Abstract
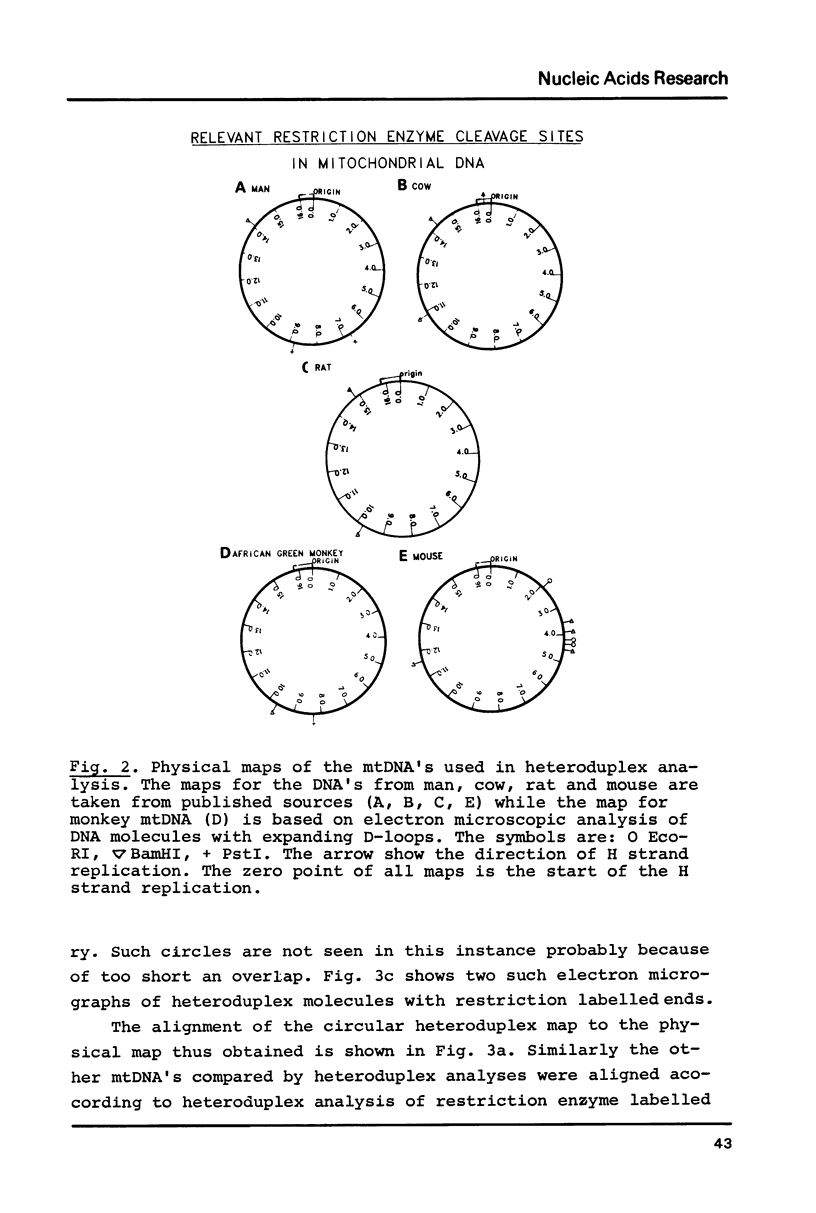
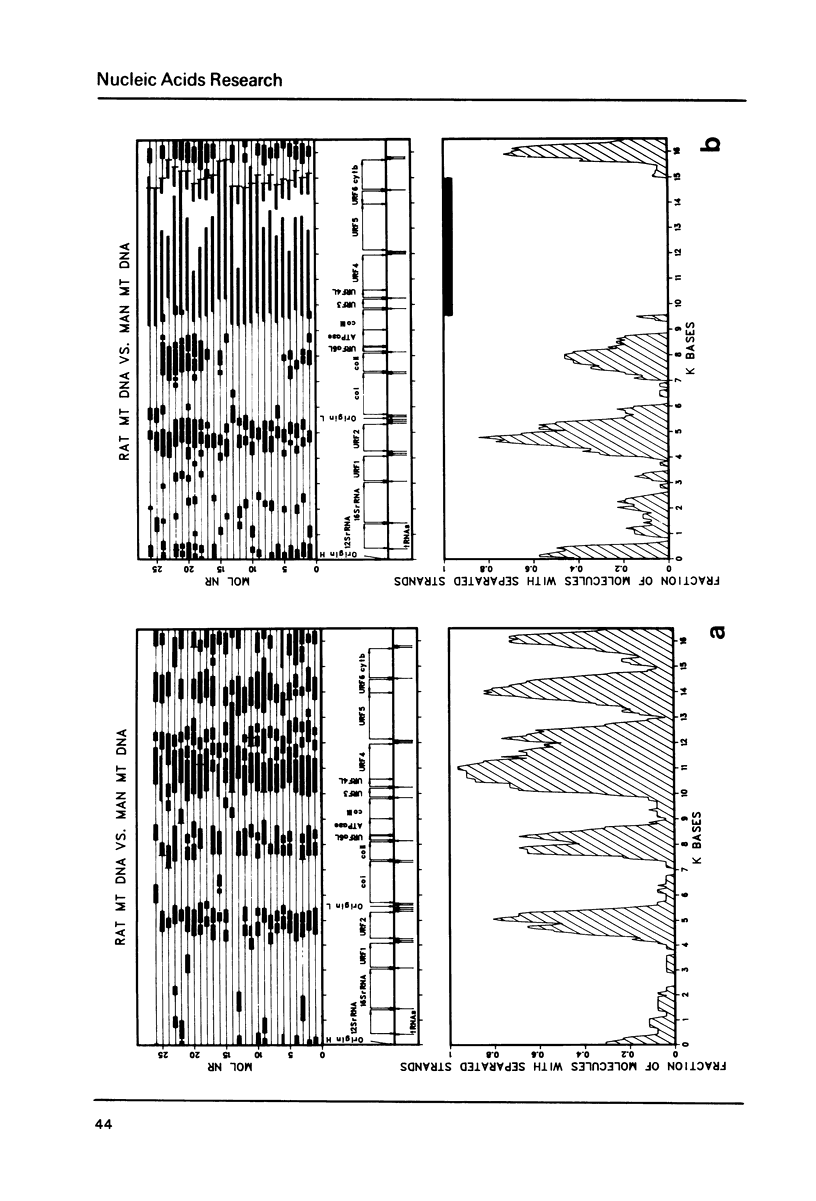
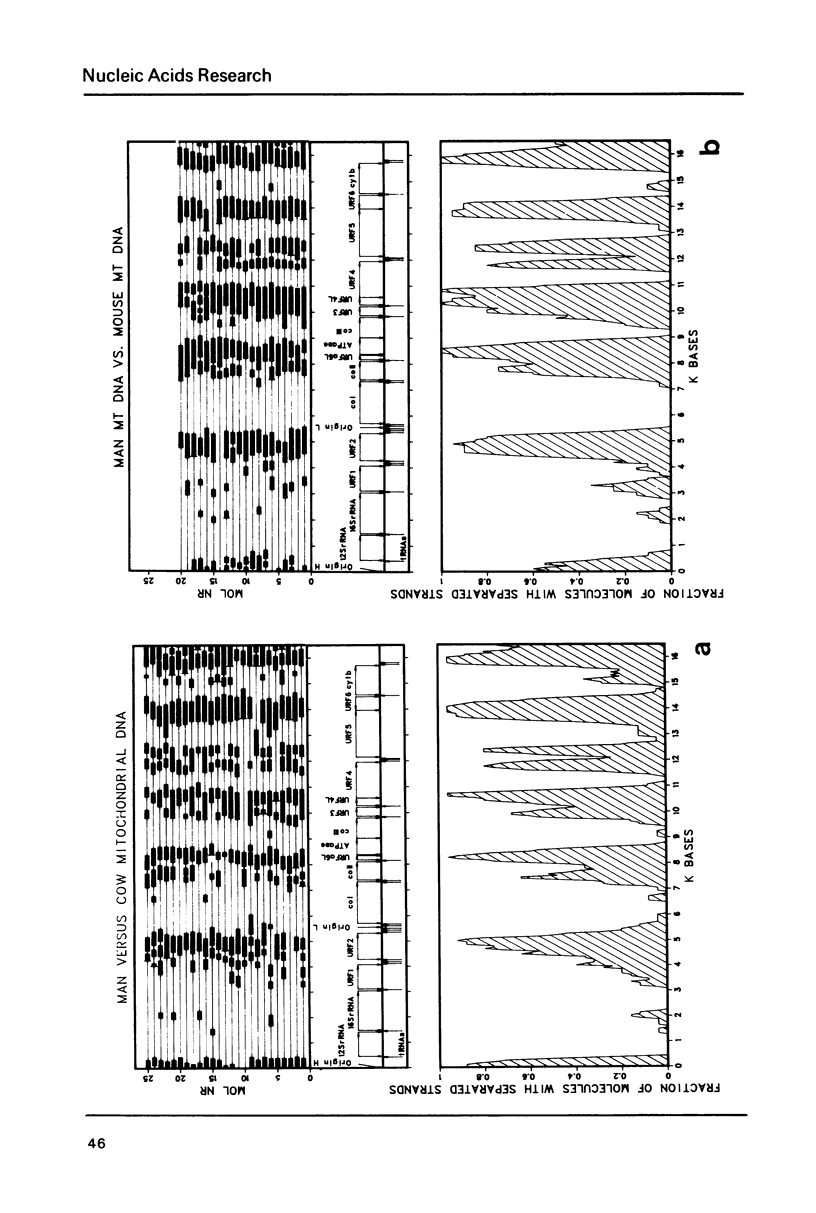
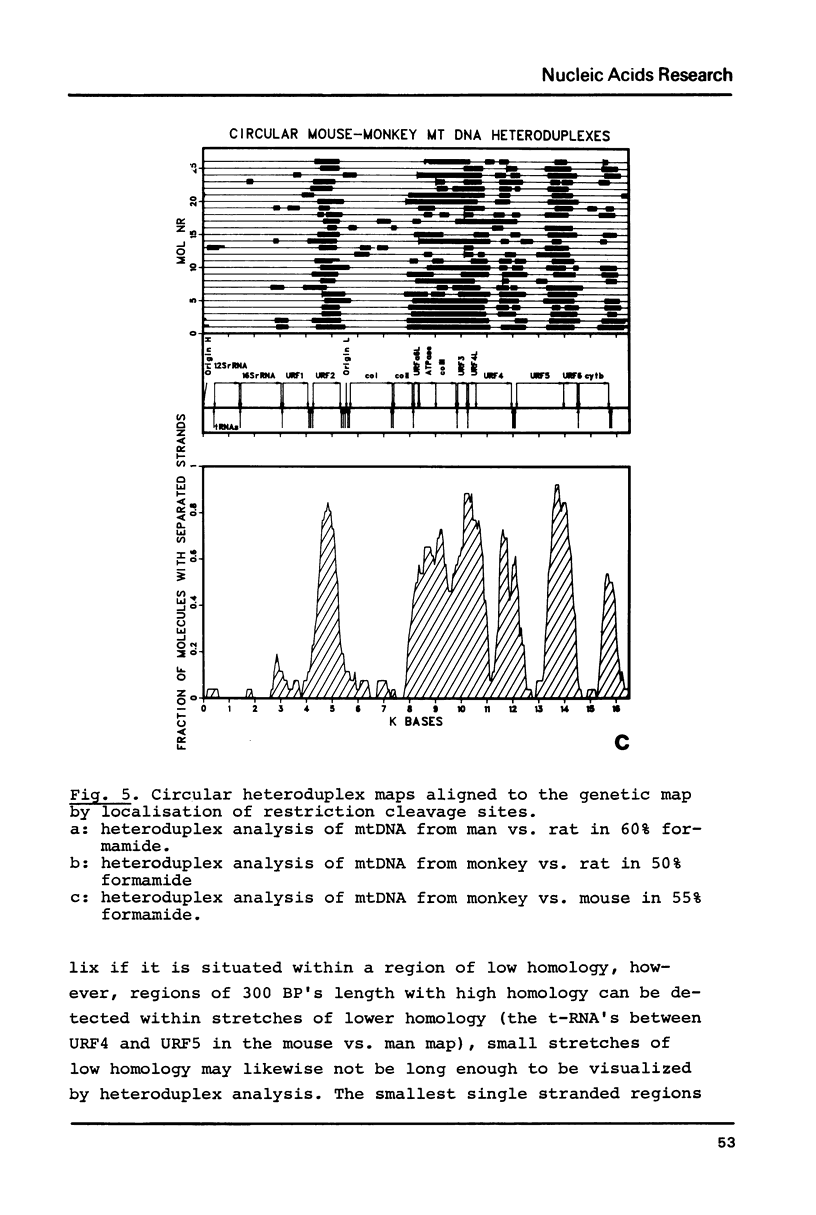
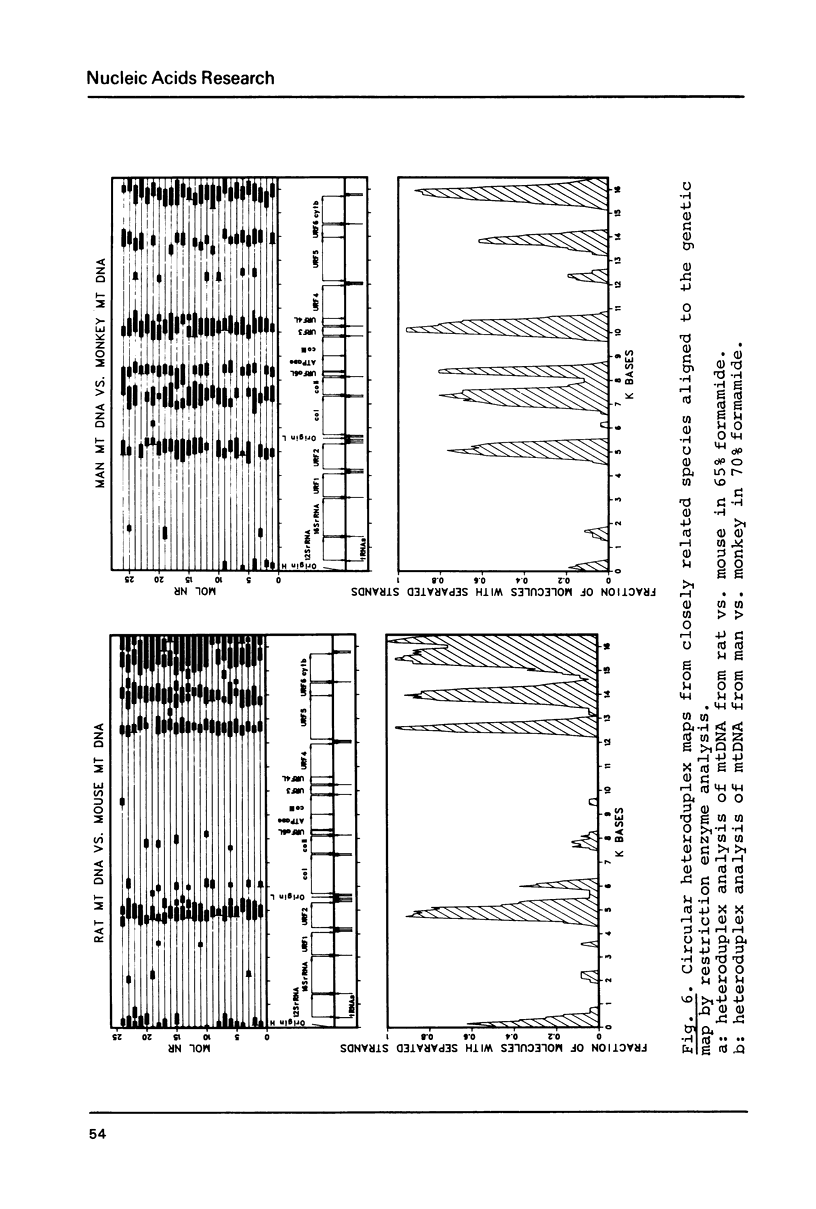
Heteroduplex analysis of mitochondrial DNA (mtDNA) from evolutionary closely related mammals (rat vs. mouse, man vs. monkey) are analyzed and compared to heteroduplex analysis of mt-DNA from more distantly related mammals (rat vs. man, rat vs. monkey, mouse vs. man, mouse vs. monkey and man vs. cow). Each analysis is transformed into a heteroduplex map and all maps are aligned to restriction enzyme maps and to genetic maps and where possible compared with the known sequence. We show that early evolutionary changes are seen mainly in URF2, URFA6L, URF6 and the D-loop region. The regions of rRNA, URF1, COI and COIII are generally very conserved regions but areas with some evolutionary activity can be localized. Heteroduplex analysis between distantly related species show much more heterology than do closely related species and the heteroduplex maps between all the distantly related species show a common pattern of heterology. Comparisons between the DNA sequence of mtDNA from man, cow and mouse and the equivalent heteroduplex maps show that base pair homologies higher than 73% are displayed as homologous regions. In the heteroduplex analysis of mtDNA's from more closely related species very few heterologies are displayed at 50% formamide but an increase in formamide concentration to 65-70% demonstrate also in these instances general heterologous regions.
Full text
PDF














Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Van Etten R. A., Wright C. T., Walberg M. W., Clayton D. A. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981 Oct;26(2 Pt 2):167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Brown W. M., George M., Jr, Wilson A. C. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1967–1971. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen G., Christiansen C. Comparison of the fine structure of mitochondrial DNA from Saccharomyces cerevisiae and S. carlsbergensis: electron microscopy of partially denatured molecules. Nucleic Acids Res. 1976 Feb;3(2):465–476. doi: 10.1093/nar/3.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D. A., Vinograd J. Complex mitochondrial DNA in leukemic and normal human myeloid cells. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1077–1084. doi: 10.1073/pnas.62.4.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Hyman R. W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971 Dec 14;62(2):287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- Forsheit A. B., Davidson N., Brown D. D. An electron microscope heteroduplex study of the ribosomal DNAs of Xenopus laevis and Xenopus mulleri. J Mol Biol. 1974 Dec 5;90(2):301–314. doi: 10.1016/0022-2836(74)90375-1. [DOI] [PubMed] [Google Scholar]
- Laipis P. J., Hauswirth W. W., O'Brien T. W., Michaels G. S. A physical map of bovine mitochondrial DNA from a single animal. Biochim Biophys Acta. 1979 Nov 22;565(1):22–32. doi: 10.1016/0005-2787(79)90080-7. [DOI] [PubMed] [Google Scholar]
- Moore K. H., Johnson P. H., Chandler S. E., Grossman L. I. A restriction endonuclease cleavage map of mouse mitochondrial DNA. Nucleic Acids Res. 1977;4(5):1273–1289. doi: 10.1093/nar/4.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojala D., Attardi G. A detailed physical map of HeLa cell mitochondria DNA and its alignment with the positions of known genetic markers. Plasmid. 1977 Nov;1(1):78–105. doi: 10.1016/0147-619x(77)90010-5. [DOI] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M. Restriction endonuclease cleavage maps of rat and mouse mitochondrial DNAs. Nucleic Acids Res. 1977;4(5):1291–1299. doi: 10.1093/nar/4.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson D. L., Clayton D. A. Replication of mitochondrial DNA in mouse L cells and their thymidine kinase - derivatives: displacement replication on a covalently-closed circular template. Proc Natl Acad Sci U S A. 1972 Dec;69(12):3810–3814. doi: 10.1073/pnas.69.12.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upholt W. B., Dawid I. B. Mapping of mitochondrial DNA of individual sheep and goats: rapid evolution in the D loop region. Cell. 1977 Jul;11(3):571–583. doi: 10.1016/0092-8674(77)90075-7. [DOI] [PubMed] [Google Scholar]
- Upholt W. B. Estimation of DNA sequence divergence from comparison of restriction endonuclease digests. Nucleic Acids Res. 1977;4(5):1257–1265. doi: 10.1093/nar/4.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakour R. A., Bultmann H. Evoluation of Drosophila mitochondrial DNAs. Analysis of heteroduplex molecules. Biochim Biophys Acta. 1979 Sep 27;564(2):342–351. doi: 10.1016/0005-2787(79)90231-4. [DOI] [PubMed] [Google Scholar]





