Background: Antibodies that recognize glycolipids presented by the antigen-presenting molecule CD1d are useful tools in studying natural killer T cell biology.
Results: The CD1d-α-galactosylceramide specific antibody L363 has TCR-like binding properties.
Conclusion: Glycolipid-reactive antibodies are more antigen-specific than the TCR of iNKT cells.
Significance: This is the first crystal structure of a glycolipid-reactive TCR-like antibody.
Keywords: Antibodies, Antigen Presentation, Glycolipids, Immunology, T Cell, CD1d, TCR-like Antibody
Abstract
Natural killer T (NKT) cells express a semi-invariant Vα14 T cell receptor (TCR) and recognize structurally diverse antigens presented by the antigen-presenting molecule CD1d that range from phosphoglycerolipids to α- and β-anomeric glycosphingolipids, as well as microbial α-glycosyl diacylglycerolipids. Recently developed antibodies that are specific for the complex of the prototypical invariant NKT (iNKT) cell antigen αGalCer (KRN7000) bound to mouse CD1d have become valuable tools in elucidating the mechanism of antigen loading and presentation. Here, we report the 3.1 Å resolution crystal structure of the Fab of one of these antibodies, L363, bound to mCD1d complexed with the αGalCer analog C20:2, revealing that L363 is an iNKT TCR-like antibody that binds CD1d-presented αGalCer in a manner similar to the TCR. The structure reveals that L363 depends on both the L and H chains for binding to the glycolipid-mCD1d complex, although only the L chain is involved in contacts with the glycolipid antigen. The H chain of L363 features residue Trp-104, which mimics the TCR CDR3α residue Leu-99, which is crucial for CD1d binding. We characterized the antigen-specificity of L363 toward several different glycolipids, demonstrating that whereas the TCR can induce structural changes in both antigen and CD1d to recognize disparate lipid antigens, the antibody L363 can only induce the F′ roof formation in CD1d but fails to reorient the glycolipid headgroup necessary for binding. In summary, L363 is a powerful tool to study mechanism of iNKT cell activation for structural analogs of KRN7000, and our study can aid in the design of antibodies with altered antigen specificity.
Introduction
Type I or semi-invariant natural killer T (iNKT)3 cells are a population of T lymphocytes that express both markers of natural killer cells and T cells. Because iNKT cells rapidly produce cytokines after antigen encounter (within 2–4 h), they have been termed innate-like immune cells, or cells that bridge the innate and adaptive immune system (1). iNKT cells express a semi-invariant αβ T cell receptor (TCR) that recognizes glycolipid antigens (Ag) presented by the nonclassical MHC class I-like molecule CD1d (2). NKT cells are characterized by expression of a conserved TCR α chain rearrangement (Vα14Jα18 in mouse and Vα24Jα18 in humans) that pairs predominantly with the TCR β chain Vβ8.2 (and to a lesser extend Vβ7 and Vb2), whereas in humans the most common combination is Vα24Vβ11 (1, 3). The prototypical antigen α-galactosylceramide (αGalCer, KRN7000) has been identified in a screen for anti-B16 melanoma compounds and is extensively studied as a potent iNKT Ag because of its robust and rapid activation of iNKT cells leading to the production of both pro (Th1) and anti-inflammatory (Th2) cytokines (4). Because of the opposing effects of Th1 and Th2 cytokines, synthetic KRN7000 analogs have been developed to skew the cytokine response to either a Th1- or Th2-biased phenotype, which in turn is more promising in controlling the disease progression in various animal models. For example, the glycolipids OCH, PBS-25, and C20:2 were found to induce Th2-biased cytokine production (5–7), which is beneficial for the control of Th1-driven autoimmune diseases, whereas α-C-GalCer and NU-αGalCer were found to stimulate Th1-biased cytokine production, leading to superior antitumor response compared with KRN7000, likely through increased transactivation of natural killer cells (8, 9). The mechanism leading to the induction of Th1- or Th2-biased responses by changing the glycolipid antigen structure is currently under intense investigation and takes advantage of a series of recently developed mCD1d-KRN7000 specific antibodies. The antibodies have been raised against mCD1d-KRN7000 complexes and do not bind to unloaded mCD1d or mCD1d that has been loaded with the self-antigen iGb3 (7). The antibodies also recognize many structural analogs of KRN7000, including OCH and α-C-GalCer, which makes them valuable tools to study differences in presentation of Th1 and Th2 biasing antigens. One particular antibody, L363, has since been successfully used by several labs to analyze CD1d-glycolipid stability (8, 10), the localization of CD1d-glycolipid complexes inside APCs by confocal microscopy (11), or the association of CD1d-bound Th1-biasing antigens in lipid rafts (12).
To account for its specificity for both the antigen-presenting molecule mCD1d and the glycolipid of interest, the complex-specific L363 antibody must bind to both mCD1d and the glycolipid, similar to the structurally related iNKT TCR, which it thus mimics. The crystal structures of CD1d with bound αGalCer, as well as in complex with the Vα14Vβ8.2 TCR and Vα14Vβ7 TCR of mouse iNKT cells, have been determined and illustrated the general binding of glycolipids to mCD1d, as well as a conserved docking mode of the TCR onto the mCD1d molecule (13, 14). αGalCer is bound to CD1d with its ceramide lipid backbone inserted in the deep and hydrophobic binding groove (composed of the two main pockets A′ and F′) of CD1d, with the phytosphingosine chain inserted in the F′ pocket and the fatty acid chain in the A′ pocket. The NKT TCR docks parallel to the CD1d-binding cleft with TCR α being the only chain interacting with the lipid Ag, whereas TCR α and β chains form conserved contacts with CD1 residues, mainly through CDR3α, CDR2β, and CDR3β (13, 15). Although the iNKT TCR can also bind to structurally distinct microbial Ags, such as α-glycuronosylceramides (GalA-GSL) from Sphingomonas (17), α-galactosyl-diacylglycerols (BbGL-2c) from Borrelia burgdorferi (18), and α-glucosyl-diacylglycerols (Glc-DAG-s2) from Streptococcus pneumoniae (19), the recognition of those glycolipids by the L363 antibody has not been addressed yet.
To elucidate the structural basis for the recognition of αGalCer by L363, as well as the antigen specificity of L363, we determined the binding kinetics of the Fab portion to different glycolipids, as well as determined the crystal structure of L363 Fab-mCD1d-C20:2-αGalCer complex to a resolution of 3.1Å.
EXPERIMENTAL PROCEDURES
Glycolipid Ags
Synthesis of NU-αGalCer, C20:2-αGalCer, GalA-GSL, BbGL-2c, and Glc-DAG-s2 has been reported previously (7, 19–22). KRN7000 was kindly provided by Kyowa Hakko Kirin (Japan). Bovine brain sulfatides were purchased from Avanti Polar Lipids Inc. OCH and α-C-GalCer were obtained from the National Institutes of Health tetramer core facility.
Cell Line and Cell Culture
The L363 expressing hybridoma cell line and iNKT hybridoma cell lines were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10 mm HEPES, pH 7.5, 1% l-glutamine, 1% nonessential amino acids, 1% sodium pyruvate, 55 μm 2-mercaptoethanol, 20 μg/ml gentamicin (Invitrogen), and 10% heat-inactivated FCS. The cell lines were maintained in an incubator with a humidified atmosphere containing 5% CO2 at 37 °C.
Antibody Production and Purification
For milligram scale mAb production, hybridomas were gradually adapted to culture in protein-free hybridoma medium (PFHM-II; Invitrogen), supplemented as indicated above. Cells from two T175 tissue culture flasks were transferred into one 2.8-liter roller bottle, filled up to 1.5 liters with supplemented PFHM-II. Roller bottles were equilibrated with CO2 by placing in the 37 °C + 5% CO2 incubator with the lid loosened for 0.5–1 h and then grown with closed lid in 37 °C room while rolling for ∼2 weeks or until the medium turned yellow. The cells were spun down (1000 × g for 6 min), and supernatant was filtered (0.22 μm) and concentrated to 300 ml using a tangential flow through filtration unit (Millipore; Pellicon 2) while exchanging buffer to PBS. IgG was collected from supernatant using affinity chromatography using a 5-ml HiTrap Protein G column (GE Healthcare). IgG was eluted from the column with 0.1 m glycine, pH 2.6, whereas 0.7-ml fractions were collected in 1.5-ml test tubes containing 0.3 ml of 1 m Tris, pH 8.5, for neutralization. IgG-containing fractions were pooled, and buffer was exchanged against PBS using centrifugal filtration devices (Amicon Ultra; Millipore). Final yield of purified IgG was 10 mg/liters of hybridoma culture.
Cloning and Sequencing of L363 VH and VL Genes
Total RNA was isolated from 5 × 106 hybridoma cells using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. First strand cDNA synthesis of 5′-rapid amplification of cDNA ends was done according to the protocol based on Clontech SMART-RACE using the Clontech cDNA amplification kit and the Invitrogen SuperScript II reverse transcriptase at 42 °C for 50 min in a 20-μl reaction volume containing: 500 ng of total RNA, 0.6 μm 5′-rapid amplification of cDNA ends CDS Primer A, 0.6 μm SMART II A oligonucleotide, 1× RT buffer (20 mm Tris-HCl, pH 8.4, 50 mm KCl), 5 mm MgCl2, 0.01 m DTT, and 4 units of RNase out recombinant RNase inhibitor. After the reverse transcriptase reaction was complete and cDNA was generated from the RNA, standard PCR (second strand reaction) was performed in a 50.5-μl final volume. For the PCR, 48 μl of PCR mix containing 1× PCR buffer, 1.5 mm MgCl2, 0.2 mm dNTP,1 μm Universal Primer A mix (sense primers, supplied in Clontech kit), 1 μm gene-specific (constant heavy and light chain genes) antisense primer, and 1.5 units of high fidelity Taq DNA polymerase were added to 2.5 μl of cDNA (from the first strand reaction). The VH gene was amplified with 5′-GGT CAA GGT CAC TGG CTC AGG GAA ATA AC-3′ antisense primer, whereas the light chain variable region (VL) gene was amplified with 5′-GTC GTT CAC TGC CAT CAA TCT TCC ACT TG-3′ antisense primer. The cycling profile used for the PCR was: denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s, and extension at 72 °C for 3 min. At the end of the 30 cycles, a 10-min incubation at 72 °C was done to complete elongation. The resulting PCR products were cloned into the pGEM-T easy vector (Promega) for sequencing. The V gene rearrangement was determined as IGKV13–84*01 (light chain) and IGHV9–4*02 (heavy chain) using the IMGT server (23). A schematic representation of the variable domains including sequence alignment of the CDR is illustrated in supplemental Fig. S1.
Fab Digestion and Purification
Purified L363 mAb (mouse IgG2a) at 1 mg/ml in 100 mm Tris, pH 7.0, was incubated with 1% (w/w) activated papain (P3125; Sigma) and for 3 h at 37 °C in digestion buffer. Papain was activated by incubating 20.80 μl of papain with 100 μl of 10× papain buffer (1 m NaOAc, pH 5.5, 12 mm EDTA) and 100 μl of cysteine (12.2 mg/ml) for 15 min at 37 °C. The papain digestion was stopped by adding 20 mm iodoacetamide. Digestion mix was concentrated and adjusted to 3 m NaCl, 1.5 m glycine, pH 8.9, for subsequent protein A purification to remove undigested IgG and Fc. The protein A flow-through containing Fab was dialyzed against 20 mm NaOAc, pH 5.5, overnight and purified to homogeneity by cation exchange chromatography using MonoS (GE Healthcare). Final yield of pure Fab was 0.27 mg/1 mg of IgG.
Mouse CD1d/β2m Expression and Purification
The expression and purification methods of fully glycosylated mouse CD1d/β2m heterodimer proteins using the baculovirus expression system were reported previously (8, 24, 25). Mouse CD1d mutants were generated using site-directed mutagenesis or kindly provided by Dr. Kronenberg (La Jolla Institute for Allergy & Immunology).
Glycolipid Loading and Ternary Complex Formation
Mouse CD1d was loaded overnight with 3–6 molar excess of C20:2-αGalCer (1 mg/ml in Me2SO). Purified mCD1d-C20:2-αGalCer was incubated with equimolar amount of L363 Fab for 1 h at room temperature. L363-C20:2-αGalCer-mCD1d ternary complex was isolated and separated from individual components by size exclusion chromatography using Superdex S200 HR16/60 (supplemental Fig. S2) and further concentrated to 6 mg/ml in 10 mm HEPES, pH 7.5, 30 mm NaCl for subsequent crystallization.
Crystallization and Structure Determination
Crystals of L363-C20:2-αGalCer-mCD1d complexes were grown at 4 °C by sitting drop vapor diffusion while mixing 0.5 μl of protein with 0.5 μl of precipitant (4% Tacsimate, pH 7.0, 12% PEG 3350). The crystals were flash-cooled at 100 K in mother liquor containing 20% glycerol. The diffraction data were collected at the Stanford Synchrotron Radiation Laboratory Beamline 7-1 and processed with the iMosflm software (26). The L363-C20:2-αGalCer-mCD1d crystal belongs to space group P63 with cell parameters a = 234.948 Å; b = 234.948 Å; c = 99.172 Å. The asymmetric unit contains two L363-C20:2-αGalCer-mCD1d molecule with an estimated solvent content of 68.87%. The crystal structure was determined by molecular replacement using MOLREP as part of the CCP4 suite (27, 28). Protein coordinates from the mCD1d-iGb3 structure (Protein Data Bank code 2Q7Y) (29) and a model of the L363 Fab obtained with the Swiss-Model server (30) (template obtained from Protein Data Bank structure 2BDN) (31) were used as the search models for mCD1d and the L363 Fab, respectively. After the MR solution was obtained containing both mCD1d and Fab, the model was rebuilt into σA-weighted 2Fo − Fc and Fo − Fc difference electron density maps using the program COOT (32). The final refinement steps were performed using the TLS procedure in REFMAC (33) with five anisotropic domains (α1-α2 domain of CD1d, including carbohydrates and lipid, α3-domain, β2m, variable and constant domains of Fab). The L363-C20:2-αGalCer-mCD1d structure was refined to 3.1 Å to Rcryst and Rfree values of 20.2% and 23.0%, respectively. The quality of the model was examined with the program Molprobity (34). The data collection and refinement statistics are presented in Table 1.
TABLE 1.
Comparison of Fab/TCR-mCD1d-glycolipid contacts
| Individual buried surface areas within the ternary complexes (buried surface area in Å2) | L363-C20:2-αGalCer-mCD1d | Vα14Vβ8.2TCR-C20:2-αGalCer-mCD1da | Vα14Vβ7TCR KRN7000-mCD1db |
|---|---|---|---|
| mCD1d | 710.3 | 676.3 | 716.1 |
| Glycolipid | 163.4 | 148.8 | 146.7 |
| mCD1d-glycolipid | 873.7 | 825.1 | 862.8 |
| Fab VL/TCR Vα | 406 | 659 | 526.7 |
| Fab VH/TCR Vβ | 467.7 | 166.1 | 336.1 |
| Total combined surfaces | 1612.8 | 1598.7 | 1754.3 |
| SC value (protein atoms only) | 0.764 | 0.707 | 0.614 |
Surface Plasmon Resonance Studies
The interaction between L363 mAb and the CD1d-glycolipid complexes was analyzed by surface plasmon resonance (SPR) with a Biacore 3000 (GE Healthcare) instrument according to the methods described previously (20). Briefly, recombinant mCD1d protein containing a birA tag (LHHILDAQKMVWNHR) was first expressed, biotinylated, and purified according to the methods published before (24, 25). Biotinylated mCD1d was loaded with glycolipids overnight, and ≈400 RU of mCD1d-glycolipid complex were immobilized on a streptavidin sensor chip (GE Healthcare). A series of increasing concentrations (0.004–32 μm) of the L363 in duplicate were passed over the mCD1d-glycolipid complex at 25 °C with a flow rate of 30 μl/min. To control for glycolipid-independent background binding, unloaded mCD1d molecules were immobilized on flow channel 1 of the sensor chip, and the binding response of L363 Fab to flow channel 1 was subtracted from the other flow channels. The experiments were performed at least twice, and kinetic parameters were calculated using a simple Langmuir 1:1 model in the BIA evaluation software version 4.1.
Fab Docking Models
The details for Fab docking models are available in the supplemental materials.
Coated Plate ELISA Assay
The interactions between L363 and KRN7000 complexes with wild type or mutant forms of mCD1d were measured by an antigen-presenting ELISA assay. Briefly, 1-μg quantities of soluble wild type or mutant mCD1d were coated in a 96-well flat-bottomed plate at 4 °C overnight. The plates were blocked by PBS and 10% FBS for 1 h and washed, and KRN7000 was added to each well to final concentrations of 0.5 and 2.0 μg/ml and incubated for 24 h at 37 °C. After washing, L363 mAb (4.8 mg/ml) at 1:1000 dilution was added to each well and incubated 1 h at room temperature. The plates were then washed and incubated with HRP-conjugated goat anti-mouse antibody secondary (Jackson ImmunoResearch Laboratories, Inc.) at 1:5000 dilution for an additional hour. Color was developed by 3,3′,5,5′-tetramethylbenzidine two-component microwell peroxidase substrate kit (KPL Inc.), and plates were read at 450-nm wavelength.
APC Free Antigen Presenting Assay
APC free antigen presenting assay for stimulation of mouse iNKT cell hybridoma by soluble mCD1d was carried out following published protocols (17, 18). IL-2 release in the APC free antigen-presenting assay was measured after 16 h of culture in a sandwich ELISA according to standard protocol.
Model Building
The models illustrating L363 Fab binding to GalA-GSL or its inability to bind BbGL-2c as well as Glc-DAG-s2 were prepared by superimposing the α1 and α2 helixes of mCD1d of the L363-C20:2-αGalCer-mCD1d complex structure with that of the mCD1d-GalA-GSL structure (Protein Data Bank code 2FIK) (35), the mCD1d-BbGL-2c structure (Protein Data Bank code 3ILQ) (20), and the mCD1d-GlcDAG-s2 structure (Protein Data Bank code 3T1F) (19), respectively.
RESULTS
L363 Antigen Specificity and Binding Kinetics
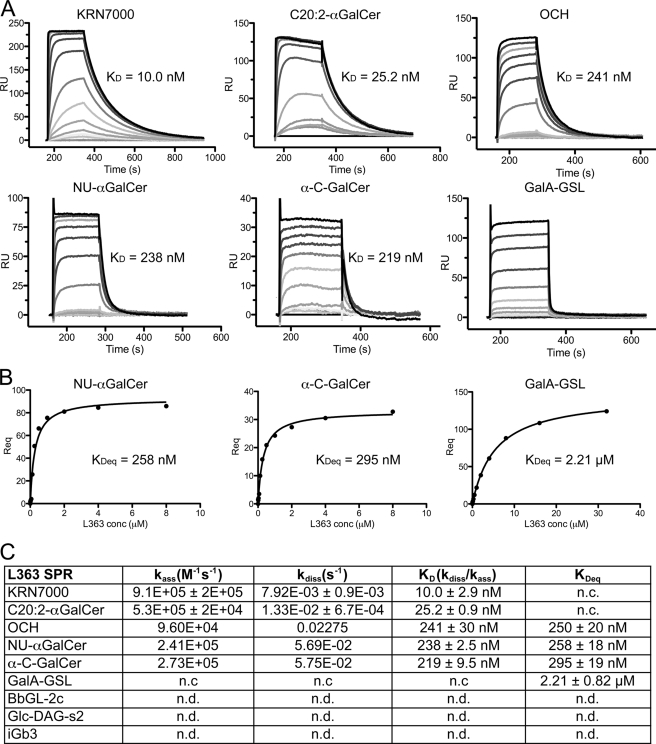
The antibody L363 was originally raised against the prototypical form of αGalCer (KRN7000, containing a C26:0 fatty acid) bound to mCD1d. Not surprisingly, the antibody also recognizes various structural analogs of KRN7000, including OCH and C20:2 (36), but the extent of chemical modifications tolerated by L363 has not been fully assessed. Using SPR, we show that L363 is able to bind to the glycolipids KRN7000, C20:2-αGalCer, NU-αGalCer, OCH, α-C-GalCer, as well as the bacterial antigen GalA-GSL from Sphingomonas spp., whereas in contrast to the TCR, other microbial glycolipids that are based on a diacylglycerol backbone rather than a ceramide backbone are not recognized (e.g. B. burgdorferi BbGL-2c and S. pneumoniae GlcDAG-s2) (Fig. 1). The affinity of the purified Fab of L363 has an almost identical equilibrium binding constant toward mCD1d-KRN7000 complexes, comparable with the Vα14Vβ8.2 TCR (CDR3β: CASGDEGYTQYF) of iNKT cells (KD = 10 nm versus 11 nm from Ref. 20), with both a 7-fold faster association rate (9.1 × 105 m−1 s−1 versus 1.3 × 105 m−1 s−1) and a 5.5-fold faster dissociation rate (7.9 × 10−3 s−1 versus 1.45 × 10−3 s−1 (Fig. 1). The binding kinetic to C20:2-αGalCer is very similar to that of KRN7000, indicating that the modification of the fatty acid moiety does not result in a different binding mode of the glycolipid to mCD1d. The KRN7000 analogs OCH, NU-αGalCer, and α-C-GalCer are bound with weaker affinities, similar to the binding kinetics by the Vα14Vβ8.2 TCR (8, 20, 37). However, L363 associated faster than both Vα14Vβ8.2 and Vα14Vβ7 TCR and dissociated faster than Vα14Vβ8.2 TCR but much slower than Vα14Vβ7 TCR, indicating that the Vα14Vβ8.2 TCR has an optimal binding surface for the mCD1d-KRN7000 complex and the highest stability (illustrated by the slowest dissociation rate). Interestingly, whereas the TCR is also able to bind various self and microbial antigens, such as iGb3, GalA-GSL, BbGl-2c, and GlcDAG-s2 (14, 19, 20, 38), L363 only binds to the sphingomonas antigen GalA-GSL with an affinity of KD = 2.21 ± 0.82 μm (Fig. 1). The restricted antigen selectivity observed for L363 raises the question as to what the structural basis for the binding specificity is compared with the more promiscuous TCR.
FIGURE 1.
Binding of L363 Fab to different glycolipids as assessed by SPR. A, sensorgrams of L363 Fab binding to mCD1d bound iNKT cell antigens. B, steady state analysis of Fab binding to Nu-αGalCer, α-C-GalCer, and GalA-GSL loaded mCD1d. The affinities derived by equilibrium analysis (KDeq) were equivalent to those derived by kinetic analysis. C, overview of kinetic data. L363 Fab binds to KRN7000 and its analogs (OCH, Nu-αGalCer, and α-C-GalCer) with affinities comparable with the Vα14Vβ8.2 TCR, whereas L363 binding to GalA-GSL is much weaker (KD = 2.21 μm). No L363 binding was detected for the microbial diacylglycerol antigens BbGL-2c and Glc-DAG-s2 or the self-antigen iGb3. n.c, not calculated; n.d., not detected.
L363 Fab-C20:2-αGalCer-mCD1d Crystal Structure
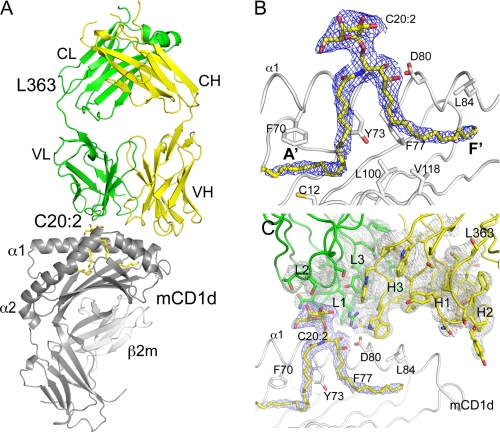
To address the question of why L363 only recognizes some of the iNKT cell antigens, we have determined the crystal structure of the L363-C20:2-αGalCer-mCD1d complex to 3.1 Å resolution (supplemental Table S1 and Fig. 2). Two highly similar Fab-C20:2-αGalCer-mCD1d complexes occupy the asymmetric unit, and unless specified, the analysis presented here focuses on one representative complex (chains A-B and H-L). Well ordered electron density was observed at the interface between Fab and mCD1d after molecular replacement, allowing for confident model building and accurate interpretation of the contacts at the binding interface, despite the relatively low resolution (Fig. 2). The C20:0-αGalCer glycolipid is bound with its lipid backbone in a deep and hydrophobic groove formed between the two anti-parallel α-helices that sit above a eight-stranded β-sheet platform. The two main pockets A′ and F′ each occupy one alkyl chain, with the sphingosine inserted in the F′ pocket and the C20:2 acyl chain accommodated in the A′ pocket. The α-galactosyl headgroup is presented at the opening of the binding groove for interaction with both polar CD1d residues, as well as the Fab. The Fab sits above the CD1d binding groove, contacting both the glycolipid as well as CD1d, thereby having specificity for both antigen and antigen-presenting molecule, similar to the TCR of iNKT cells. The Fab light chain is situated above the antigen, whereas the H chain sits above mCD1d, at the C-terminal end of the α1-helix.
FIGURE 2.
Structure of the L363-C20:2-αGalCer-mCD1d complex. A, overall structure of the ternary complex illustrates the TCR-like docking mode above the α1 and α2 helixes of mCD1d. L363 light chain (VL and CL) is shown in green, and heavy chain (VH and CH) is shown in yellow; C20:2-αGalCer is shown as yellow sticks; mCD1d heavy chain and β2m are in gray. B, final 2Fo − Fc electron density map for the glycolipid C20:2-αGalCer in a side (left panel) and top view (right panel). C, stereo view of electron density around CDR loops (L1–3 and H1–3). The 2Fo − Fc map is contoured at 1 σ and shown as blue mesh for C20:2-αGalCer and gray mesh for L363. Several mCD1d residues interacting with the glycolipid are depicted.
TCR-like Footprint of L363
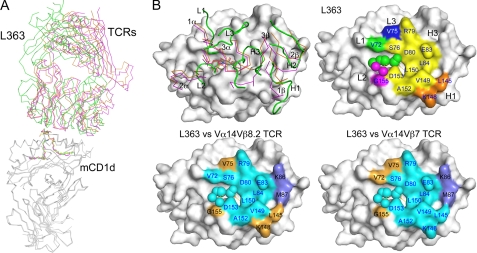
Compared with the mCD1d-KRN700-TCR structure, the L363 Fab binds overall highly similar to the iNKT TCR in the mCD1d-KRN7000-TCR complex. The Fab binds in a parallel docking orientation and offset to the C-terminal half of the α1-helix; however, L363 Fab is not tilted as much compared with both Vα14Vβ8.2 and Vα14Vβ7 TCR (Fig. 3A). The general L363 footprint on mCD1d is also relatively conserved with the TCRs, despite the fact that the individual CDR loops of L363 (L1–3 and H1–3) do not superimpose well with that of the TCR (1–3α and 1–3β). However, the general binding mode of the L363 Fab and iNKT cell TCR is similar, with the L363 L chain following the TCR α chain and the L363 H chain matching the TCRβ chain. Both L3 and H3 loops of L363 diverge the most from the TCR CDR3α and 3β (Fig. 3B). Most of the mCD1d contact residues are shared by L363 Fab and mTCRs (Val-72, Ser-76, Arg-79, Asp-80, Glu-83, Leu-84, Leu-150, Val-149, Ala-152, Leu-145, and Lys-148), whereas a few mCD1d residues differ between L363 (Val-75 and Gly-155), Vα14Vβ8.2 TCR (Lys-86, Met-87, and Asp-153), as well as the Vα14Vβ7 TCR (Arg-21, Lys-86, Met-87, and Asp-153) (supplemental Table S2). The similar footprint also correlates well with the calculated buried surface areas of the Fab/TCR-glycolipid-mCD1d complexes (Table 1). All of the three complexes have similar total buried surface areas, with similar contributions from both Fab or TCR, as well as mCD1d. However, although the TCR footprint on CD1d-KRN7000 is dominated by the invariant Vα14 chain (61–80% of total buried surface area), the L363 footprint is equally contributed by both the H and L chains (54 and 46%, respectively; Table 1). As expected, the most surface area is buried between Fab or TCR and mCD1d (676–716 Å2), whereas the surface buried between Fab or TCR and the glycolipid antigen is considerably lower (146–163 Å2) yet determines specificity (see below).
FIGURE 3.
Comparison of the antibody and TCR footprints. A, superposition of the ternary αGalCer-mCD1d complexes with bound L363 Fab (green), Vα14Vβ8.2 (Protein Data Bank ID 3ARF, orange), and Vα14Vβ7 (Protein Data Bank ID 3HE7, magenta) illustrates a binding orientation between L363 and TCRs. B, the CDR loops of L363 (L1–3 and H1–3 in green) do not overlay well with those of the two TCRs (left panel, CDR1–3α and 1–3β, colored as in A) but result in a similar overall footprint. L363 footprint is colored by individual CDR loops (top right panel). Green, L1; magenta, L2; blue, L3; orange, H1; yellow, H3. The lower panels depict shared footprint with the TCR (cyan), L363-only contacts (orange), and TCR-only contacts (light blue).
Antigen Recognition and Specificity
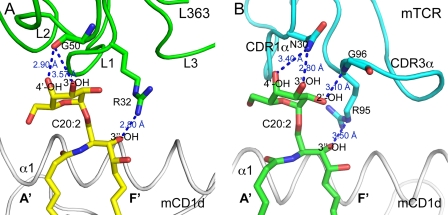
Similar to the TCR, all the contacts between L363 Fab and the glycolipid antigen are formed by one chain, VL, which mimics the TCR Vα domain. The L2 residue Gly-50 forms two potential H-bonds with the 3″-OH and 4″-OH of the galactose, similar to TCR CDR1α residue Asn-30, whereas L1 residue Arg-32 contacts the 3-OH of the ceramide backbone (Fig. 4). Because the backbone oxygen of Gly-50 is slightly shifted toward the A′ pocket of CD1d, in contrast to the Asn-30 side chain of the TCR, binding of L363 to C20:2 results in a slight tilt of the galactose toward the CD1d-binding groove (Fig. 4). A major difference between L363 and TCR is that the TCR CDR3α contacts both galactose and lipid backbone, whereas for L363 Fab, L1 exclusively contacts the lipid, whereas L2 only contacts the galactose. Additionally, the Fab and mTCRs not only share many contact residues with mCD1d (Val-72, Ser-76, Arg-79, Asp-80, Glu-83, Leu-84, Leu-150, Val-149, Ala-152, Leu-145, and Lys-148 highlighted in green in Fig. 3B) but also form similar numbers of H-bonds including H-bonds to the same residues (Ser-76, Glu-83, and Asp-80 in Fig. 5A). Three of the four contact points on the glycolipid are conserved between the TCR and L363 and form the basis for antigen selectivity; however, L363 recognition of αGalCer appears less stringent because it lacks the contact with the 2″-OH of galactose that is recognized by the TCR (CDR3α, Gly-96). Thus, the conserved contacts of L363 with the antigen determine the specificity of Fab in recognizing αGalCer and many of its structural analogs. The similar binding footprint also enables the recognition of 6″-galactose-modified KRN7000 analogs because those modifications are generally not contacted by the Fab, similar to the TCR and are facing toward the A′ pocket, whereas L363 is centered above the F′ pocket of mCD1d (Fig. 3).
FIGURE 4.
L363 contacts the ligand with different recognition logic. Similar to the TCRs, L363 interacts with C20:2-αGalCer solely through one chain, VL (mimicking TCR Vα chain). Three conserved H-bonds are formed by L363 Fab and the TCR with C20:2-αGalCer. A, L363 binding of C20:2-αGalCer. L363 VL chain in green, C20:2-αGalCer in yellow. B, Vα14Vβ8.2 TCR binding of C20:2-αGalCer (Protein Data Bank ID 3ARF), TCR Vα chain in cyan, and C20:2-αGalCer in green.
FIGURE 5.
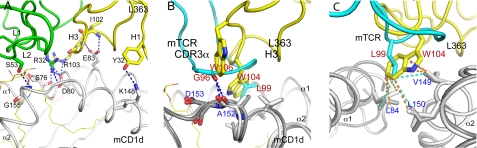
L363 binding to mCD1d. A, L363 forms similar numbers of H-bonds including H-bonds to the same residues (Ser-76, Glu-83, and Asp-80) compared with mTCR. B, superimposing the structures of the L363-C20:2-αGalCer-mCD1d and Vα14Vβ8.2 TCR-C20:2-αGalCer-mCD1d complexes indicate that Trp-106 (W106) and Trp-104 (W104) of L363 H3 mimic residues Gly-96 (G96) and Leu-99 (L99) of TCR CDR3α loop, because they form similar contacts with mCD1d residues above the F′ pocket. Cyan, TCR Vα chain; yellow, L363 Fab VH chain; gray, mCD1d. C, Fab CDR3 VH residue Trp-104 and TCR CDR3α residue Leu-99 interact closely with the hydrophobic residues Leu-84, Val-149, and Leu-150, which form the F′ roof.
L363 also binds to the Sphingomonas glycolipid GalA-GSL but not to the borrelial glycolipid BbGl-2c or the streptococcal antigen GlcDAG-s2. Our modeling studies indicate that L363 can simply dock onto the mCD1d-GalA-GSL complex, because GalA-GSL and αGalCer are presented very similarly by CD1d (supplemental Fig. S3A). However, BbGl-2c and GlcDAG-s2 are both tilted and would clash with L363 residue Arg-32 of L1. Because the TCR can reorient both BbGl-2c and GlcDAG-s2 to allow for the conserved TCR footprint (25, 39), failure of L363 binding to either BbGl-2c or GlcDAG-s2 indicates that the Fab does not have the ability to induce such structural changes in the microbial antigens, which is a prerequisite for binding to CD1d. Thus, although L363 and TCR share a similar footprint above CD1d and the glycolipid, the recognition logic differs slightly, with L2 mimicking CDR1α and L1 partially mimicking CDR3α.
Importance of the H Chain in CD1d Binding
The TCR residue Leu-99 of CDR3α is critical in binding to the F′ roof of mCD1d (25, 40), because mutation to alanine results in loss of NKT cell activation (41). In the L363 Fab structure, this residue is replaced by Trp-104 of H3, because it forms similar contacts with the F′ roof forming CD1d residues Leu-84, Val-149, and Leu-150 (Fig. 5, B and C). Interestingly, in L363, Trp-104 is not located on the light chain that recognizes the ligand but on the heavy chain, reflecting the increased contribution of the H chain in the overall Fab footprint (Table 1). This is in contrast to the TCR, where Leu-99 is on the invariant TCR α chain that also recognizes the antigen. Therefore, for L363 both the L and H chains are selected for efficiently binding to the CD1d-glycolipid complex, whereas for iNKT cells the TCR β chain is more interchangeable and mostly important for modulating the overall binding affinity (25, 40, 42).
In particular, TCR CDR3α residue Leu-99 and Fab VH CDR3 residue Trp-104 interacts closely with the hydrophobic residues Leu-84, Val-149, and Leu-150, and this hydrophobic interaction stabilizes the F′ roof above the F′ pocket on mCD1d, as well as the overall binding of TCR/L363 Fab to the mCD1d-C20:2-αGalCer complex.
In addition, although Gly-96 of mTCR forms one H-bond with Ala-152, L363 binds to the hydrophobic part of Ala-152 through van der Waals interaction using Trp-106 of H3. As a result, the Fab uses two aromatic residues on the H chain (Trp-104 and Trp-106) to mimic important contact of the TCRα chain (Gly-96α and Leu-99α).
TCR-like Binding Properties of L363 Fab
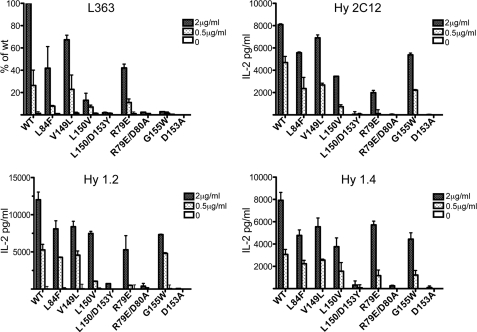
Next we compared the binding of L363 to CD1d-αGalCer with functional responses of three different iNKT cell hybridomas, expressing either Vβ8.2 (1.2 and 2C12) or Vβ10 (1.4) (Fig. 6). Wild type and several different mutant forms of mCD1d were used in this analysis. Mutations were introduced around the opening to the CD1d-binding groove, because both TCR and antibody bind directly above the groove, and we expected similar binding of the L363 antibody based on the crystal structure. Although the pattern of responses by iNKT cells (reflecting TCR avidity) and the binding of TCR-like antibody L363 was mostly similar, antibody binding appeared slightly more susceptible to mutations involved in the F′ roof formation of mCD1d (L84F, V149L, and L150V). The L84F mutation reflects the human residue at that position, and binding was reduced by 60% for the antibody, whereas responses of the iNKT hybridomas were reduced by only ∼25–30%. In addition, the G155W mutation, which is also a mutation to the equivalent human amino acid, abrogated antibody binding, as expected by the failure of the antibody to bind to human CD1d-KRN7000 complexes (36), while having little effect on TCR-mediated responses, which was consistent with the previously documented cross-species reactivity of iNKT cells (43). As such, the antibody is specific for the mouse CD1d protein-glycolipid complex against which it was raised, whereas the TCR is more promiscuous in that it can also bind to human CD1d molecules presenting iNKT cell antigens.
FIGURE 6.
L363 Fab binds similarly to mCD1d and CD1d mutants loaded with KRN7000 compared with NKT hybridoma cells. A coated plate ELISA assay was used to measure L363 binding, and an APC-free NKT cell stimulation assay was used to assess TCR recognition based on IL-2 secretion. Similar patterns of recognition were observed of various CD1d mutants by both L363 Fab and iNKT cells hybridomas expressing TCRs containing Vβ8.2 (2C12 and 1.2) or Vβ10 (1.4). With the exception of the G155W mutation of mCD1d that blocks the binding of L363 Fab, the observed binding pattern correlates with the shared footprint illustrated in Fig. 3. The data represent the means ± range of two independent experiments.
DISCUSSION
Antibodies with TCR-like binding properties exist for all MHC molecules that present antigens to T cells, including MHC class I (44–47), MHC class II (48), as well as CD1d (36, 49), and are widely used to study or modulate T cell function by specifically blocking antigen-mediated TCR activation. High affinity antibodies that exhibit superior binding affinity are especially useful to block T cell activation and as such have therapeutic potential in various T cell-mediated diseases (46).
In this study, we have determined the crystal structure of the Fab L363 bound to the complex of mouse CD1d presenting the KRN7000 glycolipid analog C20:2. The L363 Fab binds to mCD1d in a parallel orientation and centered above the F′ pocket, similar to the TCR of iNKT cells (8, 13, 25, 37). The TCR-like binding properties are also reflected in a similar recognition of C20:2-αGalCer, where most of the specific polar interactions with the presented galactose epitope and the lipid backbone are conserved. The conserved antigen recognition explains the exquisite specificity of L363 for KRN7000 and its structural analogs, whereas microbial iNKT cell antigens that are presented differently than KRN7000 by mCD1d are not recognized. It thus appears that L363, in contrast to the TCR, does not have ability to reorient the glycolipid headgroup of iNKT cell antigen to allow for CD1d binding if they are not presented nearly identical to KRN7000. The antibody therefore binds with a lock and key mechanism, which is also reflected in the higher protein shape complementarity (shape complementarity = 0.76, whereas 1 is a perfect fit of two protein surfaces and 0 is no fit), as well as in the higher Fab association rate, compared with the TCR, which still associates very rapidly, yet 7-fold slower. Because the equilibrium binding affinity of L363 and TCR toward CD1d-αGalCer are nearly identical (KD = ∼10 nm), the antibody also dissociates ∼7-fold faster from CD1d compared with the TCR (20). This is in agreement with the formation of only three of four polar contacts with C20:2-αGalCer, and possibly less stable interaction with CD1d, to which the Fab binds overall with a similar yet not identical footprint onto CD1d. However, the antibody likely retained the ability to induce the F′ roof formation in CD1d, indicated by its ability to bind to the ligand GalA-GSL, which does not have a preformed F′ roof when bound to CD1d (25, 35). In comparison, the F′ roof of CD1d is always formed upon TCR binding and believed to be a key component in determining the stability of the ternary complexes (39). In analogy, we postulate that the F′ roof is also formed upon binding of the TCR like antibody L363. The most notable difference between L363 and the TCR lies in the recognition logic of the CD1d-KRN7000 or CD1d-C20:2-αGalCer complex. Although the invariant TCR α-chain both makes crucial contacts with KRN7000 and the F′ roof of CD1d, the equivalent L chain of the antibody predominantly determines specificity for the antigen, whereas the H chain interacts with the F′ roof of CD1d, especially with the residue Trp-104, which mimics the TCR CDR3α residue Leu-99. In conclusion, the structural, biochemical, and biophysical data presented here reveal the TCR-like properties and specificity of L363 in recognition of mCD1d bound KRN7000, as well as closely related Ags, and reveal the structural basis as well as the limits of L363 to be used as a tool to study glycolipid antigen presentation and iNKT cell function.
Supplementary Material
Acknowledgments
We thank the Stanford Synchrotron Radiation Lightsource, especially Beamline 7-1, for remote data collection. The borellial glycolipid BbGl-2c was kindly provided by Dr. Gavin Painter (Industrial Research Limited), and the streptococcal glycolipid Glc-DAG-s2 was a gift from Dr. Petr Illarionov (University of Birmingham).
This work was supported, in whole or in part, by National Institutes of Health Grants AI074952 (to D. M. Z.) and AI45889 (to S. A. P.). This work was also supported by an investigator award from the Cancer Research Institute (to D. M. Z.).

This article contains supplemental Table S1 and Figs. S1–S3.
The atomic coordinates and structure factors (code 3UBX) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- iNKT
- invariant natural killer T
- TCR
- T cell receptor
- Ag
- antigen
- APC
- antigen-presenting cell.
REFERENCES
- 1. Bendelac A., Savage P. B., Teyton L. (2007) The biology of NKT cells. Annu. Rev. Immunol. 25, 297–336 [DOI] [PubMed] [Google Scholar]
- 2. Godfrey D. I., MacDonald H. R., Kronenberg M., Smyth M. J., Van Kaer L. (2004) NKT cells: what's in a name? Nat. Rev. Immunol. 4, 231–237 [DOI] [PubMed] [Google Scholar]
- 3. Godfrey D. I., Rossjohn J., McCluskey J. (2008) The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity 28, 304–314 [DOI] [PubMed] [Google Scholar]
- 4. Kawano T., Cui J., Koezuka Y., Toura I., Kaneko Y., Motoki K., Ueno H., Nakagawa R., Sato H., Kondo E., Koseki H., Taniguchi M. (1997) CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science 278, 1626–1629 [DOI] [PubMed] [Google Scholar]
- 5. Goff R. D., Gao Y., Mattner J., Zhou D., Yin N., Cantu C., 3rd, Teyton L., Bendelac A., Savage P. B. (2004) Effects of lipid chain lengths in alpha-galactosylceramides on cytokine release by natural killer T cells. J. Am. Chem. Soc. 126, 13602–13603 [DOI] [PubMed] [Google Scholar]
- 6. Miyamoto K., Miyake S., Yamamura T. (2001) A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature 413, 531–534 [DOI] [PubMed] [Google Scholar]
- 7. Yu K. O., Im J. S., Molano A., Dutronc Y., Illarionov P. A., Forestier C., Fujiwara N., Arias I., Miyake S., Yamamura T., Chang Y. T., Besra G. S., Porcelli S. A. (2005) Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of alpha-galactosylceramides. Proc. Natl. Acad. Sci. U.S.A. 102, 3383–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aspeslagh S., Li Y., Yu E. D., Pauwels N., Trappeniers M., Girardi E., Decruy T., Van Beneden K., Venken K., Drennan M., Leybaert L., Wang J., Franck R. W., Van Calenbergh S., Zajonc D. M., Elewaut D. (2011) Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis. EMBO J. 30, 2294–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujii S., Shimizu K., Hemmi H., Fukui M., Bonito A. J., Chen G., Franck R. W., Tsuji M., Steinman R. M. (2006) Glycolipid alpha-C-galactosylceramide is a distinct inducer of dendritic cell function during innate and adaptive immune responses of mice. Proc. Natl. Acad. Sci. U.S.A. 103, 11252–11257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sullivan B. A., Nagarajan N. A., Wingender G., Wang J., Scott I., Tsuji M., Franck R. W., Porcelli S. A., Zajonc D. M., Kronenberg M. (2010) Mechanisms for glycolipid antigen-driven cytokine polarization by Valpha14i NKT cells. J. Immunol. 184, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bai L., Sagiv Y., Liu Y., Freigang S., Yu K. O., Teyton L., Porcelli S. A., Savage P. B., Bendelac A. (2009) Lysosomal recycling terminates CD1d-mediated presentation of short and polyunsaturated variants of the NKT cell lipid antigen alphaGalCer. Proc. Natl. Acad. Sci. U.S.A. 106, 10254–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Im J. S., Arora P., Bricard G., Molano A., Venkataswamy M. M., Baine I., Jerud E. S., Goldberg M. F., Baena A., Yu K. O., Ndonye R. M., Howell A. R., Yuan W., Cresswell P., Chang Y. T., Illarionov P. A., Besra G. S., Porcelli S. A. (2009) Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity 30, 888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pellicci D. G., Patel O., Kjer-Nielsen L., Pang S. S., Sullivan L. C., Kyparissoudis K., Brooks A. G., Reid H. H., Gras S., Lucet I. S., Koh R., Smyth M. J., Mallevaey T., Matsuda J. L., Gapin L., McCluskey J., Godfrey D. I., Rossjohn J. (2009) Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity 31, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zajonc D. M., Cantu C., Mattner J., Zhou D., Savage P. B., Bendelac A., Wilson I. A., Teyton L. (2005) Nat. Immunol. 6, 810–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Borg N. A., Wun K. S., Kjer-Nielsen L., Wilce M. C., Pellicci D. G., Koh R., Besra G. S., Bharadwaj M., Godfrey D. I., McCluskey J., Rossjohn J. (2007) CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448, 44–49 [DOI] [PubMed] [Google Scholar]
- 16. Lawrence M. C., Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 17. Kinjo Y., Wu D., Kim G., Xing G. W., Poles M. A., Ho D. D., Tsuji M., Kawahara K., Wong C. H., Kronenberg M. (2005) Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434, 520–525 [DOI] [PubMed] [Google Scholar]
- 18. Kinjo Y., Tupin E., Wu D., Fujio M., Garcia-Navarro R., Benhnia M. R., Zajonc D. M., Ben-Menachem G., Ainge G. D., Painter G. F., Khurana A., Hoebe K., Behar S. M., Beutler B., Wilson I. A., Tsuji M., Sellati T. J., Wong C. H., Kronenberg M. (2006) Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat. Immunol. 7, 978–986 [DOI] [PubMed] [Google Scholar]
- 19. Kinjo Y., Illarionov P., Vela J. L., Pei B., Girardi E., Li X., Li Y., Imamura M., Kaneko Y., Okawara A., Miyazaki Y., Gómez-Velasco A., Rogers P., Dahesh S., Uchiyama S., Khurana A., Kawahara K., Yesilkaya H., Andrew P. W., Wong C. H., Kawakami K., Nizet V., Besra G. S., Tsuji M., Zajonc D. M., Kronenberg M. (2011) Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat. Immunol. 12, 966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J., Li Y., Kinjo Y., Mac T. T., Gibson D., Painter G. F., Kronenberg M., Zajonc D. M. (2010) Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proc. Natl. Acad. Sci. U.S.A. 107, 1535–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Trappeniers M., Van Beneden K., Decruy T., Hillaert U., Linclau B., Elewaut D., Van Calenbergh S. (2008) 6'-derivatised alpha-GalCer analogues capable of inducing strong CD1d-mediated Th1-biased NKT cell responses in mice. J. Am. Chem. Soc. 130, 16468–16469 [DOI] [PubMed] [Google Scholar]
- 22. Wu D., Xing G. W., Poles M. A., Horowitz A., Kinjo Y., Sullivan B., Bodmer-Narkevitch V., Plettenburg O., Kronenberg M., Tsuji M., Ho D. D., Wong C. H. (2005) Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proc. Natl. Acad. Sci. U.S.A. 102, 1351–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lefranc M. P., Giudicelli V., Ginestoux C., Bodmer J., Müller W., Bontrop R., Lemaitre M., Malik A., Barbié V., Chaume D. (1999) IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 27, 209–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zajonc D. M., Maricic I., Wu D., Halder R., Roy K., Wong C. H., Kumar V., Wilson I. A. (2005) Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J. Exp. Med. 202, 1517–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y., Girardi E., Wang J., Yu E. D., Painter G. F., Kronenberg M., Zajonc D. M. (2010) The Vα14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. J. Exp. Med. 207, 2383–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 27. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 28. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 29. Zajonc D. M., Savage P. B., Bendelac A., Wilson I. A., Teyton L. (2008) Crystal structures of mouse CD1d-iGb3 complex and its cognate Valpha14 T cell receptor suggest a model for dual recognition of foreign and self glycolipids. J. Mol. Biol. 377, 1104–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 31. Reid C., Rushe M., Jarpe M., van Vlijmen H., Dolinski B., Qian F., Cachero T. G., Cuervo H., Yanachkova M., Nwankwo C., Wang X., Etienne N., Garber E., Bailly V., de Fougerolles A., Boriack-Sjodin P. A. (2006) Structure activity relationships of monocyte chemoattractant proteins in complex with a blocking antibody. Protein Eng. Des. Sel. 19, 317–324 [DOI] [PubMed] [Google Scholar]
- 32. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 33. Winn M. D., Isupov M. N., Murshudov G. N. (2001) Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 57, 122–133 [DOI] [PubMed] [Google Scholar]
- 34. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 35. Wu D., Zajonc D. M., Fujio M., Sullivan B. A., Kinjo Y., Kronenberg M., Wilson I. A., Wong C. H. (2006) Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proc. Natl. Acad. Sci. U.S.A. 103, 3972–3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu K. O., Im J. S., Illarionov P. A., Ndonye R. M., Howell A. R., Besra G. S., Porcelli S. A. (2007) Production and characterization of monoclonal antibodies against complexes of the NKT cell ligand alpha-galactosylceramide bound to mouse CD1d. J. Immunol. Methods 323, 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wun K. S., Cameron G., Patel O., Pang S. S., Pellicci D. G., Sullivan L. C., Keshipeddy S., Young M. H., Uldrich A. P., Thakur M. S., Richardson S. K., Howell A. R., Illarionov P. A., Brooks A. G., Besra G. S., McCluskey J., Gapin L., Porcelli S. A., Godfrey D. I., Rossjohn J. (2011) A molecular basis for the exquisite CD1d-restricted antigen specificity and functional responses of natural killer T cells. Immunity 34, 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu E. D., Girardi E., Wang J., Zajonc D. M. (2011) Cutting Edge: Structural basis for the recognition of β-linked glycolipid antigens by invariant NKT cells. J. Immunol. 187, 2079–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Girardi E., Yu E. D., Li Y., Tarumoto N., Pei B., Wang J., Illarionov P., Kinjo Y., Kronenberg M., Zajonc D. M. (2011) Unique Interplay between Sugar and Lipid in Determining the Antigenic Potency of Bacterial Antigens for NKT Cells. PLoS Biol. 9, e1001189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wun K. S., Borg N. A., Kjer-Nielsen L., Beddoe T., Koh R., Richardson S. K., Thakur M., Howell A. R., Scott-Browne J. P., Gapin L., Godfrey D. I., McCluskey J., Rossjohn J. (2008) A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J. Exp. Med. 205, 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Scott-Browne J. P., Matsuda J. L., Mallevaey T., White J., Borg N. A., McCluskey J., Rossjohn J., Kappler J., Marrack P., Gapin L. (2007) Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat. Immunol. 8, 1105–1113 [DOI] [PubMed] [Google Scholar]
- 42. Matulis G., Sanderson J. P., Lissin N. M., Asparuhova M. B., Bommineni G. R., Schümperli D., Schmidt R. R., Villiger P. M., Jakobsen B. K., Gadola S. D. (2010) Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 8, e1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brossay L., Chioda M., Burdin N., Koezuka Y., Casorati G., Dellabona P., Kronenberg M. (1998) CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J. Exp. Med. 188, 1521–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Denkberg G., Reiter Y. (2006) Recombinant antibodies with T-cell receptor-like specificity: novel tools to study MHC class I presentation. Autoimmun. Rev. 5, 252–257 [DOI] [PubMed] [Google Scholar]
- 45. Cohen C. J., Denkberg G., Lev A., Epel M., Reiter Y. (2003) Recombinant antibodies with MHC-restricted, peptide-specific, T-cell receptor-like specificity: new tools to study antigen presentation and TCR-peptide-MHC interactions. J. Mol. Recognit. 16, 324–332 [DOI] [PubMed] [Google Scholar]
- 46. Stewart-Jones G., Wadle A., Hombach A., Shenderov E., Held G., Fischer E., Kleber S., Nuber N., Stenner-Liewen F., Bauer S., McMichael A., Knuth A., Abken H., Hombach A. A., Cerundolo V., Jones E. Y., Renner C. (2009) Rational development of high-affinity T-cell receptor-like antibodies. Proc. Natl. Acad. Sci. U.S.A. 106, 5784–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mareeva T., Martinez-Hackert E., Sykulev Y. (2008) How a T cell receptor-like antibody recognizes major histocompatibility complex-bound peptide. J. Biol. Chem. 283, 29053–29059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dahan R., Tabul M., Chou Y. K., Meza-Romero R., Andrew S., Ferro A. J., Burrows G. G., Offner H., Vandenbark A. A., Reiter Y. (2011) TCR-like antibodies distinguish conformational and functional differences in two- versus four-domain auto reactive MHC class II-peptide complexes. Eur. J. Immunol. 41, 1465–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Denkberg G., Stronge V. S., Zahavi E., Pittoni P., Oren R., Shepherd D., Salio M., McCarthy C., Illarionov P. A., van der Merwe A., Besra G. S., Dellabona P., Casorati G., Cerundolo V., Reiter Y. (2008) Phage display-derived recombinant antibodies with TCR-like specificity against alpha-galactosylceramide and its analogues in complex with human CD1d molecules. Eur. J. Immunol. 38, 829–840 [DOI] [PubMed] [Google Scholar]
- 50. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.