physiology.
FIGURE 4.
Morphology of slow and fast muscle is unperturbed in cx39.9 mutants. A–D, labeling with anti-slow muscle myosin at 26 hpf (A and B) and 48 hpf (C and D) showed that the distribution of slow myosin is normal in mutants. E–H, labeling with anti-fast muscle myosin at 26 hpf (E and F) and 48 hpf (G and H) showed that the distribution of fast myosin is normal in mutants. I–L, immunolabeling of myosepta with anti-laminin at 26 hpf (I and J) and 48 hpf (K and L) showed that the myotomes had normal morphology in mutants.
cx39.9mi264 Mutants Have Defects in Slow Muscle Contraction
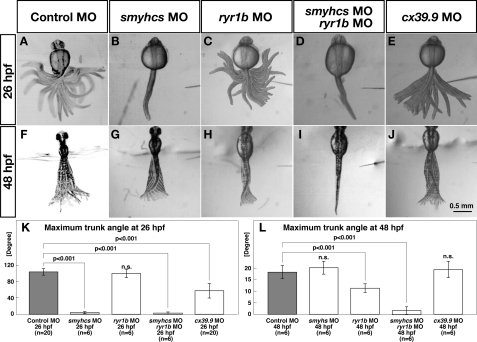
Touch-evoked coiling behavior at 26 hpf is executed by slow muscles, whereas burst swimming at 48 hpf is mediated primarily by fast muscles (37). Because cx39.9 mutants show defects in coiling at 26 hpf but not in burst swimming at 48 hpf, it is likely that cx39.9 mutants have slow but not fast muscle defects. One prediction of this hypothesis is that embryos with impaired slow muscles but not fast muscles should exhibit touch-evoked coiling similar to that shown by cx39.9-deficient embryos. To test this possibility, we replicated knockdown of slow myosins and fast muscle-specific Ca2+ release channels selectively to impair slow and fast muscles, respectively (23, 37, 55), and measured the amplitudes of the trunk movement during touch-evoked coiling and burst swimming. Control MO-injected embryos exhibited normal coiling at 26 hpf (104.1 ± 8.1°, n = 20, Fig. 5A) and normal burst swimming at 48 hpf (18.2 ± 2.8°, n = 6, Fig. 5F). Injection of a mixture of smyhc1 MO and smyhc2 MO (referred to as smyhcs MO) that is effective in inactivating slow muscles resulted in embryos that exhibited no motility in response to touch at 26 hpf (3.3 ± 3.8°, n = 6, Student's t test, p < 0.001, Fig. 5, B and K) but showed normal burst swimming at 48 hpf (20.2 ± 2.0°, n = 6, Fig. 5, G and L), confirming that touch-evoked coiling at 26 hpf is mediated by slow muscle. To selectively inactivate fast muscles, we injected MO against ryr1b, which encodes for the fast muscle-specific ryanodine receptor Ca2+ release channel that is essential for fast muscle contraction (23). The ryr1b morphants displayed normal touch-evoked coiling at 26 hpf (100.2 ± 11.4°, n = 6; Fig. 5C), but their burst swimming at 48 hpf was significantly compromised (11.3 ± 2.0°, n = 6; Student's t test, p < 0.001, Fig. 5H). Moreover, injection of both smyhcs and ryr1b MOs virtually eliminated both coiling (2.8 ± 3.4°, n = 6; Student's t test, p < 0.001, Fig. 5D) and burst swimming (1.7 ± 2.0°, n = 6, Student's t test, p < 0.001, Fig. 5I), confirming that coiling and burst swimming are mediated by slow and fast muscle, respectively. Embryos injected with cx39.9 MO exhibited weak coiling at 26 hpf (57.6 ± 19.0°, n = 20, Student's t test, p < 0.001, Fig. 5E) similar to cx39.9 mutants, whereas their burst swimming was unaffected (19.3 ± 3.8°, n = 6, Fig. 5J). These results indicate that the compromised coiling behavior seen in cx39.9 mutants is attributable to slow muscle defects.
FIGURE 5.
cx39.9 mutants have defects in slow muscle contraction. A–J, superimposed images of the trunk and tail movement during touch-evoked coiling and burst swimming. Control (A) and ryr1b (C) morphants exhibited normal coiling at 26 hpf. smyhcs morphants (B) and smyhcs-ryr1b double morphants (D) did not move in response to touch at 26 hpf. cx39.9 morphants (E) displayed weak coiling at 26 hpf. Control (F), smyhcs (G), and cx39.9 (J) morphants showed normal burst swimming at 48 hpf. The burst swimming was compromised in ryr1b morphants (H), and smyhcs-ryr1b double morphants (I) did not respond. K–L, Histograms showing that the amplitude of coiling was decreased in smyhcs, smyhcs/ryr1b, and cx39.9 morphants but not in ryr1b morphants (K) and that burst swimming amplitude was decreased in ryr1b and smyhcs/ryr1b morphants but not in either smyhcs or cx39.9 morphants (L).
Electrical Coupling Is Missing in cx39.9 Mutant Slow Muscle
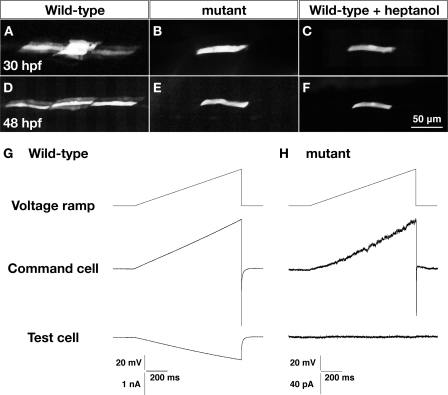
One possible explanation of why Cx39.9 is necessary for slow muscle contractions is that gap junction-mediated electrical coupling enables the sharing of synaptic currents in slow muscle tissue to coordinate coiling behavior. To assess intercellular coupling in slow muscle, we first assayed dye coupling using patch electrodes filled with recording solution containing sulforhodamine B (559 Da). At 30 hpf, the dye spread to 8.8 ± 1.5 (n = 6) slow muscle cells within 30 min in wild-type embryos (Fig. 6A). The dye was also found to have spread in both the dorsoventral and rostrocaudal directions but did not spread from slow to fast muscle cells. In cx39.9 mutants, however, there was no dye coupling of slow muscle cells (n = 4, Fig. 6B). To ensure that the dye was transferring to neighboring fibers through gap junctions, we exposed wild-type embryos to heptanol, a gap junction blocker. Application of heptanol was found to block dye migration in wild-type slow muscle (n = 3, Fig. 6C). Similarly, dye coupling was observed at 48 hpf in wild-type slow muscle (6.8 ± 1.7 cells, n = 6, Fig. 3D) but not in either mutant slow muscle (n = 3, Fig. 6E) or heptanol-treated wild-type slow muscle (n = 3, F).
FIGURE 6.
Dye and electrical coupling is absent in cx39.9 mutant slow muscle. A–D, sulforhodamine dye was loaded into a single slow muscle cell. Dye coupling was seen in wild-type (A and D) but not cx39.9 mutant (B and E) nor wild-type fibers exposed to the gap junction blocker heptanol (C and F). G and H, a voltage ramp from −50 mV to 0 mV over 1 s was generated in a slow muscle cell (command cell). Membrane currents in the command cell and an adjacent slow muscle cell were recorded in wild-type embryos (G) and cx39.9 mutants (H). No electrical coupling was detected in mutants. Note that currents are considerably smaller in mutants as a result of the higher input resistance of the mutant cells, which is also consistent with the absence of coupling in slow muscles.
Another indicator of coupling is the input resistance of muscle fibers, which is inversely related to the amount of current leaking from the cell (e.g. via gap junctions). We found that the input resistance of wild-type slow muscle fibers was significantly lower than slow muscle from cx39.9 mutants at both 30 hpf (wild-type, 25.7 ± 8.8 MΩ, n = 6, mutant: 739 ± 249 MΩ, n = 6, Student's t test, p < 0.001) and 48 hpf (wild-type, 22.3 ± 4.3 MΩ, n = 18; mutant, 820 ± 341 MΩ, n = 22, Student's t test, p < 0.001). These results indicate a severe reduction in intercellular coupling in cx39.9 mutant slow muscle.
To further examine this reduction of coupling electrophysiologically, we performed in vivo paired voltage clamp recordings between neighboring slow muscle cells. We delivered a voltage ramp in one slow muscle fiber (command cell) and recorded the corresponding current response in an adjacent slow muscle fiber (test cell) that was held at a constant membrane potential. In wild-type slow muscles, the voltage ramp generated outward currents in the command cell along with an inward current in the adjacent test cell (Fig. 6G). The inward current measured in the coupled adjacent test cell is due to current flowing from the command cell through gap junctions. The extent of current flow is proportional to the amount of coupling, which can be plotted as the ratio of the maximum currents in adjacent test cells versus command cells. In wild-type slow muscle, this ratio was found to be 0.49 ± 0.07 (n = 9). In mutants, however, voltage ramps in command cells failed to elicit currents in adjacent test cells (n = 11, Fig. 6H). The outward currents in command cells were smaller in slow muscle fibers from cx39.9 mutants because of the higher input resistance. These data, along with the lack of dye coupling and high input resistance in mutants, indicate that electrical coupling is absent in cx39.9 mutant slow muscle.
Cx39.9 Is Necessary for Synchronous Contractions of Slow Muscle
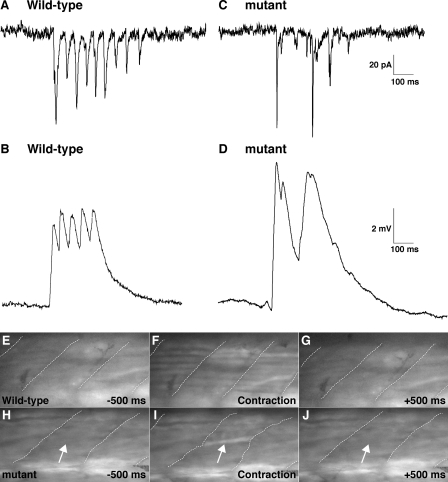
One possible explanation for why the loss of coupling between slow muscle cells leads to diminished touch-evoked coiling in cx39.9 mutants is that each slow muscle fiber in zebrafish does not receive direct synaptic input during each phase of the response but rather that synaptic inputs are shared between fibers via gap junctions. This hypothesis predicts that when coupling is eliminated, slow muscles will not reliably receive synaptic input during each phase of the touch-evoked coiling response. Indeed, in response to touch, synaptic currents and synaptic potentials in wild-type slow muscles were cyclically elicited with comparable amplitude (n = 3, Fig. 7, A and B), whereas they were sporadic in mutant slow muscles (n = 3, Fig. 7, C and D). These results indicate that gap junctions are required for reliable activation of slow muscles when synaptic inputs are not robust, at least in the coiling stage. They also suggest that coupling allows slow fibers to share synaptic currents to mediate synchronous slow muscle contractions.
FIGURE 7.
Rhythmic and synchronous activation of slow muscle is perturbed in cx39.9 mutants. A–D, electrophysiological recordings of synaptic inputs (current, A and C; membrane potential, B and D) to slow muscles. Synaptic inputs to slow muscle fibers were cyclic in wild-type (A and B) but arrhythmic in mutants (C and D). E–J, simultaneous observation of Ca2+ transients and muscle contractions. E–G, simultaneous increase in cytosolic Ca2+ was observed in many wild-type slow muscle fibers. Myotomal borders are depicted by dotted lines (H–J). A Ca2+ transient was seen only in a single mutant slow muscle cell (arrow). Contraction by the single fiber led to distortion of the myotome (wavy dotted line in I).
To further test whether coupling is required for synchronous activation of slow muscle fibers, we assessed the recruitment of slow muscle fibers by monitoring Ca2+ transients, which activate contractile machinery during muscle movement. To this end, the hspGFF28B transgenic zebrafish, which expresses a modified GAL4 transcription factor in slow muscle cells (56), were crossed with UAS:GCaMPHS4A zebrafish (57) to generate double transgenic embryos that express the Ca2+ indicator GCaMP-HS selectively in slow muscle cells. Employing these double transgenic embryos, we simultaneously assayed touch-evoked Ca2+ transients and slow muscle contractions in embryos at 28–34 hpf. In wild-type embryos, Ca2+ transients were observed simultaneously in a large number of slow muscle fibers during synchronous muscle contractions (Fig. 7, E–G and supplemental movie 3). By contrast, only a small number of slow muscle fibers in cx39.9 mutants were activated that resulted in diminished muscular contractions evident as distortions of the myotome (Fig. 7, H–J, Movie 4). These results indicate that gap junction-mediated electrical coupling is essential for the simultaneous activation of slow muscle fibers and, thereby, for the synchronous contraction of slow muscles to produce vigorous coiling behavior in response to touch.
DISCUSSION
In this study, we characterized zebrafish cx39.9 mutants that exhibit weak touch-evoked coiling behavior. cx39.9 is expressed in slow muscle and is required for electrical coupling of slow muscle fibers. In wild-type embryos, slow muscle cells share synaptic currents, and this electrical communication enables simultaneous and robust activation of a large number of slow muscle cells. In mutants, by contrast, activation of slow muscle cells was sporadic so that many muscle cells contracted asynchronously, resulting in a reduction of contractility. These findings suggest that gap junctions in differentiated skeletal slow muscle play a pivotal role in robust and orderly contractions and thus, efficient escape behavior in zebrafish. To our knowledge, this is the first demonstration of a physiological function for gap junctions in differentiated skeletal muscle of vertebrates.
Electrical Coupling in Zebrafish Skeletal Muscle
Although previous studies have established electrical and dye coupling in skeletal muscles of zebrafish (16–19), the physiological significance of coupling has been unclear. Our genetic and physiological analysis showed that electrical coupling is required for a slow muscle-dependent motor response and that this required gap junctions formed by Cx39.9. Our findings further suggest that Cx39.9-mediated coupling serves to allow sharing of synaptic inputs between neighboring slow muscle fibers because any one fiber is only sporadically activated during motor response at least by 34 hpf. Sporadic activation of slow muscle fibers could be due to sporadic activation of individual motor neurons when the motor circuit in the CNS is immature.
We assayed coupling of slow muscle cells in vivo as early as 30 hpf when zebrafish motility develops from coiling to burst swimming. Because peeling off the skin of zebrafish embryos to load fluorescent dye into an intact muscle cell is technically difficult in the earlier stage (∼30 hpf), we were unable to identify when muscle cells become coupled in zebrafish. Weak coiling behavior in cx39.9 mutants is apparent at 21 hpf. Thus, we assume that slow muscle cells are coupled at the latest by 21 hpf during early motor development.
Connexin Genes in Zebrafish
Connexin genes are named according to the predicted molecular weight of the protein in vertebrates (5, 58). Because of the suspected duplication of the entire genome during teleost evolution, zebrafish have two counterparts to many mammalian genes (54). In fact, in silico analyses of genomic databases have identified 37 connexin genes in zebrafish (5). However, the correspondence of connexin genes between teleosts and mammals appears to be complicated. Some mammalian connexins appear not to have orthologs in zebrafish, some have a single ortholog and others are found in multiple copies (5). In addition, zebrafish have 14 novel connexins that cannot be classified into any mammalian connexin subfamily. Thus, it seems likely that the connexin gene family is increasing in complexity within independently evolving lineages, leading to lineage-specific specialization of gap junction function.
Zebrafish Cx39.9 and Cx48.5 belong to the GJA3 group of the connexin superfamily that also includes human, rodent, and frog Cx46. Our phylogenetic analysis suggest that Cx48.5 is more closely related to mammalian Cx46 than Cx39.9. In mice, Cx46 is expressed in the lens and heart, and Cx46-deficient mice develop cataracts, indicating that Cx46-dependent gap junctional communication is essential for differentiation of the lens (59, 60). Likewise, zebrafish cx48.5 is expressed in the lens and heart, and antisense knockdown of Cx48.5 causes a developmental defect of the lens that is similar to cataracts, suggesting that zebrafish Cx48.5 is the functional counterpart of mammalian Cx46 (61, 62). On the other hand, cx39.9 is expressed in the skeletal muscle as well as in the lens, signifying that Cx39.9 has diverged from the GJA3 connexin subfamily. Unlike the other GJA3/Cx46 connexins, cx39.9 mutants display no defects in lens development. Thus, it appears that cx39.9 gene has evolved to be specialized for skeletal muscle function.
What is the functional counterpart of zebrafish Cx39.9 in mammals? In rodents, Cx46 is not expressed in either differentiating myoblasts or differentiated muscles. Instead, Cx39 and Cx43 are expressed in differentiating myoblasts and are involved in myogenesis (8–12). After muscle differentiation, however, expression of Cx39 and Cx43 is attenuated. Whether Cx39-, Cx43- or other connexin-mediated gap junctions are present and whether they contribute to efficient contraction in early differentiated muscle remains to be determined in mammals.
Supplementary Material
Acknowledgments
This work was supported, in whole or in part, by National Institutes of Health Grant NS-18205 (to H. W.). This work was also supported by the National BioResource Project, Zebrafish We thank Dr. Paul Brehm (Oregon Health and Science University) for assistance with dual patch recording. We also thank Dr. Yoichi Oda (Nagoya University), Dr. Shin Takagi (Nagoya University), Ms. Yurie Matsutani (Nagoya University), Kiyomi Miyazawa (National Institute of Genetics), Kazuko Ohishi (National Institute of Genetics), and Takako Ohnuki (National Institute of Genetics) for discussion, fish care, and other support.
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by Precursory Research for Embryonic Science and Technology, Japan Science and Technology Agency (Development and Function of Neural Networks), by the Takeda Science Foundation and by a career development award from the Human Frontier Science Program (to H. H.).

This article contains supplemental movies 1–4.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s)AB678409.
- Cx
- connexin
- hpf
- hours post-fertilization
- MO
- morpholino.
REFERENCES
- 1. Goodenough D. A., Paul D. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 285–294 [DOI] [PubMed] [Google Scholar]
- 2. White T. W., Paul D. L. (1999) Annu. Rev. Physiol. 61, 283–310 [DOI] [PubMed] [Google Scholar]
- 3. Yeager M., Harris A. L. (2007) Curr. Opin. Cell Biol. 19, 521–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cruciani V., Mikalsen S. O. (2006) Cell Mol. Life Sci. 63, 1125–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eastman S. D., Chen T. H., Falk M. M., Mendelson T. C., Iovine M. K. (2006) Genomics 87, 265–274 [DOI] [PubMed] [Google Scholar]
- 6. Kalderon N., Epstein M. L., Gilula N. B. (1977) J. Cell Biol. 75, 788–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rash J. E., Fambrough D. (1973) Dev. Biol. 30, 166–186 [DOI] [PubMed] [Google Scholar]
- 8. Araya R., Eckardt D., Riquelme M. A., Willecke K., Sáez J. C. (2003) Cell Commun. Adhes. 10, 451–456 [DOI] [PubMed] [Google Scholar]
- 9. Belluardo N., Trovato-Salinaro A., Mudò G., Condorelli D. F. (2005) Cell Tissue Res. 320, 299–310 [DOI] [PubMed] [Google Scholar]
- 10. Proulx A., Merrifield P. A., Naus C. C. (1997) Dev. Genet. 20, 133–144 [DOI] [PubMed] [Google Scholar]
- 11. von Maltzahn J., Euwens C., Willecke K., Söhl G. (2004) J. Cell Sci. 117, 5381–5392 [DOI] [PubMed] [Google Scholar]
- 12. Araya R., Eckardt D., Maxeiner S., Krüger O., Theis M., Willecke K., Sáez J. C. (2005) J. Cell Sci. 118, 27–37 [DOI] [PubMed] [Google Scholar]
- 13. Söhl G., Nielsen P. A., Eiberger J., Willecke K. (2003) Cell Commun. Adhes. 10, 27–36 [DOI] [PubMed] [Google Scholar]
- 14. Chatterjee B., Chin A. J., Valdimarsson G., Finis C., Sonntag J. M., Choi B. Y., Tao L., Balasubramanian K., Bell C., Krufka A., Kozlowski D. J., Johnson R. G., Lo C. W. (2005) Dev. Dyn. 233, 890–906 [DOI] [PubMed] [Google Scholar]
- 15. Gerhart S. V., Jefferis R., Iovine M. K. (2009) FEBS Lett. 583, 3419–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buckingham S. D., Ali D. W. (2004) J. Exp. Biol. 207, 841–852 [DOI] [PubMed] [Google Scholar]
- 17. Buss R. R., Drapeau P. (2000) J. Neurophysiol. 84, 1545–1557 [DOI] [PubMed] [Google Scholar]
- 18. Luna V. M., Brehm P. (2006) J. Gen. Physiol. 128, 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyen P. V., Aniksztejn L., Catarsi S., Drapeau P. (1999) J. Neurophysiol. 81, 2852–2861 [DOI] [PubMed] [Google Scholar]
- 20. Lieschke G. J., Currie P. D. (2007) Nat. Rev. Genet. 8, 353–367 [DOI] [PubMed] [Google Scholar]
- 21. Granato M., van Eeden F. J., Schach U., Trowe T., Brand M., Furutani-Seiki M., Haffter P., Hammerschmidt M., Heisenberg C. P., Jiang Y. J., Kane D. A., Kelsh R. N., Mullins M. C., Odenthal J., Nüsslein-Volhard C. (1996) Development 123, 399–413 [DOI] [PubMed] [Google Scholar]
- 22. Hirata H., Saint-Amant L., Waterbury J., Cui W., Zhou W., Li Q., Goldman D., Granato M., Kuwada J. Y. (2004) Development 131, 5457–5468 [DOI] [PubMed] [Google Scholar]
- 23. Hirata H., Watanabe T., Hatakeyama J., Sprague S. M., Saint-Amant L., Nagashima A., Cui W. W., Zhou W., Kuwada J. Y. (2007) Development 134, 2771–2781 [DOI] [PubMed] [Google Scholar]
- 24. Saint-Amant L., Sprague S. M., Hirata H., Li Q., Cui W. W., Zhou W., Poudou O., Hume R. I., Kuwada J. Y. (2008) Dev. Neurobiol. 68, 45–61 [DOI] [PubMed] [Google Scholar]
- 25. Zhou W., Saint-Amant L., Hirata H., Cui W. W., Sprague S. M., Kuwada J. Y. (2006) Cell Calcium 39, 227–236 [DOI] [PubMed] [Google Scholar]
- 26. Ono F., Higashijima S., Shcherbatko A., Fetcho J. R., Brehm P. (2001) J. Neurosci. 21, 5439–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guyon J. R., Steffen L. S., Howell M. H., Pusack T. J., Lawrence C., Kunkel L. M. (2007) Biochim. Biophys. Acta 1772, 205–215 [DOI] [PubMed] [Google Scholar]
- 28. Ingham P. W., Kim H. R. (2005) Exp. Cell Res. 306, 336–342 [DOI] [PubMed] [Google Scholar]
- 29. Ashworth R., Zimprich F., Bolsover S. R. (2001) Brain Res. Dev. Brain Res. 129, 169–179 [DOI] [PubMed] [Google Scholar]
- 30. Felsenfeld A. L., Walker C., Westerfield M., Kimmel C., Streisinger G. (1990) Development 108, 443–459 [DOI] [PubMed] [Google Scholar]
- 31. Pietri T., Manalo E., Ryan J., Saint-Amant L., Washbourne P. (2009) Dev. Neurobiol. 69, 780–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saint-Amant L., Drapeau P. (1998) J. Neurobiol. 37, 622–632 [DOI] [PubMed] [Google Scholar]
- 33. Downes G. B., Granato M. (2006) J. Neurobiol. 66, 437–451 [DOI] [PubMed] [Google Scholar]
- 34. Devoto S. H., Melançon E., Eisen J. S., Westerfield M. (1996) Development 122, 3371–3380 [DOI] [PubMed] [Google Scholar]
- 35. Ochi H., Westerfield M. (2007) Dev. Growth Differ. 49, 1–11 [DOI] [PubMed] [Google Scholar]
- 36. Moore C. A., Parkin C. A., Bidet Y., Ingham P. W. (2007) Development 134, 3145–3153 [DOI] [PubMed] [Google Scholar]
- 37. Naganawa Y., Hirata H. (2011) Dev. Biol. 355, 194–204 [DOI] [PubMed] [Google Scholar]
- 38. Fero K., Yokogawa T., Burgess H. A. (2011) in Zebrafish Models in Neurobehavioral Research (Kalueff A. V., Cachat J. M., eds.), pp. 249–291, Humana Press, New York [Google Scholar]
- 39. Müller U. K., van Leeuwen J. L. (2004) J. Exp. Biol. 207, 853–868 [DOI] [PubMed] [Google Scholar]
- 40. McLean D. L., Fan J., Higashijima S., Hale M. E., Fetcho J. R. (2007) Nature 446, 71–75 [DOI] [PubMed] [Google Scholar]
- 41. Bone Q. (1978) in Fish Physiology (Hoar W. S., Randall D. J., eds.), pp. 361–424, Academic Press, New York [Google Scholar]
- 42. Johnston I. A. (1983) in Fish Biomechanics (Webb P., Weihs D., eds.), pp. 36–67, Praeger Press, New York [Google Scholar]
- 43. Liu D. W., Westerfield M. (1988) J. Physiol. 403, 73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nüsslein-Volhard C., Dahm R. (2002) Zebrafish: A Practical Approach, Oxford University Press, Oxford [Google Scholar]
- 45. Westerfield M. (1993) The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio), M. Westerfield, Eugene, OR [Google Scholar]
- 46. Hirata H., Saint-Amant L., Downes G. B., Cui W. W., Zhou W., Granato M., Kuwada J. Y. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8345–8350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Budick S. A., O'Malley D. M. (2000) J. Exp. Biol. 203, 2565–2579 [DOI] [PubMed] [Google Scholar]
- 48. Bahary N., Davidson A., Ransom D., Shepard J., Stern H., Trede N., Zhou Y., Barut B., Zon L. I. (2004) Methods Cell Biol. 77, 305–329 [DOI] [PubMed] [Google Scholar]
- 49. Gates M. A., Kim L., Egan E. S., Cardozo T., Sirotkin H. I., Dougan S. T., Lashkari D., Abagyan R., Schier A. F., Talbot W. S. (1999) Genome Res. 9, 334–347 [PubMed] [Google Scholar]
- 50. Shimoda N., Knapik E. W., Ziniti J., Sim C., Yamada E., Kaplan S., Jackson D., de Sauvage F., Jacob H., Fishman M. C. (1999) Genomics 58, 219–232 [DOI] [PubMed] [Google Scholar]
- 51. Nasevicius A., Ekker S. C. (2000) Nat. Genet. 26, 216–220 [DOI] [PubMed] [Google Scholar]
- 52. Ogino K., Ramsden S. L., Keib N., Schwarz G., Harvey R. J., Hirata H. (2011) J. Biol. Chem. 286, 806–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakano Y., Fujita M., Ogino K., Saint-Amant L., Kinoshita T., Oda Y., Hirata H. (2010) Development 137, 1689–1698 [DOI] [PubMed] [Google Scholar]
- 54. Amores A., Force A., Yan Y. L., Joly L., Amemiya C., Fritz A., Ho R. K., Langeland J., Prince V., Wang Y. L., Westerfield M., Ekker M., Postlethwait J. H. (1998) Science 282, 1711–1714 [DOI] [PubMed] [Google Scholar]
- 55. Codina M., Li J., Gutiérrez J., Kao J. P., Du S. J. (2010) PLoS ONE 5, e8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Asakawa K., Suster M. L., Mizusawa K., Nagayoshi S., Kotani T., Urasaki A., Kishimoto Y., Hibi M., Kawakami K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Muto A., Ohkura M., Kotani T., Higashijima S. I., Nakai J., Kawakami K. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 5425–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Söhl G., Willecke K. (2004) Cardiovasc Res. 62, 228–232 [DOI] [PubMed] [Google Scholar]
- 59. Gong X., Li E., Klier G., Huang Q., Wu Y., Lei H., Kumar N. M., Horwitz J., Gilula N. B. (1997) Cell 91, 833–843 [DOI] [PubMed] [Google Scholar]
- 60. Paul D. L., Ebihara L., Takemoto L. J., Swenson K. I., Goodenough D. A. (1991) J. Cell Biol. 115, 1077–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cheng S., Christie T., Valdimarsson G. (2003) Dev. Dyn. 228, 709–715 [DOI] [PubMed] [Google Scholar]
- 62. Cheng S., Shakespeare T., Mui R., White T. W., Valdimarsson G. (2004) J. Biol. Chem. 279, 36993–37003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.