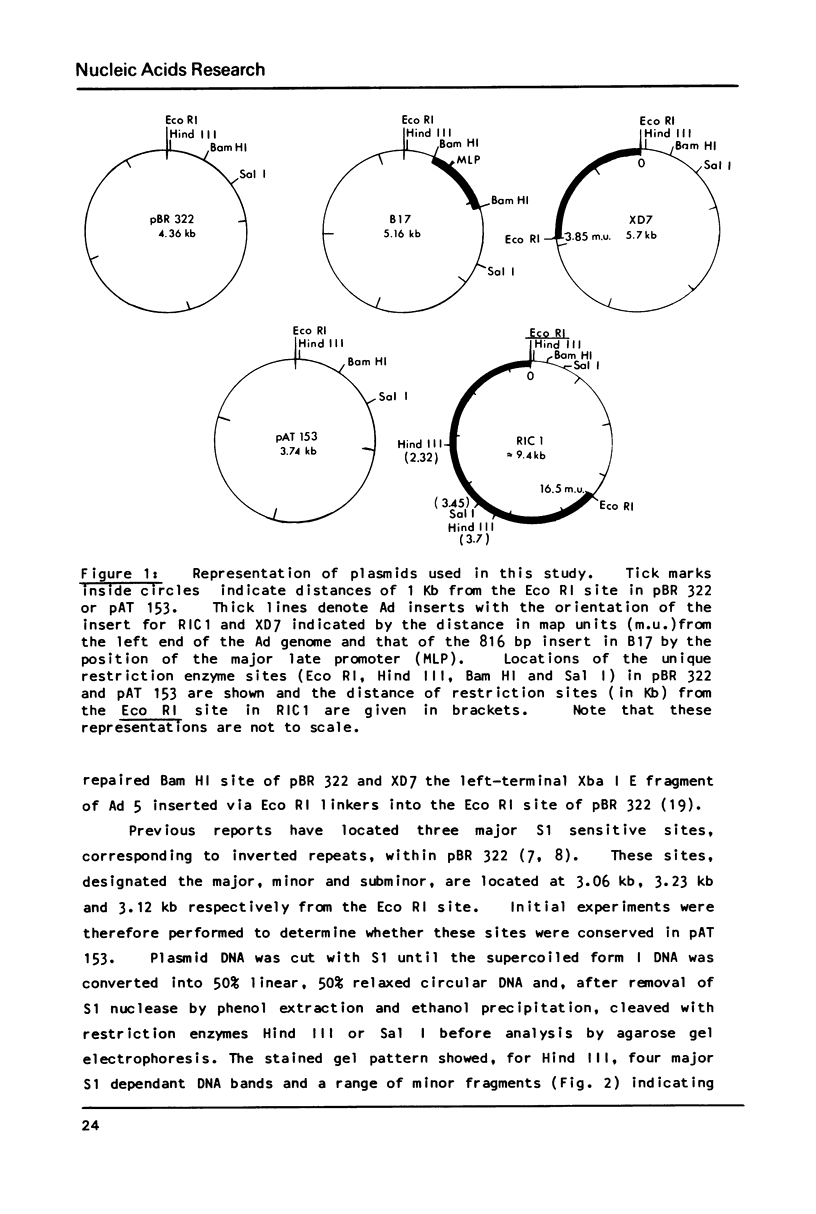
Abstract
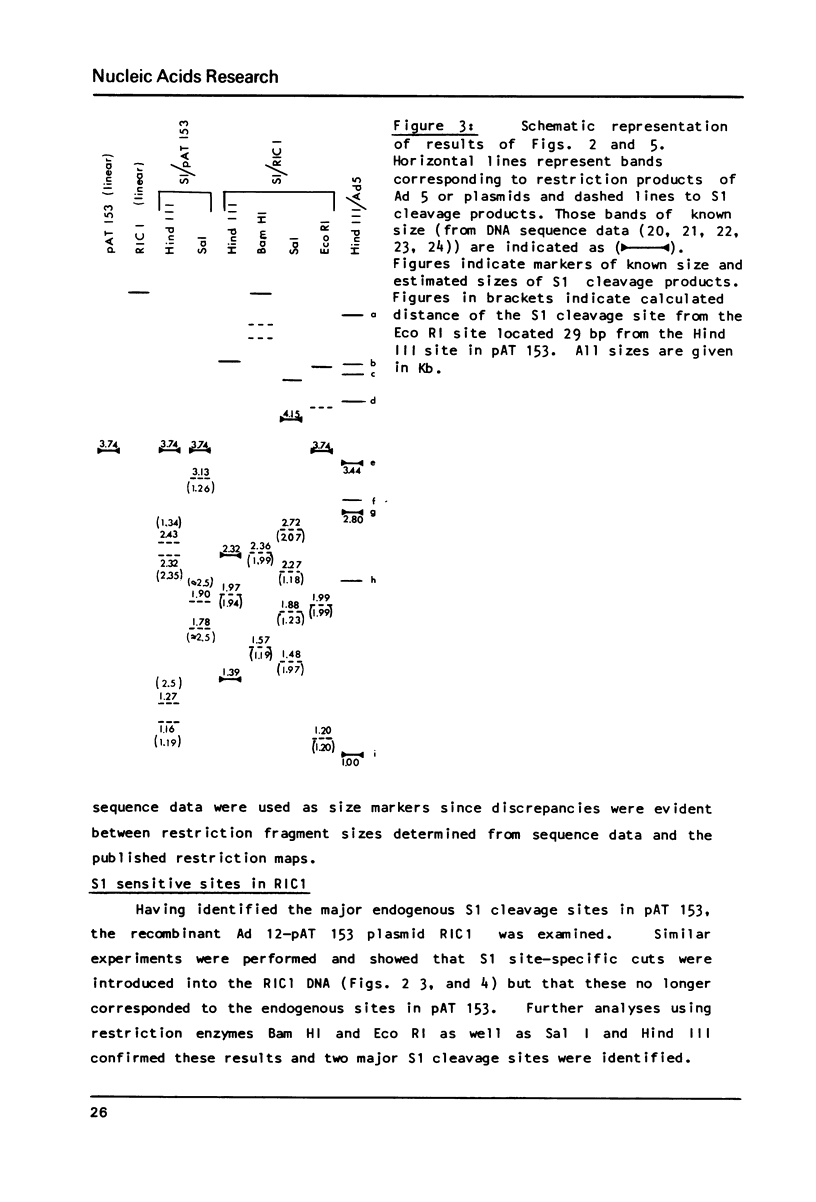
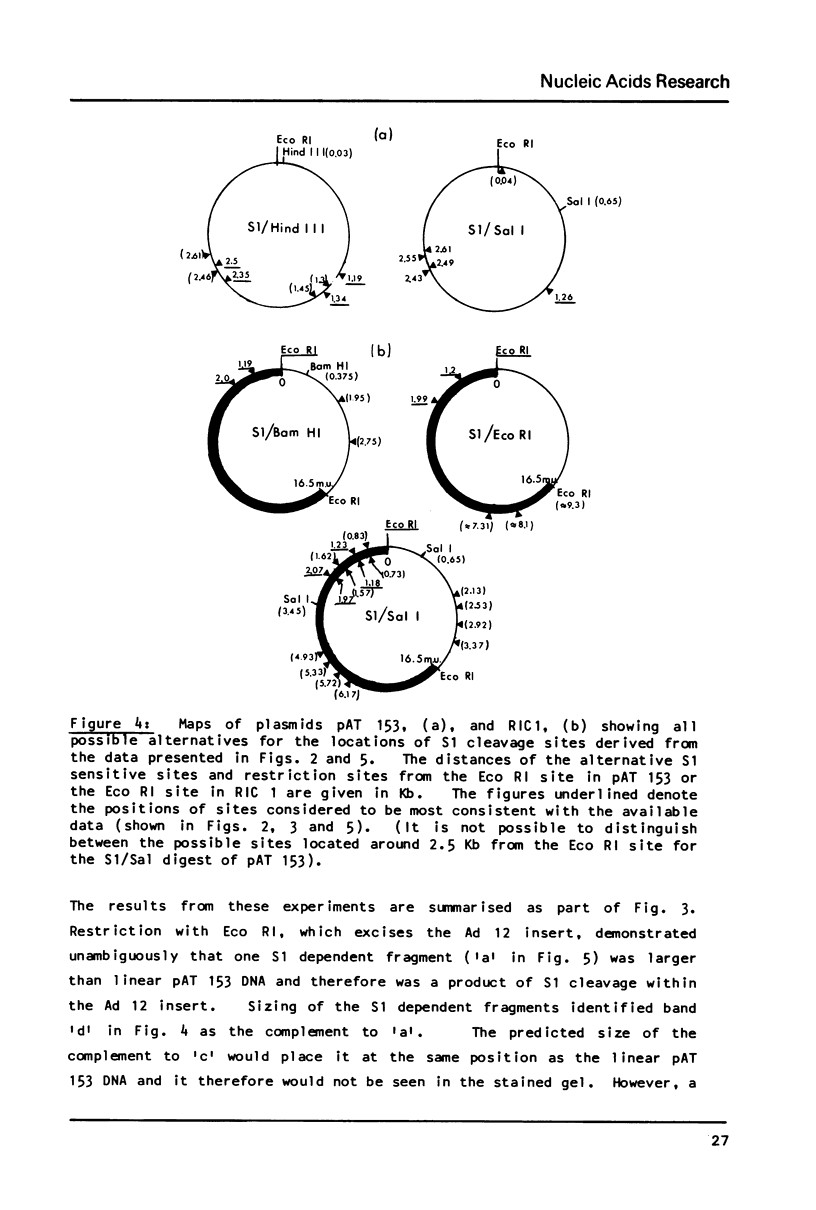
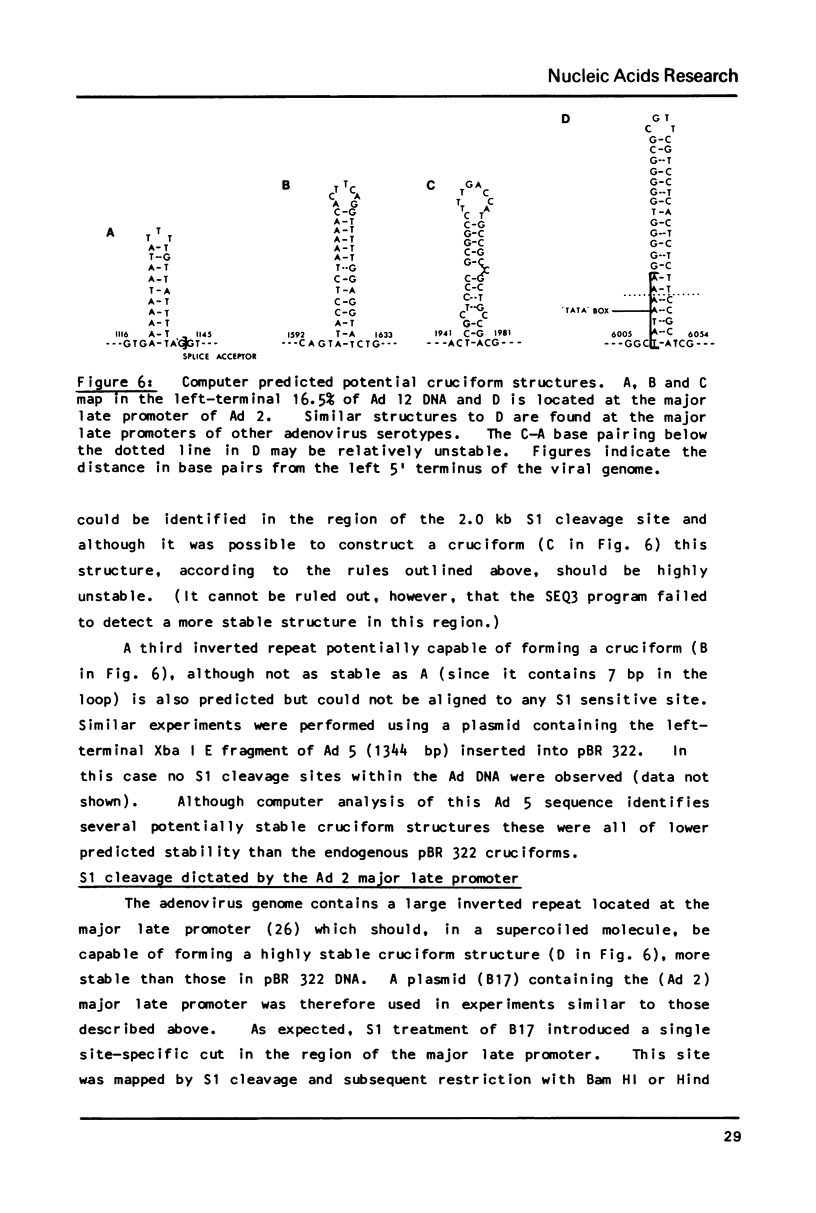
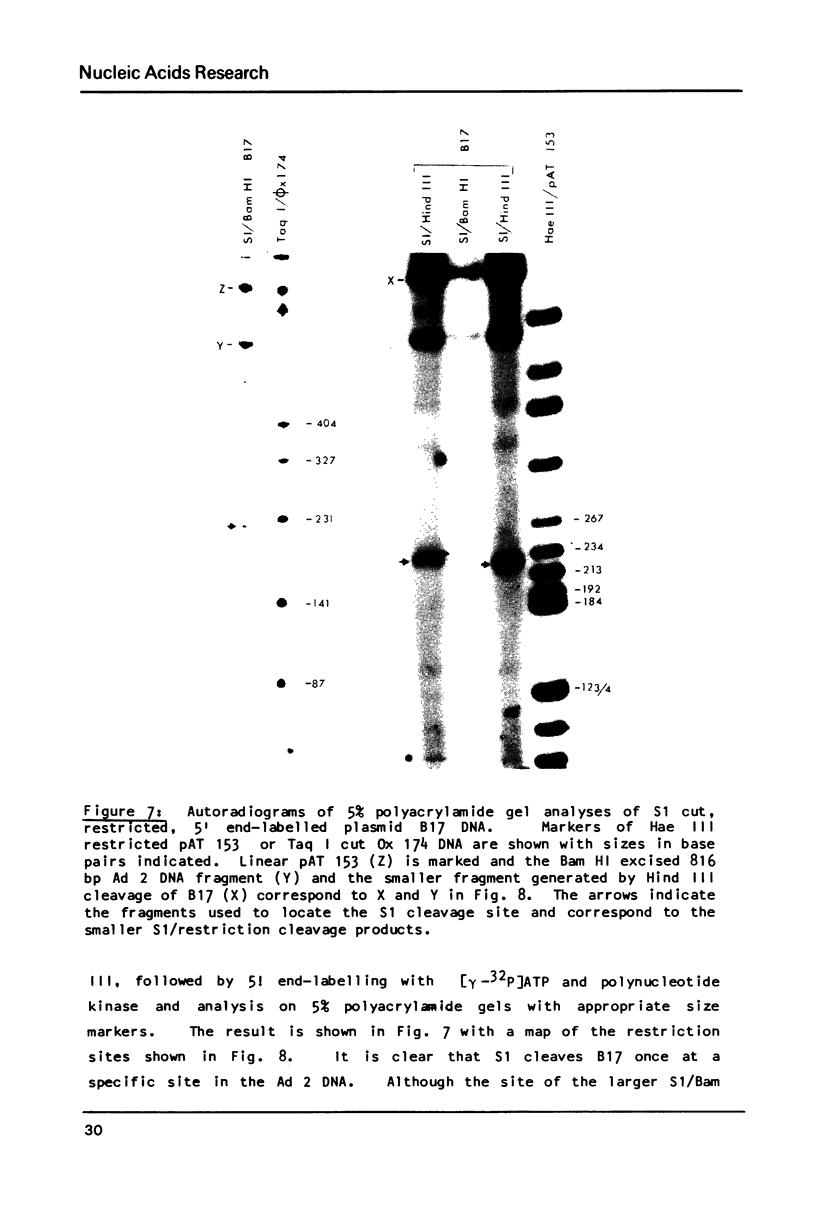
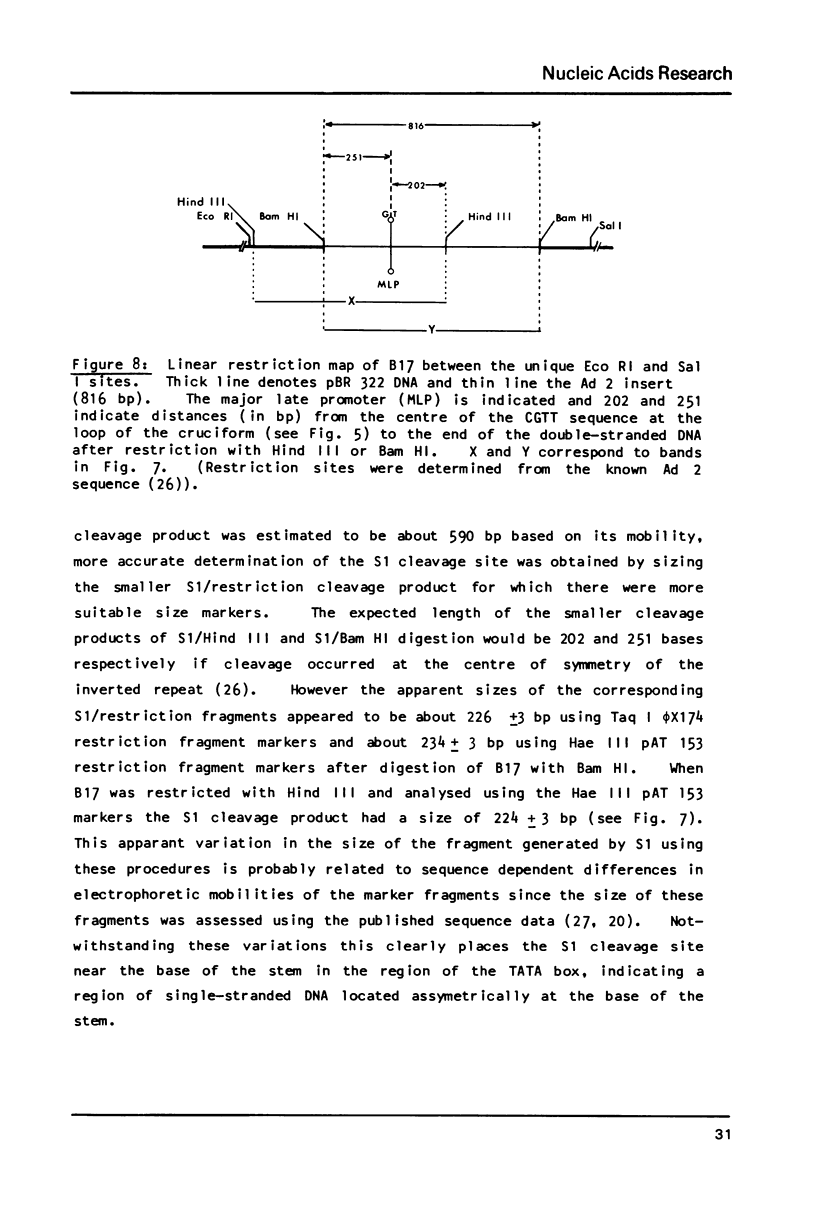
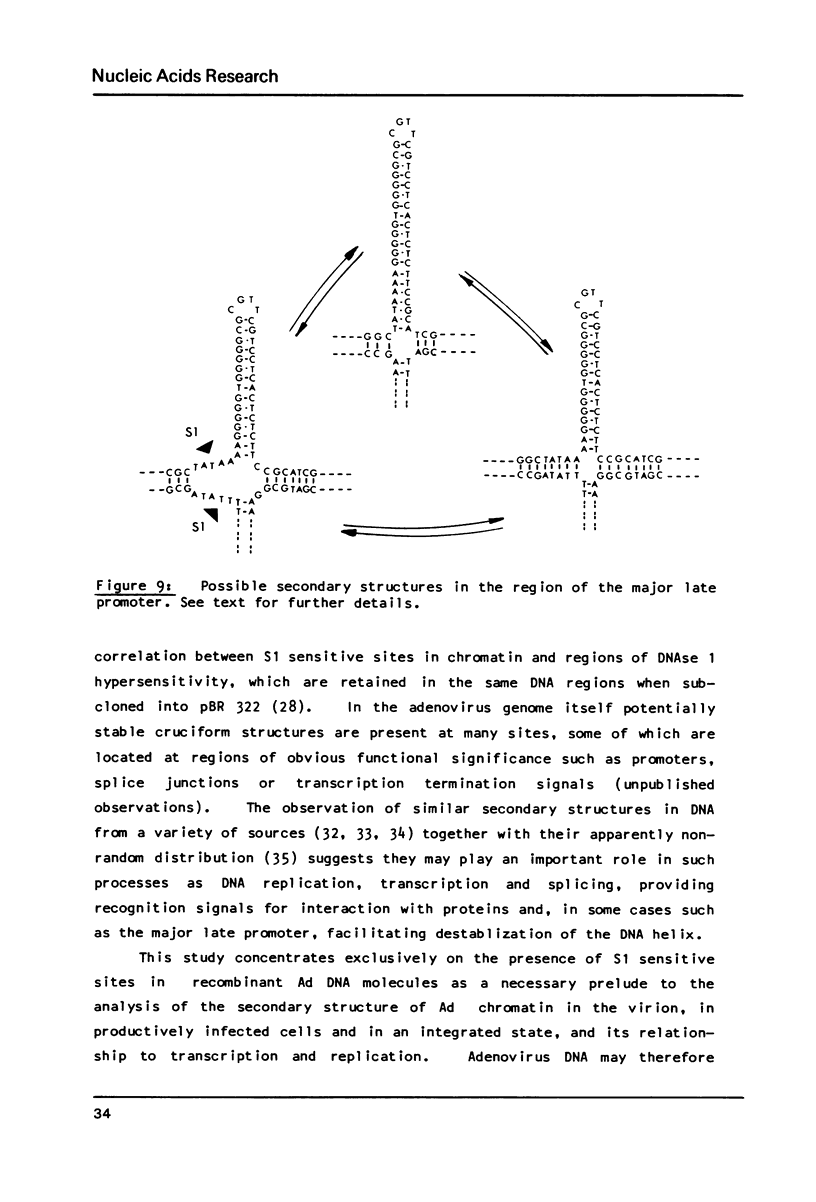
S1 nuclease has been used as a probe for regions of DNA secondary structure in supercoiled recombinant plasmids containing adenovirus (Ad) DNA sequences. In the sequences examined two S1 sensitive sites were identified in the left-terminal 16.5% of Ad 12 DNA, one of which aligned approximately with an inverted repeat region in the DNA sequence. In addition an S1 sensitive site was dictated by a potential cruciform structure in the region of the Ad 2 major late promoter. In contrast to the expected cleavage site at the loop of the cruciform, cleavage occurred at the base of the stem in the region of the TATA box. All three S1 sensitive sites identified were more sensitive to S1 than the endogenous sites in the parent plasmids.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L., Clayton J., Friedland P., Kedes L. H. SEQ: a nucleotide sequence analysis and recombination system. Nucleic Acids Res. 1982 Jan 11;10(1):279–294. doi: 10.1093/nar/10.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodgson J. B., Wells R. D. Action of single-strand specific nucleases on model DNA heteroduplexes of defined size and sequence. Biochemistry. 1977 May 31;16(11):2374–2379. doi: 10.1021/bi00630a010. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Flint S. J. Expression of adenoviral genetic information in productively infected cells. Biochim Biophys Acta. 1982 Apr 29;651(2-3):175–208. doi: 10.1016/0304-419x(82)90011-7. [DOI] [PubMed] [Google Scholar]
- GREEN M., PINA M. Biochemical studies on adenovirus multiplication. IV. Isolation, purification, and chemical analysis of adenovirus. Virology. 1963 May;20:199–207. doi: 10.1016/0042-6822(63)90157-0. [DOI] [PubMed] [Google Scholar]
- Garel A., Axel R. Selective digestion of transcriptionally active ovalbumin genes from oviduct nuclei. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3966–3970. doi: 10.1073/pnas.73.11.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C. C. Homocopolymer sequences in the spacer of a sea urchin histone gene repeat are sensitive to S1 nuclease. Nature. 1982 Feb 25;295(5851):714–716. doi: 10.1038/295714a0. [DOI] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat J., Van Ormondt H. The nucleotide sequence of the transforming HindIII-G fragment of adenovirus type 5 DNA. The region between map positions 4.5 (HpaI site) and 8.0 (HindIII site). Gene. 1979 May;6(1):75–90. doi: 10.1016/0378-1119(79)90086-6. [DOI] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller U. R., Fitch W. M. Evolutionary selection for perfect hairpin structures in viral DNAs. Nature. 1982 Aug 5;298(5874):582–585. doi: 10.1038/298582a0. [DOI] [PubMed] [Google Scholar]
- Nermut M. V. The architecture of adenoviruses: recent views and problems: Brief review. Arch Virol. 1980;64(3):175–196. doi: 10.1007/BF01322699. [DOI] [PubMed] [Google Scholar]
- Nishioka Y., Leder P. The complete sequence of a chromosomal mouse alpha--globin gene reveals elements conserved throughout vertebrate evolution. Cell. 1979 Nov;18(3):875–882. doi: 10.1016/0092-8674(79)90139-9. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Shishido K. Relationship between S1 endonuclease-sensitivity and number of superhelical turns in a negatively-twisted DNA. FEBS Lett. 1980 Mar 10;111(2):333–336. doi: 10.1016/0014-5793(80)80821-0. [DOI] [PubMed] [Google Scholar]
- Shlomai J., Kornberg A. An Escherichia coli replication protein that recognizes a unique sequence within a hairpin region in phi X174 DNA. Proc Natl Acad Sci U S A. 1980 Feb;77(2):799–803. doi: 10.1073/pnas.77.2.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J. R., Loeb L. A. S1 nuclease does not cleave DNA at single-base mis-matches. Biochim Biophys Acta. 1981 Dec 28;656(2):256–264. doi: 10.1016/0005-2787(81)90094-0. [DOI] [PubMed] [Google Scholar]
- Sims J., Benz E. W., Jr Initiation of DNA replication by the Escherichia coli dnaG protein: evidence that tertiary structure is involved. Proc Natl Acad Sci U S A. 1980 Feb;77(2):900–904. doi: 10.1073/pnas.77.2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steenbergh P. H., Sussenbach J. S. The nucleotide sequence of the right-hand terminus of adenovirus type 5 DNA: implications for the mechanism of DNA replication. Gene. 1979 Aug;6(4):307–318. doi: 10.1016/0378-1119(79)90071-4. [DOI] [PubMed] [Google Scholar]
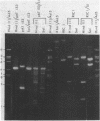
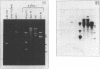
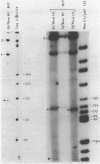
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- van Beveren C. P., Maat J., Dekker B. M., van Ormondt H. The nucleotide sequence of the gene for protein IVa2 and of the 5' leader segment of the major late mRNAs of adenovirus type 5. Gene. 1981 Dec;16(1-3):179–189. doi: 10.1016/0378-1119(81)90074-3. [DOI] [PubMed] [Google Scholar]
- van Ormondt H., Maat J., van Beveren C. P. The nucleotide sequence of the transforming early region E1 of adenovirus type 5 DNA. Gene. 1980 Nov;11(3-4):299–309. doi: 10.1016/0378-1119(80)90070-0. [DOI] [PubMed] [Google Scholar]