Background: The human pathogen Streptococcus pyogenes adapts to vascular leakage at the site of infection.
Results: S. pyogenes modifies the production of 213 in plasma determined using quantitative proteomics.
Conclusion: The results clarify the function of HSA-binding proteins in S. pyogenes.
Significance: Our data demonstrates the power of the quantitative mass spectrometry strategy to investigate bacterial adaptation to a given environment.
Keywords: Albumin, Bacterial Pathogenesis, Host-pathogen Interactions, Mass Spectrometry (MS), Proteomics, Streptococcus pyogenes, Bacterial Surface Proteins
Abstract
Streptococcus pyogenes is a major bacterial pathogen and a potent inducer of inflammation causing plasma leakage at the site of infection. A combination of label-free quantitative mass spectrometry-based proteomics strategies were used to measure how the intracellular proteome homeostasis of S. pyogenes is influenced by the presence of human plasma, identifying and quantifying 842 proteins. In plasma the bacterium modifies its production of 213 proteins, and the most pronounced change was the complete down-regulation of proteins required for fatty acid biosynthesis. Fatty acids are transported by albumin (HSA) in plasma. S. pyogenes expresses HSA-binding surface proteins, and HSA carrying fatty acids reduced the amount of fatty acid biosynthesis proteins to the same extent as plasma. The results clarify the function of HSA-binding proteins in S. pyogenes and underline the power of the quantitative mass spectrometry strategy used here to investigate bacterial adaptation to a given environment.
Introduction
Streptococcus pyogenes (or group A streptococcus), a Gram-positive bacterium infecting only humans, is a common colonizer of the upper respiratory tract and the skin where it causes relatively mild and superficial clinical conditions such as pharyngitis and impetigo (1). However, the morbidity and the costs for society of these infections are highly significant, and a study funded by the WHO in 2005 reported that S. pyogenes annually causes over 600 million cases of pharyngitis and 111 million cases of skin infections worldwide (2). Apart from these common and mostly uncomplicated infections, the bacterium is responsible for severe invasive and potentially life-threatening conditions such as necrotizing fasciitis and sepsis, and acute rheumatic fever following S. pyogenes throat or skin infections is the most frequent cause of heart disease in children. The study mentioned above reported that at least 517,000 deaths occur each year due to these conditions, emphasizing that S. pyogenes is one of the most significant bacterial pathogens in the human population. A better understanding of the biology of S. pyogenes and its interactions with the human host is required to identify novel prophylactic, diagnostic, and treatment opportunities to reduce the global burden of S. pyogenes diseases.
Several virulence factors that promote S. pyogenes colonization, immune evasion, and spread have been identified (for reviews, see Refs. 1 and 3–5), but a comprehensive view of the mechanisms operating during various phases of infection is still lacking. A characteristic property of S. pyogenes is its capacity to induce a powerful inflammatory response leading to vascular leakage at the site of infection. In this situation, and if the bacterium invades the vasculature, it will be exposed to plasma and its constituents. S. pyogenes expresses a number of surface proteins that bind several of the most abundant plasma proteins (albumin, fibrinogen, IgG, proteins of the complement and contact systems, etc.) with high affinity and specificity, underlining the intense molecular interplay between the pathogen and human plasma. These interactions could influence the gene expression in S. pyogenes with implications for bacterial adaptation and virulence, a notion supported by findings that a subset of S. pyogenes virulence proteins change their abundance levels upon contact with plasma (6, 7). However, a full picture of how the proteome is influenced by plasma is lacking, and there is presently little knowledge concerning how specific plasma proteins that bind to the surface of S. pyogenes change the gene expression of the bacterium.
Mass spectrometry-based proteomics methods have rapidly developed over the past decade to a point where almost comprehensive identification and quantification of complete bacterial proteomes is possible (8). In the present work, a label-free quantitative shotgun proteomics workflow was adapted to study the homeostasis of the S. pyogenes proteome upon exposure to increasing concentrations of human plasma. The determined S. pyogenes protein abundance profile was grouped into specific functional protein categories allowing the investigation of transcriptional regulation, protein function, and pathway organization using a targeted selected reaction monitoring (SRM)4 proteomics workflow. To analyze how a specific protein from human plasma known to bind to the surface of S. pyogenes influences gene expression, we studied the effect of albumin (HSA), the most abundant human plasma protein. The results demonstrate a profound and specific influence on the proteins of the fatty acid biosynthesis (FAB), clarifying an important function of HSA-binding surface proteins of S. pyogenes.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Proteins, Growth Conditions, and Sample Preparation
S. pyogenes strains of the M1 serotype, SF370 (ATCC 700294) and AP1 (a covS truncated strain 40/58 from the WHO Collaborating Centre for Reference and Research on Streptococci, Prague, Czech Republic), and an mga mutant of AP1 expressing low amounts of M and M-like proteins (9), were grown (37 °C; 5% CO2) in Todd-Hewitt broth (TH) (Difco Laboratories). Supplements were added at the following concentrations: 1, 5, 10, or 20% (v/v) citrated human plasma (Lund University Hospital), 4 mg/ml of HSA (Sigma), 4 mg/ml of essentially fatty acid free (∼0.005%) HSA (Sigma), 0.3 mg/ml of human fibrinogen (Sigma), or 1.2 mg/ml of human IgG (Sigma). Cells were harvested at mid-exponential phase (A620 nm = 0.5) by centrifugation, washed three times in ice-cold PBS (this will remove secreted proteins), and re-suspended in sterile ice-cold water. To isolate intracellular bacterial proteins, the samples were homogenized in a cell disruptor (FastPrep FP120, Savant Machines Inc.) three times for 20 s at level 6. Cell debris was removed by centrifugation at 10,000 × g and the homogenates were diluted in 200 mm Tris, pH 8.3, containing 6 m urea, 5 mm EDTA, and 0.2% Triton X-100. With this procedure the homogenates will contain the intracellular protein pool but also the fraction of cell wall proteins and plasma proteins bound to the bacterial surface that is released during the homogenization procedure. The protein concentration was determined using the Bradford reagent (Sigma), and the homogenates were subjected to reversed phase LC-MS/MS analysis.
Reversed Phase LC-MS/MS Analysis
The hybrid LTQ-FT-ICR mass spectrometer (Thermo Finnigan) was interfaced to a nanoelectrospray ion source. Chromatographic separation of peptides was achieved on an Agilent Series 1100 LC system (Agilent Technologies) equipped with a 11-cm fused silica emitter, 100-μm inner diameter (BGB Analytik), packed in-house with a Magic C18 AQ 5-μm resin (Michrom BioResources). Peptides were separated by a 65-min linear gradient of 5 to 40% acetonitrile in water, containing 0.1% formic acid, with a flow rate of 0.95 μl/min. Three MS/MS spectra were acquired in the linear ion trap per each FT-MS scan, which was acquired at 100,000 FWHM nominal resolution settings with an overall cycle time of ∼1 s. Charge state screening was employed to select for ions with at least two charges and rejecting ions with undetermined charge state.
The hybrid Orbitrap-LTQ XL mass spectrometer (Thermo Electron, Bremen, Germany) was coupled online to a split-less Eksigent Two-dimensional NanoLC system (Eksigent Technologies, Dublin, CA). Peptides were loaded with a constant flow rate of 10 μl/min onto a pre-column (Zorbax 300SB-C18 5 × 0.3 mm, 5 μm, Agilent Technologies, Wilmington, DE) and subsequently separated on a RP-LC analytical column (Zorbax 300SB-C18 150 mm × 75 μm, 3.5 μm, Agilent Technologies) with a flow rate of 350 nl/min. The peptides were eluted with a linear gradient from 95% solvent A (0.1% formic acid in water) and 5% solvent B (0.1% formic acid in acetonitrile) to 40% solvent B over 55 min. The mass spectrometer was operated in data-dependent mode to automatically switch between Orbitrap-MS (from m/z 400 to 2000) and LTQ-MS/MS acquisition. Four MS/MS spectra were acquired in the linear ion trap per each FT-MS scan, which was acquired at 60,000 FWHM nominal resolution settings using the lock mass option (m/z 445.120025) for internal calibration. The dynamic exclusion list was restricted to 500 entries using a repeat count of two with a repeat duration of 20 s and with a maximum retention period of 120 s. Precursor ion charge state screening was enabled to select for ions with at least two charges and rejecting ions with undetermined charge state. The normalized collision energy was set to 30%, and one microscan was acquired for each spectrum. The resulting MS/MS data were deposited in the peptide atlas.
Selected Reaction Monitoring
Identified peptides with high probability from FAB proteins were synthesized (JPT Technologies, Berlin, Germany) and the SRM transition lists were generated using a previously published method (10). The SRM measurements were performed on a TSQ Vantage triple quadropole mass spectrometer (Thermo Electron, Bremen, Germany) equipped with a nanoelectrospray ion source (Thermo Electron). Chromatographic separations of peptides were performed on an Eksigent One-dimensional NanoLC system (Eksigent Technologies) using the same chromatographic conditions as described above for the Eksigent Two-dimensional NanoLC system connected to the hybrid Orbitrap-LTQ XL mass spectrometer. The LC was operated with a flow rate of 400 nl/min. The mass spectrometer was operated in SRM mode, with both Q1 and Q3 settings at unit resolution (FWHM 0.7 Da). A spray voltage of +1700 V was used with a heated ion transfer setting of 270 °C for desolvation. Data were acquired using the Xcalibur software (version 2.1.0). The dwell time was set to 10 ms and the scan width to 0.01 m/z. All collision energies were calculated using the formula: CE = (parent m/z) × 0.034 + 3.314.
Data Analysis
The MS2 spectra were searched with the spectra identification pipeline. The pipeline combines the results of the search engines X! Tandem, version 2009.04.01.1 with k-score plugin (11), Mascot, version 2.3 (12), and OMSSA, version 2.7.1 (13), and generates a common peptide and protein list using the Trans-Proteomic pipeline, version 4.4.0 (14). All searches were performed with full tryptic cleavage specificity, up to 2 allowed missed cleavages, a precursor mass error of 15 ppm, and an error tolerance of 0.5 Da for the fragment ions. The sample preparation cysteine carbamidomethylation was defined as fixed modification in the search parameters. A protein data base with sequences for S. pyogenes (NC_002737 from NCBI) and human (Swiss-Prot, version 57.1 including known splice variants and isoforms) was used to match the individual spectra to certain peptides. The data base was extended by decoy sequences to validate the resulting peptide-spectrum matches (15). A value of 0.01 for the false-discovery rate was then used to generate the final protein list with ProteinProphet.
Label-free quantification was performed with the OpenMS software framework (16). Unless indicated otherwise, TOPP tools (17), small applications provided by OpenMS, were used for the individual processing steps. The input to the analysis pipeline consisted of 10-mzXML files containing LC-MS/MS data acquired on the LTQ-FTICR instrument in profile mode. These files were first converted to mzML format with the FileConverter tool, then centroided using the PeakPicker (“high_res” algorithm). These two steps were carried out in OpenMS 1.6; for the following steps, an early (January 2010) development version of OpenMS 1.7 was used. On the centroided mzML files, feature detection was performed using the FeatureFinder (“centroided” algorithm), producing one feature map per sample. In parallel, results from the MS2 identification pipeline were preprocessed with a custom Python script and split into individual pepXML files (one per sample and search engine). Peptide identification data from these files was read, filtered to 1% FDR, and stored in OpenMS′ idXML format (one file per sample) by a custom C++ program built on top of the OpenMS library (release version of OpenMS 1.7). To annotate the feature maps with identified peptides, the IDMapper tool was applied to the pairs of feature maps/idXML files derived from the same sample. The resulting annotated feature maps were adjusted to a common retention time scale with MapAligner (“identification” algorithm), then combined into one consensus map by grouping features across samples with the FeatureLinker (“unlabeled” algorithm). The consensus map was converted to a table in CSV format using the TextExporter. A custom R script was used to read the quantification data from the CSV file, clean up annotation conflicts, and compute peptide abundances from the intensities of annotated features. To reduce the impact of feature detection differences in the individual samples, only the charge state with the highest intensity per peptide was used for quantification. The resulting peptide abundances were exported into a data base, where protein abundances were inferred by summing up the abundances for the peptides uniquely mapping to each protein (18). The quantitative data from openMS was normalized using the total ion current from each LC/MS experiment that could be assigned to S. pyogenes and then organized in a matrix with proteins as rows and conditions (e.g. % plasma) as columns. The three first principle components resulting from applying principle component analysis (19) to the matrix were used to cluster the proteins using k-mean clustering (20) always performed in R (version 2.9.0).
Fatty Acid Uptake Assay
One vial of the BODIPY-FA probe from the QBT Fatty Acid Uptake Assay Kit (Molecular Devices) was mixed with 1 ml of 20 mg/ml of HSA-FA in 20 mm Tris-HCl, pH 7.4. The probe-HSA complex was added 1/10 (v/v) to S. pyogenes wild type (AP1) and mga mutant AP1 bacteria grown to A620 nm = 0.15 in TH broth. Triplicate samples of either living or heat-killed (80 °C, 5 min) bacteria containing probe-HSA complex were further incubated at 37 °C and 5% CO2. At time points 0, 40, 80, and 120 min, a small aliquot of the samples were removed and placed on ice. Bacteria were labeled using IVIG (polyclonal human therapeutic IgG, 5 min, 500 μg/ml) and DyLight 649-conjugated goat anti-human F(ab′)2 fragments (Jackson ImmunoResearch, 5 min, 1:100) on ice.
Fluorescence Microscopy
Acquisition of images was performed using a fluorescence microscope (Nikon Eclipse TE300 equipped with a Hamamatsu C4742–95 cooled CCD camera, using a Plan Apochromat ×100 objective with NA 1.4). Images were acquired with identical exposure time (NIS Elements 3, Nikon) and have been processed with linear brightness and contrast adjustments (Adobe Photoshop CS5) to represent the samples as they were observed in the microscope.
Flow Cytometry
The flow cytometry measurements were performed using a FacsCalibur flow cytometer (BD Biosciences) equipped with lasers tuned to 488 and 633 nm. Bacteria were gated using forward and side scatter in the logarithmic mode and bound IgG (DyLight 649-labeled) was detected in the FL4 channel. Fluorescence from the BODIPY-FA probe was collected in the FL1 channel.
RESULTS
Proteome-wide Analysis of the S. pyogenes Proteome in Response to Human Plasma
Adapting to human plasma is a critical ability for S. pyogenes with implication for pathogenesis and virulence. Human plasma is a complex mixture of proteins and nutrients constituting on one hand a rich medium for the bacterium, and on the other hand a source of host defense systems. In the adaptation process, the type and amount of proteins synthesized is tightly regulated to create a composition that is optimized to cope with the new environment. To study this process, mass spectrometry-based quantitative proteomics was used to identify and quantify the intracellular protein constituent of the S. pyogenes-translated genome. S. pyogenes (strain SF370 of the M1 serotype) was grown in TH medium supplemented with different amounts of human plasma (0, 1, 5, 10, and 20%) in two biological replicates. Following centrifugation and washing, the bacteria were homogenized. Resulting debris was spun down and proteins of the supernatants were digested to peptides and analyzed using LC-MS/MS. To maximize the number of identified proteins we extensively annotated the detected peptides using the combined outputs of multiple search engines at a stringent false-discovery rate. An analysis pipeline based on the OpenMS software framework (16) was then used to quantitatively compare the LC-MS peptide patterns from different samples resulting in an extensive quantification matrix for the S. pyogenes proteins. In this experiment, 523 S. pyogenes proteins (supplemental Table S2) were identified and quantified with an estimated false discovery rate of 5% using a reversed data base search strategy (15) and Protein and PeptideProphet (21, 22).
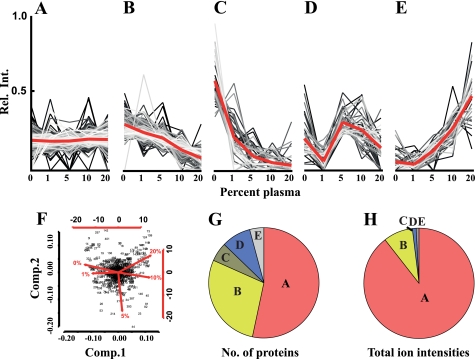
The data indicate that S. pyogenes regulates its proteome composition in a dose-dependent manner upon exposure to increasing amounts of plasma (Fig. 1), first demonstrated by using principle component analysis revealing the degree of proteome reorganization. This analysis suggests that there is a small difference in protein abundance between bacteria grown in 0 or 1% plasma, whereas the proteome of bacteria grown in 10 or 20% plasma was considerably different compared with bacteria grown in medium without plasma (Fig. 1F). Cultures grown in 5% plasma display an intermediate profile. In a second stage, the protein abundance profiles were split, using k-mean clustering, into five clusters containing the distinguishing proteins as shown in Fig. 1. The predominant cluster contains the proteins that remain constant at the plasma concentrations tested (Fig. 1A). Two clusters represent the proteins with decreasing protein abundances (Fig. 1, B and C), and two contain the proteins with increasing or fluctuating protein abundances (Fig. 1, D and E). Sixty-one proteins observed in less than four of the five growth conditions were excluded, as these proteins are present at a too low concentration to allow accurate clustering. The clustered data reveal that there are a large number of proteins that either increase or decrease in protein abundance. The largest cluster, 246 proteins (46%), contains the proteins that remain constant at the different plasma concentrations (Fig. 1G, label A), followed by the cluster containing 155 proteins (30%) where the protein abundances were decreased (Fig. 1G, labels B and C). A lower number of proteins, 58 proteins (11%), showed increased abundance (Fig. 1G, labels D and E) (supplemental Table S1 lists the proteins that were mostly influenced by plasma). However, the number of proteins that change their abundance does not necessarily reflect the degree of proteome adaptation. The disproportional amount of proteins of the cluster in Fig. 1A becomes more apparent when the total ion intensities for proteins in the respective cluster are summed up and compared with the protein count (Fig. 1H). Total ion intensities represent the recorded signal intensities for the peptides associated with a protein in the mass spectrometer, and provide a rough estimate of protein abundance (8, 23–26). The cluster in Fig. 1A can be ascribed to 89% of the ion intensities, which indicates that the abundance level for the majority of the intracellular proteins remains constant. Only 9% of ion intensities represent the proteins with decreased protein abundances (Fig. 1H, labels A and B), and 2% of the ion intensities are ascribed to proteins that increase (Fig. 1H, labels D and E). The large over-representation of the proteins with unchanged abundance indicates that the overall proteome reorganization is relatively modest in response to human plasma. Even though a considerable number of proteins change in concentration they tend to be present at relatively low amounts compared with the proteins that remain constant. There are almost three times as many proteins that display decreased protein abundances compared with those that are increased (155 versus 58), and the difference in ion intensities is close to 5-fold (9 versus 2%). The results indicate that a number of proteins become dispensable for S. pyogenes growing in plasma. It should be noted that proteins secreted by the bacteria are lost during the washing step, and despite low intracellular abundance they could be present in high amounts in the growth medium.
FIGURE 1.
Label-free quantitative mass spectrometry analysis of S. pyogenes adaptation to human plasma. Bacteria were grown at increasing concentrations of human plasma (0–20%), harvested, and lysed. Proteins of the lysates were identified and quantified by label-free LC-MS/MS. Using k-mean clustering the expression profiles were split into five different clusters. A, proteins that do not change in abundance. B and C, proteins with lower abundance. D and E, proteins with increased or fluctuating protein abundance levels when exposed to increasing amount of plasma. F, principal component analysis (for details, see Ref. 19) of S. pyogenes expression profiles. Each dot represents a protein and the spread of the dots and the red lines indicate the degree of proteome reorganization following exposure to increasing amounts of human plasma. G, distribution of proteins within the clusters when counting the protein from profile clusters in panel A–E. H, distribution of proteins within the clusters when summing up the ion intensities from profile clusters of panels A–E.
Over-represented Functional Protein Categories in Expression Clusters
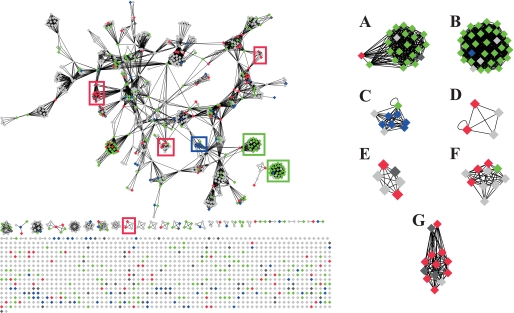
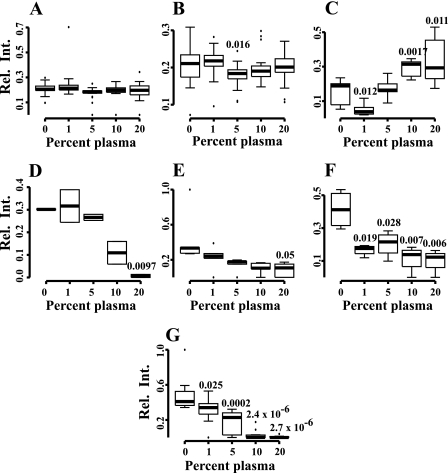
Hypothetically, the proteins that are repressed or induced belong to certain functional categories that could reflect how the bacteria adapt to the new environment. To allow visualization of the complicated expression profile data we used Cytoscape (27). Cytoscape is a network viewer showing protein annotations together with protein-protein interaction databases or functional categories to provide context, in this case subsystem classification from the National Microbial Pathogen Data Resource (NMPDR) (28). The network topology to the left in Fig. 2 outlines the overall network organization, where rectangles indicate proteins, clustered together if they belong to the same functional categories. Green rectangles indicate the proteins in Fig. 1A and represent the proteins with constant abundance. Red rectangles are the proteins in Fig. 1, B and C, showing decreased abundance, and blue rectangles the proteins in Fig. 1, D and E, that are increased. To make the view comprehensive and genome-wide, all open reading frames from the SF370 strain were loaded into the Cytoscape network and proteins that were not identified are indicated as light gray rectangles. Dark gray rectangles denote proteins that were identified but not clustered. In NMPDR every protein is assigned to a subsystem that indicates protein function, and the enrichment of certain subsystems in the different clusters is shown in Fig. 1. Subsystems with a z-score higher than 3 containing at least 3 entries were considered as enriched. Ribosomal proteins were over-represented among the proteins that remained constant in abundance, and they are displayed as predominantly green colored rectangles (Fig. 2. A and B). Proteins involved in de novo pyrimidine synthesis were over-represented among the proteins showing increased abundance; depicted as predominantly blue colored rectangles (Fig. 2C). For the proteins with decreasing protein abundances the following over-represented NMPDR subsystems (displayed as predominantly red colored dots) were identified: choline and betaine uptake, and betaine biosynthesis (Fig. 2D), control of cell elongation (Fig. 2E), and ribonucleotide reduction (Fig. 2F). Finally, all proteins involved in FAB are dramatically repressed when the bacteria are grown in the presence of plasma (Fig. 2G), demonstrating that the endogenous FAB of S. pyogenes is turned off in plasma environment. The relative protein expression profiles for the protein groups were calculated and visualized as box plots in Fig. 3.
FIGURE 2.
A Cytoscape network view of all NMPDR subsystem annotated open reading frames from S. pyogenes. The left part represents an outline of the network topology of the S. pyogenes (strain SF370) proteome displayed using Cytoscape. Rectangles indicate proteins and lines connecting the dots represent functional categories using NMPDR subsystem classification. Sets of rectangles grouped into clusters represent one functional category and rectangles connected with more than one line represent a protein present in several functional categories. The rectangle color scheme indicates changes in protein abundance as a function of increasing plasma concentration shown in Fig. 1. Proteins that show constant abundance are colored green, proteins with increasing abundance are blue, decreasing proteins are red, non-quantified proteins dark gray, and non-identified proteins are represented as light gray rectangles. In this view all hypothetical proteins and proteins without subsystem annotations are outlined at the bottom as unconnected rectangles. Several functional protein categories were significantly enriched in the clusters: A, ribosomal proteins (SSU); B, ribosomal proteins (LSU); C, de novo pyrimidine synthesis; D, choline and betaine uptake and betaine biosynthesis; E, control of cell elongation; F, ribonucleotide reduction; G, fatty acid biosynthesis, FASII.
FIGURE 3.
The protein expression profiles for the enriched functional categories. Increasing levels of plasma induces changes in protein abundances for specific functional categories shown in Fig. 2: A, ribosomal proteins (SSU); B, ribosomal proteins (LSU); C, de novo pyrimidine synthesis; D, choline and betaine uptake, and betaine biosynthesis; E, control of cell elongation; F, ribonucleotide reduction; G, fatty acid biosynthesis (FASII). The numbers indicate p values, where the relative ion intensities generated at different plasma concentrations are compared with TH alone (0% plasma).
In summary, by using quantitative label-free tandem mass spectrometry we quantified the S. pyogenes proteome after exposure to increasing amounts of plasma. The bacteria responded by considerable reorganization of the proteome. Minimally 155 proteins were lowered in concentration comprising an estimated 8.7% of the total protein ion intensities when grown in TH medium alone, compared with 4.7% in TH containing 20% plasma. This is compensated by an increased abundance of 58 proteins (increasing from 1.7 to 3% of the total protein ion intensities) following growth in 20% plasma. The grouping of the induced and repressed gene products into the NMPDR subsystem classification revealed several NMPDR subsystem that were over-represented among the proteins with reduced protein abundances, where in particular the proteins involved in FAB showed a dramatic decrease in S. pyogenes grown in the presence of plasma, suggesting that the required fatty acids might be acquired from the added plasma.
Binding of Fatty Acid-loaded HSA to Surfaces of S. pyogenes Down-regulates Fatty Acid Biosynthesis
S. pyogenes was cultivated to mid-exponential growth phase in the presence of various concentrations of plasma. The bacteria were carefully washed in buffer (this will remove secreted proteins) and homogenized. Following centrifugation to remove cell debris, the resulting supernatants were analyzed by mass spectrometry. These supernatants will, apart from the intracellular S. pyogenes protein pool, contain the fraction of bacterial cell wall proteins and plasma proteins associated with the bacterial surface that are released during the homogenization procedure. Among human plasma proteins, peptides derived from HSA, fibrinogen, and IgG gave rise to the strongest ion intensities, consuming 97.9% (82.5, 10.3, and 5.1%, respectively) of the total ion intensities for the human proteins. This was not unexpected because the binding of these proteins to S. pyogenes in the plasma environment has been known for decades (29, 30). The SF370 strain used here is of the clinically important M1 serotype expressing the M1 protein, a surface protein with specific and separate binding sites for HSA, fibrinogen, and IgG (31, 32). Whereas some of the biological consequences of the binding of fibrinogen and IgG to S. pyogenes have been clarified (for review, see Ref. 33), the function of HSA binding to the surface of S. pyogenes is unknown.
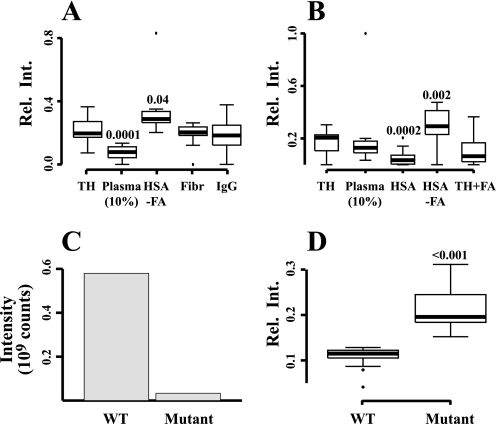
In plasma, FAs are transported by HSA, and the sharp decline in the proteins involved in FAB indicate that the bacteria can accommodate their requirement for FA in a 10–20% plasma environment. S. pyogenes specifically binds HSA via repetitive so-called C domains of the cell wall anchored M and M-like proteins (32, 34–36), and large amounts of HSA were detected in lysates following growth of strain SF370 in the plasma environment. Because this strain lacks emm-like genes (37) encoding M-like proteins, this observation suggests that the M1 protein is solely responsible for the binding of HSA to SF370 bacteria. To confirm the hypothesis that HSA-bound FA are involved in direct regulation of FAB, S. pyogenes was incubated with individual preparations of the three plasma proteins that are most abundantly present at the bacterial surface (HSA, fibrinogen, and IgG) in two biological and three technical replicates. The same quantitative LC-MS/MS strategy as described above was then used to identify and quantify proteins of the SF370 bacteria. In total we identified 684 S. pyogenes proteins at 5% false-discovery rate. Supplemental Table S3 contains the quantified proteins along with their relative protein abundance under the selected conditions. As expected, growth in 10% plasma, used as a positive control, resulted in a statistically significant decrease of the FAB proteins (Fig. 4A). Adding only HSA devoid of FA resulted in a small but statistically significant increase of these proteins, whereas fibrinogen and IgG had no effect. In the next series of experiments, S. pyogenes was exposed to HSA with or without FA, and the proteomes were investigated in three biological replicates, identifying 566 S. pyogenes proteins (supplemental Table S4). Again, incubating the bacteria with HSA devoid of FA caused a statistically significant increase of FAB proteins (Fig. 4B). In contrast, the addition of HSA containing FA resulted in a marked decrease of the biosynthesis proteins. The decrease was less prominent when FA alone was added to bacterial growth medium. In a final set of experiments we investigated whether the lack of HSA-binding M and M-like proteins influenced the expression of FAB proteins using SRM mass spectrometry. For this purpose another S. pyogenes strain (AP1) of the M1 serotype was used, because we had access to a mutant of this strain in which the regulatory gene mga is inactivated by transposon insertion, resulting in a very low (<1%) expression of M and M-like proteins (9). The wild type AP1 strain has a truncation in covS of the two-component covR/S system,5 and in relationship to the present study it is important that previous work has demonstrated that a covS mutation does not affect the level of FAB gene expression neither in vivo nor in vitro (4). Mass spectrometry analysis demonstrated that the amount of HSA released from the mutant bacteria during the homogenization procedure was more than a 100-fold lower than from the wild type strain (Fig. 4C). When growing the two strains in the presence of 10% plasma, wild type bacteria decrease their expression of FAB proteins determined by targeted SRM mass spectrometry. In 10% plasma, the levels of these proteins are on an average 2.4-fold higher in the mutant strain compared with wild type bacteria, with a p value of <0.001 (Fig. 4D). Collectively, these experiments demonstrate that the binding of HSA containing FA to the surface of S. pyogenes via M and M-like proteins down-regulates the expression of the proteins required for FAB.
FIGURE 4.
Regulation of fatty acid biosynthesis proteins in S. pyogenes. A, s. pyogenes (strain SF370) was grown in TH broth alone, TH with 10% plasma, or TH containing the plasma proteins most abundantly associated with the bacterial surface; HSA (devoid of fatty acids), fibrinogen and IgG. FAB proteins were quantified using label-free LC-MS/MS, and their relative ion intensities are shown. Significant p values are indicated and represent comparison with the TH control. B, the FAB proteins were measured in S. pyogenes (strain SF370) grown in TH alone, TH with 10% plasma, TH with HSA containing or devoid of fatty acids, or TH containing free fatty acids. p values compared with the TH control are indicated. C, wild-type S. pyogenes (strain AP1) or mutant AP1 bacteria expressing low amounts of HSA-binding surface proteins were grown in 10% plasma. The amount of HSA released from washed and homogenized bacteria was determined using MS. D, following growth in 10% plasma, the FAB proteins of wild-type AP1 and the mutant bacteria were measured by MS.
Higher Uptake of HSA-bound BODIPY-FA by Wild Type S. pyogenes
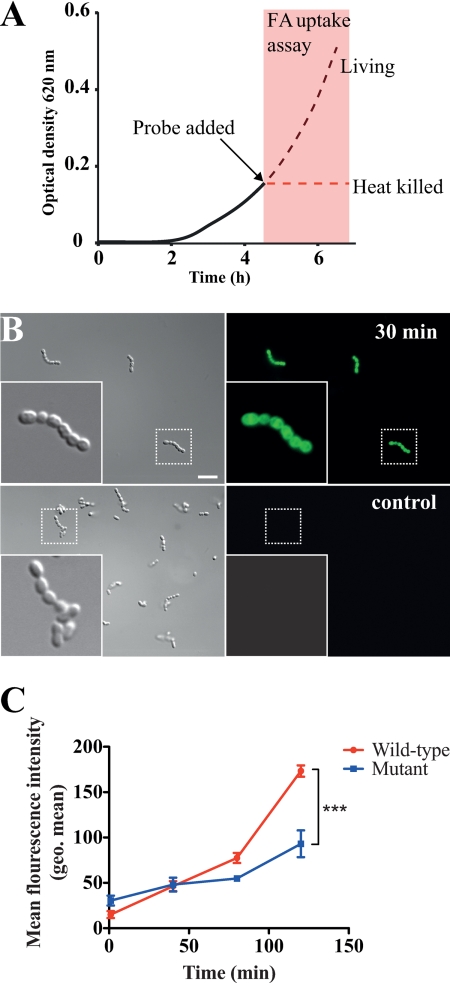
To further verify that the binding of HSA to bacterial surface proteins promotes the uptake of FA in complex with HSA, we utilized a dodecanoic acid fluorescent fatty acid analog (BODIPY-FA) coupled with a cell-impermeable quenching agent. In eukaryotic cells, this probe is used for monitoring fatty acid uptake, whereupon an increase in fluorescence signal is detected by transport into the cell (38).
The probe was complexed with FA-free HSA and added to live or heat-killed wild type (AP1) and mga mutant AP1 bacteria at the early exponential phase (A620 nm = 0.15). The samples were then further incubated and internalization of BODIPY-FA was monitored during growth (Fig. 5A). The probe had no impact on bacterial viability (data not shown), and the uptake of probe and subsequent removal of the quencher in bacteria was confirmed with fluorescence microscopy (Fig. 5B). Differences in the average probe uptake of single streptococcal chains or aggregates were quantified using flow cytometry. For both strains a time-dependent increase of probe internalization was observed, but the uptake of BODIPY-FA by the wild type strain was significantly higher compared with mutant bacteria (Fig. 5C). The internalization of BODIPY-FA by the heat-killed wild type and mutant strains was saturated after 30 min of incubation, indicating that uptake of the FA probe is dependent on active processes in living cells. The higher level of uptake of BODIPY-FA by wild type bacteria demonstrates that HSA-binding bacterial surface proteins promotes FA uptake, thereby reducing FAB protein production.
FIGURE 5.
Internalization of BODIPY-FA by S. pyogenes. A, wild-type (AP1) and mga mutant AP1 bacteria were grown to early exponential phase (A620 nm = 0.15). At this point samples were withdrawn and heat-killed. BODIPY-FA complexed with HSA was then added to living and heat-killed bacterial samples to a final concentration of 2 mg/ml of HSA (corresponding to ∼5% of the concentration in plasma) in triplicate replicates. Bacterial cultures were further incubated and aliquots were removed and analyzed at the indicated time points. The figure schematically shows the experimental procedure. B, image showing uptake of the BODIPY-FA probe (green) exemplified by wild-type bacteria and a negative control, without the probe after 30 min of incubation. C, quantification of BODIPY-FA uptake by wild-type and mutant bacteria determined by FACS. Changes in geometric mean fluorescence intensity are plotted over the 2-h time course. The error bars indicate 95% confidence interval of the three replicates. Data represent mean ± 95% confidence interval of the three replicates. Asterisks (***) indicate p < 0.0001 by two-way analysis of variance.
DISCUSSION
This study is focused on intracellular S. pyogenes proteins and the experimental approach does not allow the quantitative analysis of secreted proteins, bacterial surface proteins, or human plasma proteins bound to the bacterial surface. Our aim was to investigate how the intracellular proteome of the bacterium is influenced by human plasma, and to investigate whether quantitative proteomics could be applied also to clarify biological consequences of the binding of a specific human plasma protein to surface proteins of S. pyogenes. These questions are relevant because the pathogen has the capacity to induce powerful inflammatory responses causing vascular leakage, and is equipped with a number of surface proteins that bind some of the most abundant human plasma proteins with high affinity and specificity. The proteome of S. pyogenes strain SF370 comprises 1905 proteins (37), and in total from all experiments 842 of these could be identified and quantified following growth at different concentrations of plasma. The abundance of many of the proteins, in total 213, was affected by plasma (in most cases a decrease was recorded), but when summing up the ion intensities for these proteins, their amount corresponds to ∼10%, a surprisingly small fraction. The vast majority of the proteins do not change in abundance, demonstrating that there is no major change in the total intracellular proteome turnover but rather a specific and fine-tuned response to plasma in the environment.
In addition to proteins required for FAB, proteins involved in a wide range of cellular functions such as metabolism, protein translation, and proteolysis were affected by the presence of plasma in the growth medium. Several proteins annotated as hypothetical proteins were also among those that changed in abundance level. To objectively select regulated proteins, we used a number of on-line resources such as NMPDR and STRING, and statistical tools such as principle component analysis and k-mean clustering, to group the proteins into functional categories to estimate which protein classes are enriched in the repressed and induced clusters. This type of analysis tends to favor the selection of well characterized biological processes, because these are often better annotated in the databases. In total seven functional categories were identified, where the most pronounced were de novo pyrimidine synthesis (increased) and FAB (decreased). The subsystem de novo pyrimidine synthesis comprises the proteins necessary for the formation of the pyrimidine bases. Although not statistically significant, proteins involved in purine conversion also tended to be enriched. In contrast to 100% plasma, which inhibits the multiplication of S. pyogenes through contact system activation (39), 5–50% plasma in the growth medium promotes growth of the bacteria,6 which may explain these changes. Bacterial FAB is performed by a highly conserved set of enzymes in the so-called FASII pathway (for reviews, see Ref. 40–42). When S. pyogenes is grown in 20% plasma there is a 75-fold reduction of these enzymes compared with TH medium alone, indicating that the bacteria can turn off their synthesis of FA and rely on exogenous FA for lipid homeostasis. There was a strong co-regulatory trend of the FAB proteins. This is not surprising considering that they are part of the same operon and, according to online protein interaction databases, also physically interconnected. In contrast to the biosynthesis enzymes, the reduction of the transcriptional FAB regulator FabT is less prominent. This is particularly apparent in the final experiment where FabT was detected only in 10% plasma. FabT has previously been shown to decrease the level of FAB proteins in Streptococcus pneumoniae (43, 44). Because the abundance level is low, quantitative data concerning this protein are not reliable, and it was not possible to determine its regulation pattern. When the observed protein abundance data were plotted on top of the open reading frames in the genome, all the FAB and ribosomal proteins, expressed from their respective parts of the genome, are highly abundant when grown in TH broth without plasma. However, in contrast to the FAB proteins, the ribosomal proteins were not affected by plasma.
Gram-positive bacterial species such as Staphylococcus epidermidis, Finegoldia magna, and group C and G streptococci, express HSA-binding surface proteins (45–47), which have evolved through shuffling of an HSA-binding so-called GA module (48). Previous work has suggested that the acquisition of this module adds selective advantages to the bacteria by promoting growth and protection against anti-bacterial peptides (49, 50). As mentioned, the binding of HSA to S. pyogenes has been known for many years and the interaction was mapped to the COOH-terminal region of M and M-like proteins, important virulence determinants of S. pyogenes (for review, see Ref. 51). However, there is no homology between the GA module and the HSA-binding domains of M and M-like proteins, and the biological function of HSA-binding to S. pyogenes was unknown when the present study was initiated.
HSA is the main carrier of FA in human plasma, and the profound influence of plasma on the expression of genes related to FAB suggested that this could be due to the binding of HSA to the bacterial surface via M and M-like proteins. Growth medium containing 10–20% plasma or HSA saturated with FA results in a strong decrease in the abundance for all quantified FAB proteins. At 10% plasma or 4 mg/ml of HSA (corresponds to the concentration in 10% plasma), there is a large excess of HSA in relationship to the HSA-binding sites of M and M-like proteins. It is possible that the binding of HSA is transient and that HSA molecules from which the FA have dissociated are replaced with HSA carrying FA. Alternatively, the FA are exchanged between HSA bound to the bacterial surface and FA of the surrounding solution, resulting in a constant and high FA concentration at the bacterial surface. This hypothesis is supported by the fact that HSA carrying FA decreases the FAB proteins to an equal extent, as does 10–20% plasma. In contrast, adding HSA devoid of FA to growth medium causes increased levels of the FAB proteins. TH medium contains low amounts of free FA, which will bind to the added FA-depleted HSA. However, because only a small fraction of the total HSA, bound to the bacterial surface or in solution, will contain FA, this pool of FA is mostly inaccessible to the bacteria. When free FA were added directly to the TH medium, the decrease in FAB proteins was less pronounced, indicating that an efficient delivery of FA to the bacteria requires HSA transport. To further investigate the role of HSA binding for the observed decrease in FAB proteins, we used a mutant S. pyogenes strain in which the regulatory mga gene has been inactivated. This results in a markedly reduced production of the M and M-like proteins known to be responsible for HSA binding in S. pyogenes. Compared with the wild-type strain, the mutant possesses more than 100-fold decreased HSA binding. The decreased HSA concentration at the bacterial surface of the mga mutant strain results in significantly lower amounts of internalized FA and significantly higher levels of FAB proteins, suggesting that binding of HSA containing FA to M and M-like proteins of S. pyogenes causes an increased local concentration of FA at the bacterial surface. At an equilibrium between unbound FA and FA bound to HSA, unbound FA are taken up by the bacteria through diffusion or by active transport. In summary, by binding HSA the bacteria increase the local concentration of FA at their surface, and utilize FA of the host instead of running energy-consuming intrinsic synthesis.
The finding that S. pyogenes utilizes FA of human plasma bound to HSA for its metabolism sheds light on a well established but also puzzling property of the bacterium. During in vitro growth when S. pyogenes enters stationary phase and is running out of nutrients, a situation that in vivo probably reflects a late phase of infection, the pathogen switches on genes that induce an inflammatory response in the human host (for review, see Ref. 52). It is far from obvious that this response, resulting in the activation of various human defenses (production of antibacterial peptides, recruitment of neutrophils and monocytes, activation of the complement and contact systems, etc.), is beneficial to the bacteria. However, inflammation will also cause an influx of plasma containing HSA with FA to the site of infection, and for starving bacteria this could make the risk worth taking. The molecular interactions between a parasite and its host are in general highly complex, but there are few human pathogens which like S. pyogenes exhibit such a multitude of interactions with human plasma proteins. The present work gives a comprehensive picture of the S. pyogenes proteome in response to human plasma, and a novel way to analyze the mass spectrometry-generated data made it possible to identify functional protein categories changing their abundance in the presence of plasma. As a result the function of the interaction between HSA, the most abundant human plasma protein, and the M and M-like surface proteins, classical virulence determinants of S. pyogenes, has been clarified. We anticipate that a similar approach can be used to identify and clarify other molecular host-microbe interactions. A deeper understanding of such interactions and their significance for microbial pathogenesis and virulence is required for successful prevention, diagnosis, and treatment of infectious diseases.
Supplementary Material

This article contains supplemental Tables S1–S4.
J. Malmström, C. Karlsson, R. Ossola, H. Weisser, A. Quandt, K. Hansson, R. Aebersold, L. Malmström, and L. Björck, unpublished data.
I. M. Frick and L. Björck, unpublished data.
- SRM
- selected reaction monitoring
- HSA
- human serum albumin
- FAB
- fatty acid biosynthesis
- TH
- Todd-Hewitt broth
- FA
- fatty acid.
REFERENCES
- 1. Cunningham M. W. (2000) Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13, 470–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carapetis J. R., Steer A. C., Mulholland E. K., Weber M. (2005) The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694 [DOI] [PubMed] [Google Scholar]
- 3. Musser J. M., DeLeo F. R. (2005) Toward a genome-wide systems biology analysis of host-pathogen interactions in group A Streptococcus. Am. J. Pathol. 167, 1461–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aziz R. K., Kansal R., Aronow B. J., Taylor W. L., Rowe S. L., Kubal M., Chhatwal G. S., Walker M. J., Kotb M. (2010) Microevolution of group A streptococci in vivo. Capturing regulatory networks engaged in sociomicrobiology, niche adaptation, and hypervirulence. PLoS One 5, e9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ashbaugh C. D., Warren H. B., Carey V. J., Wessels M. R. (1998) Molecular analysis of the role of the group A streptococcal cysteine protease, hyaluronic acid capsule, and M protein in a murine model of human invasive soft-tissue infection. J. Clin. Invest. 102, 550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lange V., Malmström J. A., Didion J., King N. L., Johansson B. P., Schäfer J., Rameseder J., Wong C. H., Deutsch E. W., Brusniak M. Y., Bühlmann P., Björck L., Domon B., Aebersold R. (2008) Targeted quantitative analysis of Streptococcus pyogenes virulence factors by multiple reaction monitoring. Mol. Cell. Proteomics 7, 1489–1500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graham M. R., Virtaneva K., Porcella S. F., Barry W. T., Gowen B. B., Johnson C. R., Wright F. A., Musser J. M. (2005) Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am. J. Pathol. 166, 455–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malmström J., Beck M., Schmidt A., Lange V., Deutsch E. W., Aebersold R. (2009) Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature 460, 762–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kihlberg B. M., Cooney J., Caparon M. G., Olsén A., Björck L. (1995) Biological properties of a Streptococcus pyogenes mutant generated by Tn916 insertion in mga. Microb. Pathog. 19, 299–315 [DOI] [PubMed] [Google Scholar]
- 10. Picotti P., Rinner O., Stallmach R., Dautel F., Farrah T., Domon B., Wenschuh H., Aebersold R. (2010) High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods 7, 43–46 [DOI] [PubMed] [Google Scholar]
- 11. Craig R., Beavis R. C. (2003) A method for reducing the time required to match protein sequences with tandem mass spectra. Rapid Commun. Mass Spectrom. 17, 2310–2316 [DOI] [PubMed] [Google Scholar]
- 12. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 13. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 14. Keller A., Eng J., Zhang N., Li X. J., Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol. Syst. Biol. 1, 2005 0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 16. Sturm M., Bertsch A., Gröpl C., Hildebrandt A., Hussong R., Lange E., Pfeifer N., Schulz-Trieglaff O., Zerck A., Reinert K., Kohlbacher O. (2008) OpenMS, an open-source software framework for mass spectrometry. BMC Bioinformatics 9, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohlbacher O., Reinert K., Gröpl C., Lange E., Pfeifer N., Schulz-Trieglaff O., Sturm M. (2007) TOPP. The OpenMS proteomics pipeline. Bioinformatics 23, e191–197 [DOI] [PubMed] [Google Scholar]
- 18. Malmström L., Marko-Varga G., Westergren-Thorsson G., Laurell T., Malmström J. (2006) 2DDB. A bioinformatics solution for analysis of quantitative proteomics data. BMC Bioinformatics 7, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Venables W. N., Ripley B. D. (2002) Modern Applied Statistics with S (Chambers J., Eddy W., Hārdle W., Sheater S., Tierney L., eds) 4 Ed., Springer, New York [Google Scholar]
- 20. Hartigan J. A., Wong M. A. (1979) A k-means clustering algorithm. Appl. Statist. 28, 100–108 [Google Scholar]
- 21. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 22. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 23. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE. A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 24. Vogel C., Marcotte E. M. (2008) Calculating absolute and relative protein abundance from mass spectrometry-based protein expression data. Nat. Protoc. 3, 1444–1451 [DOI] [PubMed] [Google Scholar]
- 25. Ishihama Y., Oda Y., Tabata T., Sato T., Nagasu T., Rappsilber J., Mann M. (2005) Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics 4, 1265–1272 [DOI] [PubMed] [Google Scholar]
- 26. Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 27. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape. A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McNeil L. K., Reich C., Aziz R. K., Bartels D., Cohoon M., Disz T., Edwards R. A., Gerdes S., Hwang K., Kubal M., Margaryan G. R., Meyer F., Mihalo W., Olsen G. J., Olson R., Osterman A., Paarmann D., Paczian T., Parrello B., Pusch G. D., Rodionov D. A., Shi X., Vassieva O., Vonstein V., Zagnitko O., Xia F., Zinner J., Overbeek R., Stevens R. (2007) The National Microbial Pathogen Database Resource (NMPDR). A genomics platform based on subsystem annotation. Nucleic Acids Res. 35, D347–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tillett W. S., Garner R. L. ( 1934) The agglutination of the haemolytic streptococci by plasma and fibrinogen. A comparison of the phenomenon to serological reactions with the same organism. Bull. Johns Hopkins Hosp. 54, 145–156 [Google Scholar]
- 30. Kronvall G., Simmons A., Myhre E. B., Jonsson S. (1979) Specific absorption of human serum albumin, immunoglobulin A, and immunoglobulin G with selected strains of group A and G streptococci. Infect. Immun. 25, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schmidt K. H., Wadström T. (1990) A secreted receptor related to M1 protein of Streptococcus pyogenes binds to fibrinogen, IgG, and albumin. Zentralbl. Bakteriol. 273, 216–228 [DOI] [PubMed] [Google Scholar]
- 32. Akesson P., Schmidt K. H., Cooney J., Björck L. (1994) M1 protein and protein H. IgGFc- and albumin-binding streptococcal surface proteins encoded by adjacent genes. Biochem. J. 300, 877–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oehmcke S., Shannon O., Mörgelin M., Herwald H. (2010) Streptococcal M proteins and their role as virulence determinants. Clin. Chim. Acta 411, 1172–1180 [DOI] [PubMed] [Google Scholar]
- 34. Gubbe K., Misselwitz R., Welfle K., Reichardt W., Schmidt K. H., Welfle H. (1997) C repeats of the streptococcal M1 protein achieve the human serum albumin binding ability by flanking regions, which stabilize the coiled-coil conformation. Biochemistry 36, 8107–8113 [DOI] [PubMed] [Google Scholar]
- 35. Hong K. (2007) Characterization of group A streptococcal M23 protein and comparison of the M3 and M23 protein's ligand-binding domains. Curr. Microbiol. 55, 427–434 [DOI] [PubMed] [Google Scholar]
- 36. Retnoningrum D. S., Cleary P. P. (1994) M12 protein from Streptococcus pyogenes is a receptor for immunoglobulin G3 and human albumin. Infect. Immun. 62, 2387–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferretti J. J., McShan W. M., Ajdic D., Savic D. J., Savic G., Lyon K., Primeaux C., Sezate S., Suvorov A. N., Kenton S., Lai H. S., Lin S. P., Qian Y., Jia H. G., Najar F. Z., Ren Q., Zhu H., Song L., White J., Yuan X., Clifton S. W., Roe B. A., McLaughlin R. (2001) Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. U.S.A. 98, 4658–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liao J., Sportsman R., Harris J., Stahl A. (2005) Real-time quantification of fatty acid uptake using a novel fluorescence assay. J. Lipid Res. 46, 597–602 [DOI] [PubMed] [Google Scholar]
- 39. Frick I. M., Akesson P., Herwald H., Mörgelin M., Malmsten M., Nägler D. K., Björck L. (2006) The contact system. A novel branch of innate immunity generating antibacterial peptides. EMBO J. 25, 5569–5578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang Y. M., Rock C. O. (2008) Membrane lipid homeostasis in bacteria. Nat. Rev. Microbiol. 6, 222–233 [DOI] [PubMed] [Google Scholar]
- 41. White S. W., Zheng J., Zhang Y. M., Rock C. O. (2005) The structural biology of type II fatty acid biosynthesis. Annu. Rev. Biochem. 74, 791–831 [DOI] [PubMed] [Google Scholar]
- 42. Black P. N., DiRusso C. C. (1994) Molecular and biochemical analyses of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim. Biophys. Acta 1210, 123–145 [DOI] [PubMed] [Google Scholar]
- 43. Lu Y. J., Rock C. O. (2006) Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 59, 551–566 [DOI] [PubMed] [Google Scholar]
- 44. Schujman G. E., de Mendoza D. (2005) Transcriptional control of membrane lipid synthesis in bacteria. Curr. Opin. Microbiol. 8, 149–153 [DOI] [PubMed] [Google Scholar]
- 45. Björck L., Kastern W., Lindahl G., Widebäck K. (1987) Streptococcal protein G, expressed by streptococci or by Escherichia coli, has separate binding sites for human albumin and IgG. Mol. Immunol. 24, 1113–1122 [DOI] [PubMed] [Google Scholar]
- 46. de Château M., Björck L. (1994) Protein PAB. A mosaic albumin-binding bacterial protein representing the first contemporary example of module shuffling. J. Biol. Chem. 269, 12147–12151 [PubMed] [Google Scholar]
- 47. Christner M., Franke G. C., Schommer N. N., Wendt U., Wegert K., Pehle P., Kroll G., Schulze C., Buck F., Mack D., Aepfelbacher M., Rohde H. (2010) The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol. Microbiol. 75, 187–207 [DOI] [PubMed] [Google Scholar]
- 48. de Château M., Björck L. (1996) Identification of interdomain sequences promoting the intronless evolution of a bacterial protein family. Proc. Natl. Acad. Sci. U.S.A. 93, 8490–8495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. de Château M., Holst E., Björck L. (1996) Protein PAB, an albumin-binding bacterial surface protein promoting growth and virulence. J. Biol. Chem. 271, 26609–26615 [DOI] [PubMed] [Google Scholar]
- 50. Egesten A., Frick I. M., Mörgelin M., Olin A. I., Björck L. (2011) Binding of albumin promotes bacterial survival at the epithelial surface. J. Biol. Chem. 286, 2469–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fischetti V. A. (1989) Streptococcal M protein. Molecular design and biological behavior. Clin. Microbiol. Rev. 2, 285–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rasmussen M., Björck L. (2002) Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 43, 537–544 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.