Background: In yeast, phospholipid synthesis is regulated by zinc deficiency.
Results: PAH1-encoded phosphatidate phosphatase activity was induced in zinc-deficient cells by a transcriptional mechanism.
Conclusion: The zinc-mediated regulation of phosphatidate phosphatase affects phospholipid synthesis by controlling phosphatidate and diacylglycerol.
Significance: The transcriptional regulation of phosphatidate phosphatase plays an important role in controlling phospholipid synthesis by zinc.
Keywords: Phosphatase, Phosphatidate, Phosphatidylcholine, Phospholipid, Yeast, Zinc
Abstract
In the yeast Saccharomyces cerevisiae, the synthesis of phospholipids is coordinately regulated by mechanisms that control the homeostasis of the essential mineral zinc (Carman, G.M., and Han, G. S. (2007) Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim. Biophys. Acta 1771, 322–330; Eide, D. J. (2009) Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284, 18565–18569). The synthesis of phosphatidylcholine is balanced by the repression of CDP-diacylglycerol pathway enzymes and the induction of Kennedy pathway enzymes. PAH1-encoded phosphatidate phosphatase catalyzes the penultimate step in triacylglycerol synthesis, and the diacylglycerol generated in the reaction may also be used for phosphatidylcholine synthesis via the Kennedy pathway. In this work, we showed that the expression of PAH1-encoded phosphatidate phosphatase was induced by zinc deficiency through a mechanism that involved interaction of the Zap1p zinc-responsive transcription factor with putative upstream activating sequence zinc-responsive elements in the PAH1 promoter. The pah1Δ mutation resulted in the derepression of the CHO1-encoded phosphatidylserine synthase (CDP-diacylglycerol pathway enzyme) and loss of the zinc-mediated regulation of the enzyme. Loss of phosphatidate phosphatase also resulted in the derepression of the CKI1-encoded choline kinase (Kennedy pathway enzyme) but decreased the synthesis of phosphatidylcholine when cells were deficient of zinc. This result confirmed the role phosphatidate phosphatase plays in phosphatidylcholine synthesis via the Kennedy pathway.
Introduction
In the yeast Saccharomyces cerevisiae, the essential mineral zinc serves as a catalytic or structural cofactor for many enzymes and transcription factors (1). Accordingly, cells have developed mechanisms to maintain internal stores of zinc (e.g. induced expression of zinc transporters) when extracellular zinc is limiting (1). In addition, cells require mechanisms to adapt physiological processes to limiting amounts of zinc, and one of these processes is the synthesis of membrane phospholipids (1, 2).
The major membrane phospholipids in yeast include PC,2 PE, PI, and PS (3, 4). PC and PE are synthesized (de novo) from the phospholipid precursor PA by complementary CDP-DAG and Kennedy pathways (see Fig. 1) (4). In the CDP-DAG pathway, PC and PE are derived from PA via CDP-DAG, whereas in the Kennedy pathway, these phospholipids are derived from PA via DAG (Fig. 1). PI and PS are only derived from PA via CDP-DAG (Fig. 1). Zinc limitation causes changes in cellular phospholipid composition that include a decrease in PE content and an increase in PI content (5). The decrease in PE is attributed to reductions in both PS synthase and PS decarboxylase activities (which produce PS and PE, respectively, in the CDP-DAG pathway), whereas the increase in PI is attributed to an elevation in PI synthase activity (5, 6). Despite the fact that CDP-DAG pathway activities (including PE/phospholipid methyltransferases that convert PE to PC) are repressed in response to zinc depletion, the PC content is not significantly affected (5). Maintaining PC content in response to zinc depletion is attributed to the induced expression of choline kinase (7) and ethanolamine kinase (8),3 the enzyme activities that catalyze the committed steps in the Kennedy pathway (Fig. 1).
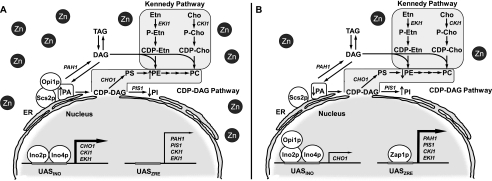
FIGURE 1.
Model for zinc-mediated regulation of phospholipid synthesis in S. cerevisiae. The CDP-DAG and Kennedy pathways shown for the synthesis of phospholipids include the relevant steps discussed in this work. A more detailed diagram of the phospholipid synthesis pathways can be found in Refs. 74 and 4. The phospholipid synthesis genes known to be regulated by zinc availability are indicated in the pathways. A, CHO1, CKI1, and EKI1 (left) and PAH1, PIS1, CKI1, and EKI1 (right) are expressed at some level when cells are grown in zinc-replete medium (depicted by numerous zinc atoms outside the nucleus). Some derepressed expression levels of CHO1, CKI1, and EKI1 (indicated by the bold arrow) are dependent on the interaction of the Ino2p-Ino4p transcriptional activation complex with the UASINO sequence in the promoter of these genes (4, 15). Under this growth condition, the repressor Opi1p is associated with the nuclear/ER membrane through interactions with PA and Scs2p (4, 15). B, when cytosolic zinc is limiting (depicted by a reduced number of zinc atoms outside the nucleus), the expression of the Zap1p transcriptional activator is induced (1), and it binds to the UASZRE sequences in the promoter of PAH1 (this work), PIS1 (6), CKI1 (7), and EKI1 (8) to increase transcription (indicated by the bold arrow). Transcription of CHO1 is attenuated in zinc-deficient cells by the interaction of Opi1p with Ino2p (5) (indicated by the thin arrow). Dissociation of Opi1p from the nuclear/ER membrane and its translocation into the nucleus are caused by a decrease in PA concentration due to an increase in PI synthesis via CDP-DAG (9) and by the induction of PA phosphatase activity (this work). Any repressive effect that Opi1p might have on the expression of CKI1 and EKI1 (not depicted in B, left side) is overcome by their Zap1p-mediated induction under this growth condition. Cho, choline; P-Cho, phosphocholine; Etn, ethanolamine; P-Etn, phosphoethanolamine.
The zinc-mediated regulation of some phospholipid synthesis enzymes has been ascribed to transcriptional mechanisms (1, 2, 4) (Fig. 1). The repression of CHO1 (for PS synthase) is mediated by the PA-regulated transcriptional repressor Opi1p (5, 9), whereas the inductions of PIS1 (for PI synthase), CKI1 (for choline kinase), and EKI1 (for ethanolamine kinase) are mediated by the zinc-sensing and zinc-inducible transcriptional activator Zap1p (6–8, 10, 11). According to the model shown in Fig. 1, Zap1p, which is induced by zinc depletion (10, 11), interacts with UASZRE sequences in the promoters of PIS1, CKI1, and EKI1 to increase the expressions of their enzyme products (6–8). A reduction in the content of PA (which tethers Opi1p and stabilizes its association with Scs2p at the nuclear/ER membrane (9, 12)), which is caused by the increased synthesis of PI via CDP-DAG, leads to the translocation of Opi1p into the nucleus where it interacts with Ino2p to attenuate the transcriptional activation of CHO1 by the Ino2p-Ino4p complex (9, 13–15). This regulation occurs in the absence of inositol supplementation, a growth condition that represses the expression of CHO1 and other UASINO-containing phospholipid synthesis genes (4, 15–17).
The yeast PAH14-encoded PA phosphatase catalyzes the dephosphorylation of PA to produce DAG and Pi (18). The DAG generated by this reaction is used for the synthesis of TAG and may also be used for the synthesis of PE and PC via the Kennedy pathway (19, 20) (Fig. 1). The activity of PA phosphatase also controls PA content and the transcriptional regulation of UASINO-containing phospholipid synthesis genes (21, 22). Given these roles in phospholipid synthesis, we hypothesized that PA phosphatase was regulated in response to zinc status. In this work, we found that the expression of PAH1-encoded PA phosphatase was induced by zinc deficiency through a mechanism that involved interaction of the Zap1p zinc-responsive transcription factor with putative UASZRE sequences in the PAH1 promoter. Moreover, loss of Pah1p PA phosphatase resulted in the derepression of both choline kinase and PS synthase activities and loss of the zinc-mediated regulation of PS synthase. A PA phosphatase activity, whose gene(s) has yet to be identified, was also induced in response to zinc depletion through a mechanism that involved Zap1p.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were reagent grade. Growth medium supplies were from Difco, and yeast nitrogen base lacking zinc sulfate was purchased from BIO 101. Restriction endonucleases, modifying enzymes, and the NEBlot kit were purchased from New England Biolabs. The YeastmakerTM yeast transformation kit was obtained from Clontech. Plasmid DNA purification and DNA gel extraction kits were from Qiagen. Oligonucleotides for PCRs and electrophoretic mobility shift assays were prepared by Genosys Biotechnology, Inc. ProbeQuant G-50 columns were purchased from GE Healthcare. DNA markers for agarose gel electrophoresis and protein assay reagents were purchased from Bio-Rad. Ampicillin, aprotinin, benzamidine, bovine serum albumin, choline, phosphocholine, CDP-choline, leupeptin, O-nitrophenyl β-d-galactopyranoside, pepstatin, phenylmethylsulfonyl fluoride, IGEPAL CA-630, and Triton X-100 were purchased from Sigma. Lipids were purchased from Avanti Polar Lipids. Silica gel 60 thin layer chromatography plates were from EM Science. Radiochemicals and scintillation counting supplies were purchased from PerkinElmer Life Sciences and National Diagnostics, respectively. Liqui-Nox detergent was from Alconox, Inc.
Strains, Plasmids, and Growth Conditions
The strains and plasmids used in this work are listed in Table 1. Plasmid pFP1 contains the PAH1 promoter fused to the coding sequence of the lacZ gene of Escherichia coli. This plasmid was constructed by replacing the DPP1 promoter in pJO2 (23) with the PAH1 promoter sequence at the KpnI/EcoRI site. The PAH1 promoter was obtained by PCR (24) with the primers 5′-GCGGTACCTAGAGTCCAAACTCAACAGCC-3′ (forward) and 5′-GCCGGAATTCATAATCGACCGATGTGTC-3′ (reverse) using strain W303-1A genomic DNA as the template. The PCR primer used in the forward direction corresponds to −1000 bp from the start codon, and the primer used in the reverse direction corresponds to +3 bp from the start codon. The correct orientation of the promoter was confirmed by restriction enzyme digestion. Plasmid pPAH1-ZRE2 was a derivative of pFP1 in which the putative UASZRE sequence ZRE2 in the PAH1 promoter was mutated to a nonconsensus sequence, 5′-AAAAAAAAAAA-3′. Plasmid and genomic DNA preparation, restriction enzyme digestion, and DNA ligations were performed by standard methods (25). Plasmid maintenance and amplification were performed in E. coli strain DH5α. Transformation of plasmids into E. coli (25) or S. cerevisiae (26) was performed as described previously.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Ref. |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−mk+) phoA supE44 l−thi-1 gyrA96 relA1 | 25 |
| S. cerevisiae | ||
| W303-1A | MATa ade2–1 can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–1 | 71 |
| GHY57 | pah1Δ::URA3 derivative of W303-1A | 18 |
| TBY1 | dpp1Δ::TRP1/Kanr lpp1Δ::HIS3/Kanr derivative of W303-1A | 43 |
| DY1457 | MATα ade6 can1–100 his3–11,15 leu2–3,112 trp1–1 ura3–52 | 72 |
| ZHY3 | zrt1Δ::LEU2 zrt2Δ::HIS3 derivative of DY1457 | 73 |
| ZHY6 | MATa ade6 can1–100oc his3 leu2 ura3 zap1Δ::TRP1 | 72 |
| Plasmids | ||
| pJO2 | PDPP1-lacZ reporter gene containing the DPP1 promoter with URA3 | 23 |
| pFP1 | PPAH1-lacZ reporter gene containing the PAH1 promoter with URA3 | This study |
| pPAH1-ZRE2 | Derivative of pFP1 with mutations in ZRE2 | This study |
S. cerevisiae cells were grown in YEPD medium (1% yeast extract, 2% peptone, 2% glucose) or in synthetic complete medium (27) containing 2% glucose at 30 °C. The appropriate amino acids of synthetic complete medium were omitted for selection purposes. Zinc-deficient medium was synthetic complete medium (27) prepared with yeast nitrogen base lacking zinc sulfate. Standard synthetic complete medium contains 1.5 μm zinc sulfate. The internal stores of zinc were depleted from cells by growth in zinc-deficient medium (28). To confirm that the intracellular levels of zinc were depleted, we made use of the PCYC1-ZRE-lacZ reporter gene assay described by MacDiarmid et al. (29). The synthetic complete medium lacked inositol supplementation to preclude the regulatory effects that this phospholipid precursor molecule has on the regulation of phospholipid metabolism (4, 30–32). For growth on plates, the growth media were supplemented with 2% agar. Yeast cell numbers in liquid medium were determined spectrophotometrically at an absorbance of 600 nm. Exponential phase cells were harvested at a density of ∼0.5 × 107 cells/ml. Glassware was washed with Liqui-Nox, rinsed with 0.1 mm EDTA, and then rinsed several times with deionized distilled water to remove zinc contamination. E. coli cells were grown in LB medium (1% tryptone, 0.5% yeast extract, 1% NaCl, pH 7.4) at 37 °C. Ampicillin (100 μg/ml) was added to bacterial cultures that carried plasmids.
Preparation of Cell Extracts, Cytosolic Fraction, and Protein Determination
All steps were performed at 4 °C. Cell extracts were prepared by disruption of yeast cells with glass beads (0.5-mm diameter) using a BioSpec Products Mini-BeadBeater-16 (33). The cell disruption buffer contained 50 mm Tris-HCl, pH 7.5, 0.3 m sucrose, 10 mm 2-mercaptoethanol, 0.5 mm phenylmethanesulfonyl fluoride, 1 mm benzamidine, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 5 μg/ml pepstatin. The cytosolic (supernatant) fraction was prepared by centrifugation at 100,000 × g for 1 h (33). Protein concentration was estimated by the Coomassie Blue dye binding assay of Bradford (34) using bovine serum albumin as the standard.
Enzyme Assays
All assays were conducted in triplicate at 30 °C. β-Galactosidase activity was measured by following the formation of O-nitrophenyl from O-nitrophenyl β-d-galactopyranoside spectrophotometrically at a wavelength of 410 nm (35). The assay mixture contained 100 mm sodium phosphate, pH 7.0, 3 mm O-nitrophenyl β-d-galactopyranoside, 1 mm MgCl2, 100 mm 2-mercaptoethanol, and enzyme protein in a total volume of 0.1 ml. PA phosphatase activity was measured by following the release of water-soluble 32Pi from chloroform-soluble [32P]PA (10,000 cpm/nmol) (33). The reaction mixture contained 50 mm Tris-HCl buffer, pH 7.5, 5 mm MgCl2, 0.2 mm PA, 2 mm Triton X-100, and enzyme protein in a total volume of 0.1 ml. Choline kinase activity was measured by following the incorporation of [methyl-14C]choline (2,000 cpm/nmol) into radiolabeled phosphocholine after precipitation of the labeled choline as a reineckate salt (36). The assay mixture contained 67 mm glycine-NaOH buffer, pH 9.5, 5 mm choline, 5 mm ATP, 10 mm MgSO4, 1.3 mm dithiothreitol, and enzyme protein in a total volume of 0.06 ml (37). PS synthase activity was measured by following the incorporation of 0.5 mm [3-3H]serine (10,000 cpm/nmol) into PS in the presence of 50 mm Tris-HCl, pH 8.0, 0.6 mm MnCl2, 3.2 mm Triton X-100, and 0.2 mm CDP-DAG in a total volume of 0.1 ml (38). All assays were linear with time and protein concentration. The average standard deviation of all assays was ±5%. The units of β-galactosidase and choline kinase activities were defined as the amount of enzymes that catalyzed the formation of 1 μmol of product/min. The units of PA phosphatase and PS synthase activities were defined in nmol of product/min. Specific activity was defined as units/mg of protein.
Electrophoretic Mobility Shift Assays
Double-stranded oligonucleotides (Table 2) were prepared, labeled with [α-32P]dATP (400–800 Ci/mmol) and Klenow fragment (5 units), and then purified by gel filtration using ProbeQuant G-50 spin columns as described previously (7). Purified recombinant GST-Zap1p(687–880) (6) was incubated with 1 pmol of radiolabeled DNA probe (2.0 × 105 cpm/pmol) for 15 min at room temperature in a total volume of 10 μl. The reaction buffer contained 10 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 50 mm KCl, 1 mm dithiothreitol, 0.025 mg/ml poly(dI-dC)·poly(dI-dC), 0.2 mg/ml bovine serum albumin, 0.04% IGEPAL CA-630, and 10% glycerol. Following incubation, the reaction mixture was resolved on a 6% polyacrylamide gel (1.5-mm thickness) in 0.5× Tris borate-EDTA buffer at 100 V for 45 min. Gels were dried onto blotting paper, and the radioactive signals were visualized by phosphorimaging analysis.
TABLE 2.
Oligonucleotides used for electrophoretic mobility shift assays
| Element | Annealed oligonucleotidesa |
|---|---|
| ZRE1 | 5′-GGTCTACCAATCTGAAGGTGTGTTagg-3′ |
| 3′-ccaGATGGTTAGACTTCCACACAATCC-5′ | |
| ZRE2 | 5′-AGCAGCGCACGGTGAGGGTAGAAGgaa-3′ |
| 5′-tcgTCGCGTGCCACTCCCATCTTCCTT-5′ | |
| ZRE3 | 5′-ACAGTTTTACCTTCTAAGAAACATaca-3′ |
| 3′-tgtCAAAATGGAAGATTCTTTGTATGT-5′ |
a Underlined sequences are putative ZRE sites. The lowercase letters indicate the nucleotides filled with the Klenow fragment.
Labeling and Analysis of Lipids and Kennedy Pathway Intermediates
Exponential phase cells were labeled for five to six generations with either 0.2 μCi/ml [methyl-14C]choline (for Kennedy pathway intermediates and PC) or 1 μCi/ml [2-14C]acetate (for neutral lipids) (39). Lipids and the Kennedy pathway intermediates were extracted from cells using chloroform/methanol/water followed by separation of the chloroform and aqueous phases (40). The chloroform and aqueous phases that contained the lipids and Kennedy pathway intermediates, respectively, were dried in vacuo and then dissolved in 100 μl of chloroform and 100 μl of methanol/water (1:1, v/v), respectively (39). The Kennedy pathway intermediates were subjected to TLC on silica gel plates using the solvent system methanol, 0.6% sodium chloride, ammonium hydroxide (10:10:1, v/v). PC was analyzed by TLC on silica gel plates using the solvent system chloroform, pyridine, 88% formic acid, methanol, water (60:35:10:5:2, v/v). Neutral lipids were analyzed by TLC on silica gel plates using the solvent system hexane/diethyl ether/glacial acetic acid (40:10:1, v/v). The identity of the labeled lipids and Kennedy pathway intermediates on TLC plates was confirmed by comparison with standards. Radiolabeled compounds were visualized by phosphorimaging analysis, and the relative quantities of labeled lipids were analyzed using ImageQuant software.
Analyses of Data
The Student's t test was used to assess statistical significance and was performed with SigmaPlot software. The p values <0.05 were taken as a significant difference.
RESULTS
Zinc Deficiency Causes Zap1p-dependent Induction of PA Phosphatase Activity
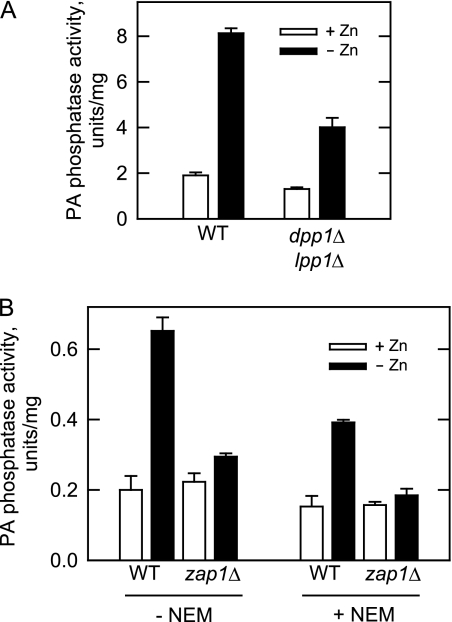
We addressed the hypothesis that PAH1-encoded PA phosphatase was regulated by zinc deficiency. Activity was first measured from wild type cells. The standard assay mixture contained Mg2+ ions that are needed for Pah1p activity (18). The concentration of Mg2+ (5 mm) used in the assay does not have a major effect on the DPP1- and LPP1-encoded PA phosphatase activities that do not have a Mg2+ requirement (41–44). Removal of zinc from the growth medium resulted in a 4.3-fold increase in PA phosphatase activity (Fig. 2A). This increase in activity could be attributed in part to Dpp1p, which is known to be induced by zinc deficiency (28). We next examined the effect of zinc deficiency on PA phosphatase activity in dpp1Δ lpp1Δ mutant cells that lack both Dpp1p and Lpp1p (Fig. 2A). In zinc-deficient medium, the activity in the double mutant was 50% lower than that of wild type cells. Furthermore, PA phosphatase activity in the dpp1Δ lpp1Δ mutant grown without zinc was 3-fold higher when compared with the same cells grown in zinc-replete medium. Thus, the attenuated expression of PA phosphatase activity in dpp1Δ lpp1Δ mutant cells deficient of zinc was consistent with the loss of DPP1. Moreover, the induced expression of PA phosphatase activity in zinc-deficient dpp1Δ lpp1Δ mutant cells indicated that the activity encoded by PAH1 was also regulated by zinc status.
FIGURE 2.
Zinc depletion causes Zap1p-dependent induction of PA phosphatase activity encoded by PAH1 and unknown gene(s). A, wild type (strain W303-1A) and dpp1Δ lpp1Δ mutant (strain TBY1) cells were grown in the absence and presence of 1.5 μm ZnSO4. Cell extracts were prepared and assayed for PA phosphatase activity under standard assay conditions. B, wild type (strain DY1457) and zap1Δ mutant (strain ZHY6) cells were grown in the absence and presence of 1.5 μm ZnSO4. The cytosolic fractions were prepared and assayed for PA phosphatase activity without and with 20 mm N-ethylmaleimide (NEM). Each data point represents the average of triplicate enzyme determinations from two independent experiments ±S.D. (error bars).
The PAH1 promoter contains three putative UASZRE sequences (see below) that are potential binding sites for the Zap1p transcription factor (11). Zap1p, which itself is induced by zinc depletion (10), interacts with UASZRE sequences in the promoters of several phospholipid synthesis genes (e.g. DPP1, CKI1, EKI1, and PIS1) to activate transcription when cells are deficient of zinc (6–8, 28). To address whether the zinc-mediated regulation of the Mg2+-dependent PA phosphatase enzymes was mediated by Zap1p, we measured activity in the zap1Δ mutant. The cytosolic fractions of wild type and zap1Δ mutant cells were utilized for this experiment. In doing so, we eliminated the PA phosphatase activities attributed to DPP1 and LPP1 because their products are associated with the membrane fraction (41, 43). Moreover, the PA phosphatase assay was performed in the presence of N-ethylmaleimide to differentiate between the Mg2+-dependent activities encoded by PAH1 and an unknown gene(s) (18). When the assay was performed in the absence of N-ethylmaleimide, the activity was a reflection of PA phosphatase encoded by PAH1 and the unknown gene(s), whereas the activity measured in the presence of N-ethylmaleimide was a reflection of the PAH1-encoded enzyme (18). Indeed, the PA phosphatase activity in the cytosolic fraction of wild type cells that were grown in zinc-replete medium was reduced by 25% when N-ethylmaleimide was present in the assay (Fig. 2B). The deficiency of zinc in the growth medium resulted in a 3.2- and 2.6-fold increase in PA phosphatase activity when the assays were performed in the absence and presence of N-ethylmaleimide, respectively (Fig. 2B). Thus, both PA phosphatase activities were regulated in response to zinc status. That this regulation was practically eliminated in the zap1Δ mutant (Fig. 2B) supported the conclusion that the PA phosphatase activities encoded by PAH1 and the unknown gene(s) were induced by zinc deficiency in a Zap1p-dependent manner.
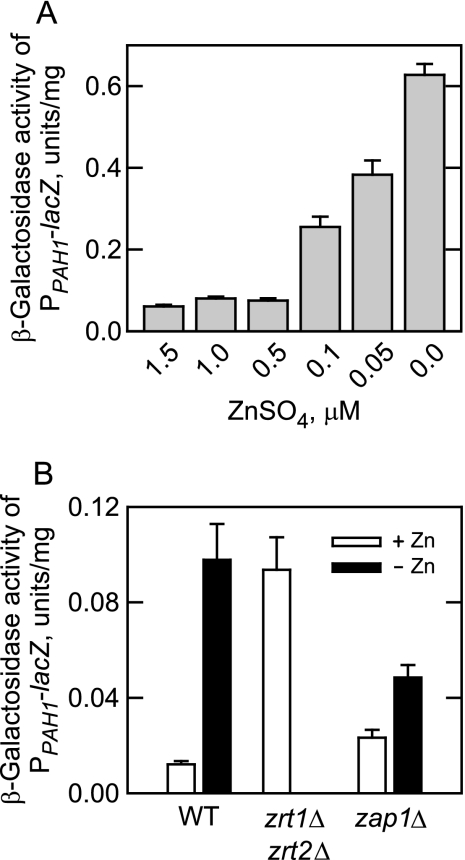
Deficiency of Cytosolic Zinc Causes Zap1p-mediated Induction of PPAH1-lacZ Reporter Gene Activity
The effect of zinc deficiency on the expression of the PAH1 gene was examined by use of a PPAH1-lacZ reporter gene in which the PAH1 promoter was fused in-frame with the coding sequence of the E. coli lacZ gene, and β-galactosidase activity was dependent on the transcription of lacZ driven by the PAH1 promoter. The analysis of β-galactosidase activity from extracts derived from wild type cells grown in the absence and presence of zinc indicated that PAH1 expression was induced in response to zinc deficiency (Fig. 3A). The β-galactosidase activity in zinc-deficient cells was 6-fold greater than the activity found in cells grown with 1.5 μm zinc. The expression of PAH1 was also examined in the zrt1Δ zrt2Δ mutant that lacks both the high (Zrt1p) and low (Zrt2p) affinity plasma membrane zinc transporters (45, 46). This mutant contains a low cytosolic level of zinc despite excess zinc supplementation in the growth medium (45, 46). That the expression of PPAH1-lacZ reporter gene in the zrt1Δ zrt2Δ mutant grown with 1.5 μm zinc was elevated to the same extent as that shown for the reporter gene from zinc-depleted wild type cells substantiated the conclusion that PAH1 expression was induced by a low cytosolic level of zinc (Fig. 3B). In addition, the zap1Δ mutation caused the attenuation of the induced expression of the PPAH1-lacZ reporter gene in response to zinc depletion (Fig. 3B). This result substantiated the conclusion that Zap1p played a role in the zinc-mediated regulation of PAH1.
FIGURE 3.
Depletion of cytosolic zinc causes Zap1p-mediated induction of PPAH1-lacZ reporter gene activity. A, wild type (strain W303-1A) cells bearing the PPAH1-lacZ reporter plasmid pFP1 were grown in the presence of the indicated concentrations of ZnSO4. B, wild type (strain DY1457), zrt1Δ zrt2Δ mutant (strain ZHY3), and zap1Δ mutant (strain ZHY6) cells bearing the PPAH1-lacZ plasmid were grown in the absence and presence of 1.5 μm ZnSO4. Cell extracts were prepared and assayed for β-galactosidase activity. Each data point represents the average of triplicate enzyme determinations from a minimum of two independent experiments ±S.D. (error bars). The differences in the β-galactosidase activities of the wild type controls in A and B were due to strain differences.
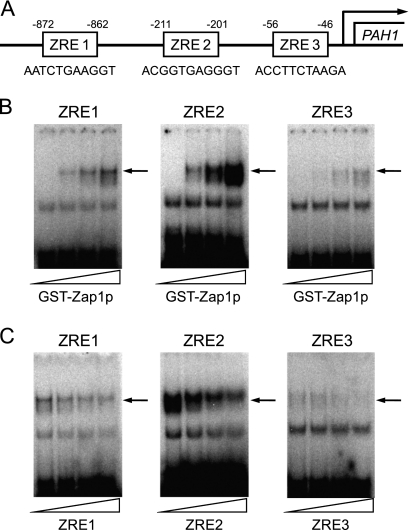
Zap1p Interacts with UASZRE Sequences in PAH1 Promoter
The PAH1 promoter contains three regions (designated ZRE1, ZRE2, and ZRE3) that share 64% sequence homology with the consensus UASZRE sequence (ACCTTNAAGGT) for Zap1p interaction (11) (Fig. 4A). To determine whether the putative ZRE sequences are Zap1p-binding sites, we performed electrophoretic mobility shift assays with oligonucleotide probes containing the putative UASZRE sequences and purified GST-Zap1p(687–880) (47). Of the three probes, ZRE2 showed the strongest interaction with GST-Zap1p(687–880) (Fig. 4, B and C). The interaction with ZRE2 was 3- and 5-fold greater, respectively, when compared with the interactions with ZRE1 and ZRE3. The specificity of GST-Zap1p(687–880) binding to the three sequences was examined further using the same assay. The formation of the complexes was dependent on the concentration of GST-Zap1p(687–880) (Fig. 4B), and the unlabeled probes competed with the labeled probes for GST-Zap1p(687–880) binding in a concentration-dependent manner (Fig. 4C). To further examine the specificity of GST-Zap1p(687–880) for interactions with ZRE1, ZRE2, and ZRE3, the sequences were changed to the nonconsensus UASZRE sequence of AAAAAAAAAAA. These changes abolished the interactions with GST-Zap1p(687–880) (data not shown). These data supported the conclusion that the Zap1p-mediated regulation of PAH1 expression occurred by a mechanism that involves the direct interaction of Zap1p with the PAH1 promoter.
FIGURE 4.
Interactions of GST-Zap1p(687–880) with putative UASZRE sequences in PAH1 promoter. A, the locations and sequences of the putative UASZRE sequences in the PAH1 promoter. B, samples (1 pmol) of radiolabeled double-stranded synthetic oligonucleotides (2.0 × 105 cpm/pmol) with sequences for ZRE1, ZRE2, and ZRE3 in the PAH1 promoter were incubated with 0, 0.15, 0.3, and 0.5 μg of purified recombinant GST-Zap1p(687–880). C, GST-Zap1p(687–880) (0. 5 μg) was incubated with 0, 25, 50, and 100 pmol of unlabeled oligonucleotide with sequences for ZRE1, ZRE2, and ZRE3, respectively. Interactions of GST-Zap1p(687–880) with the labeled oligonucleotides were determined by electrophoretic mobility shift assays using 6% polyacrylamide gels. The data shown are representative of three independent experiments. The arrow indicates the position of the GST-Zap1p(687–880)-ZRE complex.
We confirmed that the induction of PAH1 in response to zinc deficiency was dependent on the ZRE2 sequence in the promoter. The ZRE2 sequence within the PPAH1-lacZ reporter gene was mutated to AAAAAAAAAAA. Cells (wild type strain DY1457) bearing the wild type or mutant PPAH1-lacZ reporter genes were grown in the presence and absence of zinc, and cell extracts were prepared and assayed for β-galactosidase activity. As discussed above, zinc deficiency caused the induced expression of the wild type reporter gene activity. However, the ZRE2 mutation in the reporter gene caused the loss of PAH1 induction in response to zinc deficiency. The β-galactosidase activities in cells grown with and without zinc were 0.018 ± 0.002 and 0.022 ± 0.002 μmol/min/mg, respectively.
pah1Δ Mutation Causes Derepression and Loss of Zinc-mediated Regulation of PS Synthase Activity
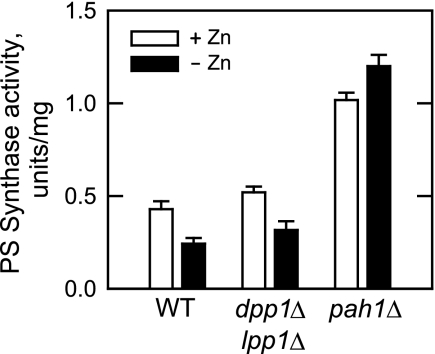
The CHO1-encoded PS synthase catalyzes the committed step in the synthesis of PC via the CDP-DAG pathway (Fig. 1). The expression of the enzyme is governed by the interaction of the Ino2p-Ino4p activation complex with the UASINO element in the CHO1 promoter to drive transcription and by the Opi1p repressor that interacts with Ino2p to attenuate transcription (15) (Fig. 1). PS synthase is repressed in response to zinc deficiency by a mechanism that involves the Opi1p-mediated repression of CHO1 (5) (Fig. 1). Because of the facts that the repressor function of Opi1p is attenuated by its association with PA (9) and that the pah1Δ (18, 48) and dpp1Δ lpp1Δ (43) mutations cause an elevation in PA content, we examined the effect of zinc depletion on the expression of PS synthase activity in these mutants (Fig. 5). As described previously (5), depletion of zinc from the growth medium resulted in a 50% reduction in the expression of PS synthase activity. The dpp1Δ lpp1Δ mutations did not have major effects on the expression of PS synthase activity in cells grown with zinc or on the repression of activity in response to zinc deficiency. The pah1Δ mutation, however, caused the derepression of PS synthase activity (2.3-fold), and the derepressed level of activity was no longer regulated in response to zinc deficiency (Fig. 5).
FIGURE 5.
pah1Δ mutation causes derepression of PS synthase activity and ablates zinc-mediated regulation of enzyme. Wild type (strain W303-1A), dpp1Δ lpp1Δ mutant (strain TBY1), and pah1Δ mutant (strain GHY57) cells were grown in the absence and presence of 1.5 μm ZnSO4. Cell extracts were prepared and assayed for PS synthase activity. Each data point represents the average of triplicate enzyme determinations from two independent experiments ±S.D. (error bars).
pah1Δ Mutation Causes Derepression of Choline Kinase Activity but Decreases Synthesis of PC via Kennedy Pathway When Cells Are Deficient of Zinc
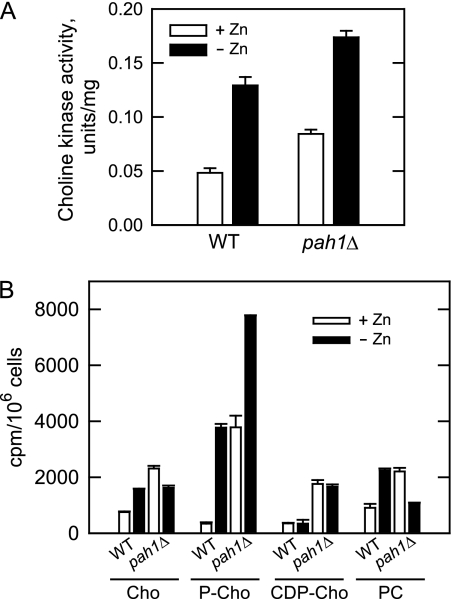
Choline kinase, the enzyme that catalyzes the committed step in PC synthesis via the Kennedy pathway, is induced by zinc depletion through the interaction of Zap1p with UASZRE sequences in the CKI1 promoter (7) (Fig. 1B). Like PS synthase, the expression of choline kinase is governed by the regulatory circuit that includes the Ino2p-Ino4p activation complex, the Opi1p repressor, and the UASINO element in the CKI1 promoter (7, 15) (Fig. 1A). Because Opi1p repressor function is mediated by PA content, we questioned what effect the pah1Δ mutation would have on choline kinase expression and its regulation in response to zinc deficiency. For cells grown in zinc-replete medium, the choline kinase activity in the pah1Δ mutant was 1.75-fold higher than that found in wild type cells (Fig. 6A). As described previously (7), depletion of zinc from wild type cells resulted in the induced expression (2.6-fold) of choline kinase activity (Fig. 6A). For pah1Δ mutant cells, the depletion of zinc still caused the induction (2-fold) in choline kinase activity, and thus, for zinc-deficient cells, the activity in the pah1Δ mutant was elevated (1.4-fold) when compared with the wild type control (Fig. 6A).
FIGURE 6.
pah1Δ mutation causes derepression of choline kinase activity but inhibits synthesis of PC via Kennedy pathway when cells are deficient of zinc. Wild type (strain W303-1A) and pah1Δ mutant (strain GHY57) cells were grown in the absence and presence of 1.5 μm ZnSO4. A, cell extracts were prepared and assayed for choline kinase activity. Each data point represents the average of triplicate enzyme determinations from two independent experiments ±S.D. B, the cultures were incubated with [methyl-14C]choline (0.2 μCi/ml) to uniformly label the Kennedy pathway intermediates and PC. The water-soluble intermediates and PC were extracted and analyzed separately by TLC. The 14C-labeled compounds were visualized by phosphorimaging, and their amounts in cpm were determined from a standard curve using ImageQuant software. The values reported are the average of three separate experiments ±S.D. (error bars). Cho, choline; P-Cho, phosphocholine.
As discussed above, PA phosphatase activity may provide DAG that is used for the synthesis of PC via the Kennedy pathway (19, 20) (Fig. 1). Because of the fact that PAH1 expression was induced by zinc deficiency, we questioned what effect the pah1Δ mutation would have on the zinc-mediated regulation of the pathway. Cells were labeled to steady state with [methyl-14C]choline followed by the extraction and analyses of the water-soluble Kennedy pathway intermediates and PC. As described previously (7), zinc depletion of wild type cells caused an increase in the label that was incorporated into phosphocholine (10-fold) and PC (2.5-fold) but no change in the amount of CDP-choline (Fig. 6B). Consistent with the induced expression of choline kinase activity, pah1Δ mutant cells exhibited elevated levels of phosphocholine when grown in zinc-replete (10-fold) and in zinc-deficient (2-fold) growth media (Fig. 6B). The amounts of CDP-choline, which did not change in response to zinc depletion, were also elevated (5-fold) in the pah1Δ mutant. In contrast to wild type cells, the depletion of zinc did not result in an increase in PC content of pah1Δ mutant cells, and in fact, the mutation caused a 50% decrease in PC content (Fig. 6B). Thus, PAH1-encoded PA phosphatase activity was important for the synthesis of PC via the Kennedy pathway when cells were deficient of zinc.
Neutral Lipid Composition Is Affected in pah1Δ Mutant Cells Deficient of Zinc
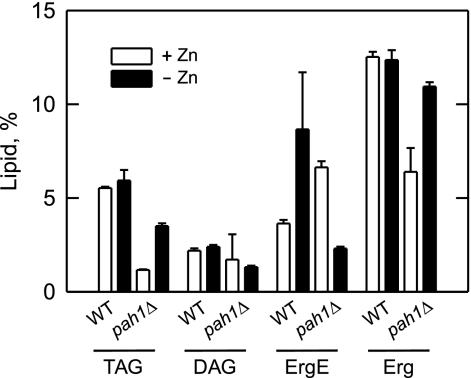
Because the Pah1p PA phosphatase was induced in response to zinc depletion, we questioned what effect the pah1Δ mutation would have on the TAG content of zinc-deficient cells. Wild type and pah1Δ mutant cells were labeled to steady state with [2-14C]acetate followed by the extraction and analysis of lipids by TLC (Fig. 7). For wild type cells, zinc depletion did not affect TAG content, but it did cause an increase (2.4-fold) in the content of ergosterol esters. As described previously for zinc-replete cells (18), the pah1Δ mutation caused a decrease in the levels of TAG (87%), DAG (20%), and ergosterol (49%) but caused an increase in the amount of ergosterol esters (1.8-fold). The increased amount of ergosterol esters in the pah1Δ mutant has been attributed to the decreased ability of the mutant to utilize fatty acids for TAG synthesis (18). Interestingly, the TAG content of the pah1Δ mutant increased (3.2-fold) when the cells were deficient of zinc (Fig. 7). In addition, the zinc-deficient pah1Δ mutant exhibited changes in the relative amounts of ergosterol (1.7-fold increase) and ergosterol esters (65% decrease) that indicated a switch in the synthesis of TAG at the expense of ergosterol esters.
FIGURE 7.
Neutral lipid composition is affected in pah1Δ mutant cells deficient of zinc. Wild type (strain W303-1A) and pah1Δ mutant (strain GHY57) cells were grown in the absence and presence of 1.5 μm ZnSO4. The cultures were incubated with [2-14C]acetate (1 μCi/ml) to uniformly label cellular lipids. Lipids were extracted and separated by one-dimensional TLC. The 14C-labeled lipids were visualized by phosphorimaging and quantified by ImageQuant analysis. The percentages shown for the individual lipids were normalized to the total 14C-labeled chloroform-soluble fraction. The values reported are the average of three separate experiments ±S.D. (error bars). ErgE, ergosterol esters; Erg, ergosterol.
DISCUSSION
Zinc is an essential nutrient in S. cerevisiae and in higher eukaryotic organisms (1, 49). It serves as a cofactor for hundreds of enzymes and is a structural component of many proteins (49–51). Thus, the cellular concentrations of zinc must be tightly regulated (1, 52). Cellular zinc status is primarily controlled by plasma membrane zinc transporters (45, 46, 53) as well as by the zinc transporters found in the membranes of the vacuole (29, 54–56), endoplasmic reticulum (51, 57), and mitochondria (58). The expression of many of these transporters (e.g. plasma membrane-associated Zrt1p) is induced (via Zap1p) when zinc is deficient, and interestingly, this regulation is coordinated with changes in membrane phospholipid composition that are brought about by the transcriptional regulation of several phospholipid synthesis genes (1, 2, 52). Although the expression of some phospholipid synthesis genes (e.g. PIS1, CKI1, and EKI1) is induced (via Zap1p), some genes (e.g. CHO1) are repressed (via Opi1p) (2) (Fig. 1). This genetic regulation, coupled to biochemical mechanisms that occur through the availability of phospholipid synthesis substrates and intermediates, causes the PI content to increase, the PE content to decrease, and the PC content to remain unchanged (2, 4, 59). Stabilization of PC content under zinc-limiting conditions results from a balance between the inhibition of PC synthesis via the CDP-DAG pathway and the activation of PC synthesis via the Kennedy pathway (2, 4). In this work, we addressed the hypothesis that PAH1-encoded PA phosphatase activity plays an important role in the zinc-mediated regulation of phospholipid synthesis. The impetus for this work stems from the fact that the PA phosphatase enzyme controls cellular levels of PA (a precursor to both CDP-DAG and Kennedy pathways and a regulator of Opi1p repressor function) and may provide the DAG used for the synthesis of PC via the Kennedy pathway (19, 20).
Examination of the effects of zinc status on PA phosphatase activity was not straightforward because there are at least four genes in S. cerevisiae that encode the enzyme activity (18–20, 41, 43). A bona fide PA phosphatase enzyme that is involved in the synthesis of lipids is encoded by PAH1 (18). The reaction catalyzed by this enzyme is dependent on Mg2+ ions and is based on a DXDX(T/V) catalytic motif within a haloacid dehalogenase-like domain in the enzyme (18, 22). This enzyme is distinguished in catalytic activity from DPP1- and LPP1-encoded lipid phosphate phosphatase enzymes that dephosphorylate a broad spectrum of substrates (e.g. PA, lysoPA, DAG pyrophosphate, sphingoid base phosphates, and dolichol phosphates) by a distinct catalytic mechanism that does not require divalent cations (19, 41, 43, 60). The DPP1- and LPP1-encoded phosphatases are not involved in de novo lipid synthesis (18) but instead are thought to be involved in lipid signaling (19, 20, 61). Although Pah1p is primarily localized to the cytosol and associates with the nuclear/ER membrane to interact with its substrate PA (18), Dpp1p and Lpp1p are integral membrane enzymes localized to the vacuole and Golgi compartments, respectively (18, 28, 41, 43, 62, 63). There is yet a fourth PA phosphatase activity that shares some properties with those of Pah1p (e.g. it requires Mg2+ for activity, and its membrane association is peripheral in nature) but differs from the enzyme in that its activity is sensitive to inhibition by N-ethylmaleimide (18).
Our studies showed that Pah1p PA phosphatase and the activity whose gene(s) has yet to be identified were induced in response to zinc deficiency. This analysis also confirmed that the vacuole-associated Dpp1p activity was also induced by zinc deficiency (28). Although the zinc-mediated regulation of DPP1 does affect the concentration of PA in the vacuole membrane (64), this regulation does not occur in the location (i.e. nuclear/ER membrane) that would affect the role of Opi1p in the regulation of phospholipid synthesis (15). A similar argument about location holds for the Golgi-associated Lpp1p phosphatase. In addition, there is no evidence that LPP1 expression is regulated by zinc. In fact, the dpp1Δ lpp1Δ mutations did not affect the zinc-mediated regulation of PS synthase that is mediated by Opi1p (5). Because of the fact that neither Dpp1p nor Lpp1p phosphatases play roles in the de novo synthesis of phospholipids and neutral lipids (18), we posit that the Mg2+-dependent PA phosphatases encoded by PAH1 and the unknown gene(s) are the enzymes involved in the regulation of phospholipid synthesis in response to zinc deficiency. At least for Pah1p, we know that the enzyme does associate with the nuclear/ER membrane (65) and that its activity is intimately involved in the synthesis of phospholipids and TAG (18, 22, 48, 66).
The analysis of the PPAH1-lacZ reporter gene indicated that the induction of Pah1p PA phosphatase activity in response to zinc depletion was due to a transcriptional mechanism. Moreover, this regulation was sensitive to cytosolic zinc levels and mediated by the Zap1p zinc-responsive transcription factor. In addition, the interaction of Zap1p with the PAH1 promoter was confirmed by electrophoretic mobility shift assays using DNA probes containing sequences with homology to the consensus UASZRE found in the promoters of highly regulated Zap1p target genes (e.g. ZRT1, ZRT2, ZRT3, and DPP1) (67). Of the three putative UASZRE sequences, the strongest interaction was with ZRE2, and mutations in ZRE2 obviated the induction of PPAH1-lacZ reporter gene activity in response to zinc deficiency. Deviations from the consensus UASZRE sequence are known to reduce interactions of Zap1p (6–8), and this provides an explanation why the induction of PAH1 in response to zinc deficiency was not as great as other Zap1p targets that contain the consensus sequence (28, 29, 45, 46) and why PAH1 was not identified as a Zap1p target in a genome-wide microarray analysis of genes induced by zinc deficiency (67). Nonetheless, that zinc-mediated regulation of Pah1p PA phosphatase occurred by a transcriptional mechanism was a novel finding for this enzyme. Previous work has shown that the biochemical regulation of Pah1p by phosphorylation/dephosphorylation is a major mechanism for controlling enzyme function in vivo (20, 21, 65, 66, 68). Phosphorylation inhibits activity and favors a cytosolic location where the enzyme is not physiologically active, whereas dephosphorylation favors membrane association and its physiological functions (21, 66, 68).
The level of induced PA phosphatase activity was not as great as the induced level of the reporter gene. Based on its primary sequence, Pah1p may be classified as an unstable protein (69), and this instability may provide an explanation for the differences in the expressed amounts of PA phosphatase activity and the reporter gene. For reasons that are not yet clear, we could not detect Pah1p by immunoblotting when PAH1 was induced by zinc deficiency. It is known that excess PA phosphatase activity is detrimental to cell physiology (68), and preliminary studies indicate that Pah1p is subject to proteolytic degradation for control of its cellular activity.
The derepression of the CHO1-encoded PS synthase and the CKI1-encoded choline kinase activities in zinc-replete pah1Δ mutant cells was consistent with that observed for other UASINO-containing genes (e.g. OPI3, INO1, and INO2) (21). The basis for the derepression of these genes is the attenuation of Opi1p repressor function due to elevated PA content caused by the pah1Δ mutation (4, 15, 22, 68). That the pah1Δ mutation eliminated the zinc-mediated regulation of PS synthase supported the notion that PA phosphatase activity controlled Opi1p repressor function. On the other hand, the pah1Δ mutation did not eliminate the zinc-mediated regulation of choline kinase, indicating that the repressive effect that Opi1p would have on expression was overcome by the derepression of CKI1 by Zap1p (7).
In addition to its role in regulating the PA-mediated control of Opi1p repressor function, the choline labeling studies confirmed that Pah1p PA phosphatase plays a role in PC synthesis via the Kennedy pathway. For wild type cells deficient of zinc, the increased levels of phosphocholine and PC were consistent with the inductions of both choline kinase and PA phosphatase activities. However, in pah1Δ mutant cells (that already had an elevated PC content with zinc), the depletion of zinc did not cause an increase in PC, but instead there was a decrease in PC, a result consistent with the loss of PA phosphatase activity. With respect to neutral lipids, the loss of Pah1p caused a dramatic decrease in TAG (87%) content in zinc-replete cells (18). However, the level of TAG in pah1Δ mutant cells increased by over 3-fold when zinc was removed from the growth medium. This result was consistent with the induction of the PA phosphatase activity encoded by the unknown gene(s). However, this interpretation, as well as any conclusions about the role this other PA phosphatase enzyme plays in PC synthesis, cannot be confirmed until its gene(s) is identified and a mutant becomes available for analysis.
A question that has yet to be resolved is why phospholipid composition is regulated in response to zinc deficiency. Although many genes in cellular metabolism are either induced or repressed depending on zinc status, the genes that are most highly regulated are those encoding zinc transporters (1, 67). Thus, changes in membrane phospholipid composition in response to zinc might be important to the structure and/or function of the zinc transporters that are embedded in the membrane bilayer. The availability of mutants defective in the regulation and/or synthesis of membrane phospholipids should facilitate defined studies on the importance of membrane phospholipid composition to zinc transport function in S. cerevisiae.
Acknowledgments
Gil-Soo Han is acknowledged for helpful discussions during the course of this work and comments on the preparation of the manuscript. We also acknowledge David J. Eide for plasmids and mutants used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grants GM-28140 (to G. M. C.) and GM-75378 (to A. S.-C.) from the USPHS.
Although the induction of ethanolamine kinase results in elevated amounts of ethanolamine-containing Kennedy pathway intermediates, the steady-state PE content does not change because PE is methylated to form PC (8).
PAH1 was previously known by the alias SMP2 (70).
- PC
- phosphatidylcholine
- PA
- phosphatidate
- DAG
- diacylglycerol
- TAG
- triacylglycerol
- PE
- phosphatidylethanolamine
- PI
- phosphatidylinositol
- PS
- phosphatidylserine
- DAG
- diacylglycerol
- ZRE
- zinc-responsive element
- UASZRE
- upstream activating sequence zinc-responsive element
- UASINO
- upstream activating sequence inositol-responsive element
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Eide D. J. (2009) Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J. Biol. Chem. 284, 18565–18569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carman G. M., Han G. S. (2007) Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc depletion. Biochim. Biophys. Acta 1771, 322–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rattray J. B., Schibeci A., Kidby D. K. (1975) Lipids of yeasts. Bacteriol. Rev. 39, 197–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carman G. M., Han G. S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iwanyshyn W. M., Han G. S., Carman G. M. (2004) Regulation of phospholipid synthesis in Saccharomyces cerevisiae by zinc. J. Biol. Chem. 279, 21976–21983 [DOI] [PubMed] [Google Scholar]
- 6. Han S. H., Han G. S., Iwanyshyn W. M., Carman G. M. (2005) Regulation of the PIS1-encoded phosphatidylinositol synthase in Saccharomyces cerevisiae by zinc. J. Biol. Chem. 280, 29017–29024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Soto A., Carman G. M. (2008) Regulation of the Saccharomyces cerevisiae CKI1-encoded choline kinase by zinc depletion. J. Biol. Chem. 283, 10079–10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kersting M. C., Carman G. M. (2006) Regulation of the Saccharomyces cerevisiae EKI1-encoded ethanolamine kinase by zinc depletion. J. Biol. Chem. 281, 13110–13116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. (2004) Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science 304, 1644–1647 [DOI] [PubMed] [Google Scholar]
- 10. Zhao H., Eide D. J. (1997) Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 5044–5052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao H., Butler E., Rodgers J., Spizzo T., Duesterhoeft S., Eide D. (1998) Regulation of zinc homeostasis in yeast by binding of the ZAP1 transcriptional activator to zinc-responsive promoter elements. J. Biol. Chem. 273, 28713–28720 [DOI] [PubMed] [Google Scholar]
- 12. Loewen C. J., Roy A., Levine T. P. (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bailis A. M., Lopes J. M., Kohlwein S. D., Henry S. A. (1992) Cis and trans regulatory elements required for regulation of the CHO1 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 20, 1411–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wagner C., Dietz M., Wittmann J., Albrecht A., Schüller H. J. (2001) The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 41, 155–166 [DOI] [PubMed] [Google Scholar]
- 15. Carman G. M., Henry S. A. (2007) Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poole M. A., Homann M. J., Bae-Lee M. S., Carman G. M. (1986) Regulation of phosphatidylserine synthase from Saccharomyces cerevisiae by phospholipid precursors. J. Bacteriol. 168, 668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bailis A. M., Poole M. A., Carman G. M., Henry S. A. (1987) The membrane-associated enzyme phosphatidylserine synthase is regulated at the level of mRNA abundance. Mol. Cell. Biol. 7, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han G. S., Wu W. I., Carman G. M. (2006) The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carman G. M., Han G. S. (2006) Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem. Sci. 31, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carman G. M., Han G. S. (2009) Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J. Biol. Chem. 284, 2593–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24, 1931–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Han G. S., Siniossoglou S., Carman G. M. (2007) The cellular functions of the yeast lipin homolog Pah1p are dependent on its phosphatidate phosphatase activity. J. Biol. Chem. 282, 37026–37035 [DOI] [PubMed] [Google Scholar]
- 23. Oshiro J., Rangaswamy S., Chen X., Han G. S., Quinn J. E., Carman G. M. (2000) Regulation of the DPP1-encoded diacylglycerol pyrophosphate (DGPP) phosphatase by inositol and growth phase. Inhibition of DGPP phosphatase activity by CDP-diacylglycerol and activation of phosphatidylserine synthase activity by DGPP. J. Biol. Chem. 275, 40887–40896 [DOI] [PubMed] [Google Scholar]
- 24. Innis M. A., Gelfand D. H. (1990) in PCR Protocols. A Guide to Methods and Applications (Innis M. A., Gelfand D. H., Sninsky J. J., White T. J., eds) pp. 3–12, Academic Press, Inc., San Diego [Google Scholar]
- 25. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 26. Ito H., Fukuda Y., Murata K., Kimura A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rose M. D., Winston F., Heiter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Han G. S., Johnston C. N., Chen X., Athenstaedt K., Daum G., Carman G. M. (2001) Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J. Biol. Chem. 276, 10126–10133 [DOI] [PubMed] [Google Scholar]
- 29. MacDiarmid C. W., Gaither L. A., Eide D. (2000) Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19, 2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Greenberg M. L., Lopes J. M. (1996) Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol. Rev. 60, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henry S. A., Patton-Vogt J. L. (1998) Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog. Nucleic Acid Res. Mol. Biol. 61, 133–179 [DOI] [PubMed] [Google Scholar]
- 32. Carman G. M., Henry S. A. (1999) Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 33. Carman G. M., Lin Y. P. (1991) Phosphatidate phosphatase from yeast. Methods Enzymol. 197, 548–553 [DOI] [PubMed] [Google Scholar]
- 34. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 35. Craven G. R., Steers E., Jr., Anfinsen C. B. (1965) Purification, composition, and molecular weight of the β-galactosidase of Escherichia coli K12. J. Biol. Chem. 240, 2468–2477 [PubMed] [Google Scholar]
- 36. Porter T. J., Kent C. (1992) Choline/ethanolamine kinase from rat liver. Methods Enzymol. 209, 134–146 [DOI] [PubMed] [Google Scholar]
- 37. Kim K. H., Voelker D. R., Flocco M. T., Carman G. M. (1998) Expression, purification, and characterization of choline kinase, product of the CKI gene from Saccharomyces cerevisiae. J. Biol. Chem. 273, 6844–6852 [DOI] [PubMed] [Google Scholar]
- 38. Carman G. M., Bae-Lee M. (1992) Phosphatidylserine synthase from yeast. Methods Enzymol. 209, 298–305 [DOI] [PubMed] [Google Scholar]
- 39. Fakas S., Konstantinou C., Carman G. M. (2011) DGK1-encoded diacylglycerol kinase activity is required for phospholipid synthesis during growth resumption from stationary phase in Saccharomyces cerevisiae. J. Biol. Chem. 286, 1464–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 41. Toke D. A., Bennett W. L., Dillon D. A., Wu W. I., Chen X., Ostrander D. B., Oshiro J., Cremesti A., Voelker D. R., Fischl A. S., Carman G. M. (1998) Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding diacylglycerol pyrophosphate phosphatase. J. Biol. Chem. 273, 3278–3284 [DOI] [PubMed] [Google Scholar]
- 42. Wu W. I., Liu Y., Riedel B., Wissing J. B., Fischl A. S., Carman G. M. (1996) Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 271, 1868–1876 [DOI] [PubMed] [Google Scholar]
- 43. Toke D. A., Bennett W. L., Oshiro J., Wu W. I., Voelker D. R., Carman G. M. (1998) Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem. 273, 14331–14338 [DOI] [PubMed] [Google Scholar]
- 44. Furneisen J. M., Carman G. M. (2000) Enzymological properties of the LPP1-encoded lipid phosphatase from Saccharomyces cerevisiae. Biochim. Biophys. Acta 1484, 71–82 [DOI] [PubMed] [Google Scholar]
- 45. Zhao H., Eide D. (1996) The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. U.S.A. 93, 2454–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhao H., Eide D. (1996) The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271, 23203–23210 [DOI] [PubMed] [Google Scholar]
- 47. Bird A., Evans-Galea M. V., Blankman E., Zhao H., Luo H., Winge D. R., Eide D. J. (2000) Mapping the DNA binding domain of the Zap1 zinc-responsive transcriptional activator. J. Biol. Chem. 275, 16160–16166 [DOI] [PubMed] [Google Scholar]
- 48. Fakas S., Qiu Y., Dixon J. L., Han G. S., Ruggles K. V., Garbarino J., Sturley S. L., Carman G. M. (2011) Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 286, 29074–29085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vallee B. L., Falchuk K. H. (1993) The biochemical basis of zinc physiology. Physiol. Rev. 73, 79–118 [DOI] [PubMed] [Google Scholar]
- 50. Schwabe J. W., Klug A. (1994) Zinc mining for protein domains. Nat. Struct. Biol. 1, 345–349 [DOI] [PubMed] [Google Scholar]
- 51. Ellis C. D., Wang F., MacDiarmid C. W., Clark S., Lyons T., Eide D. J. (2004) Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell Biol. 166, 325–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eide D. J. (2003) Multiple regulatory mechanisms maintain zinc homeostasis in Saccharomyces cerevisiae. J. Nutr. 133, 1532S–1535S [DOI] [PubMed] [Google Scholar]
- 53. Waters B. M., Eide D. J. (2002) Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277, 33749–33757 [DOI] [PubMed] [Google Scholar]
- 54. MacDiarmid C. W., Milanick M. A., Eide D. J. (2003) Induction of the ZRC1 metal tolerance gene in zinc-limited yeast confers resistance to zinc shock. J. Biol. Chem. 278, 15065–15072 [DOI] [PubMed] [Google Scholar]
- 55. Miyabe S., Izawa S., Inoue Y. (2001) The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 282, 79–83 [DOI] [PubMed] [Google Scholar]
- 56. Devirgiliis C., Murgia C., Danscher G., Perozzi G. (2004) Exchangeable zinc ions transiently accumulate in a vesicular compartment in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 323, 58–64 [DOI] [PubMed] [Google Scholar]
- 57. Ellis C. D., Macdiarmid C. W., Eide D. J. (2005) Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J. Biol. Chem. 280, 28811–28818 [DOI] [PubMed] [Google Scholar]
- 58. Mühlenhoff U., Stadler J. A., Richhardt N., Seubert A., Eickhorst T., Schweyen R. J., Lill R., Wiesenberger G. (2003) A specific role of the yeast mitochondrial carriers Mrs3/4p in mitochondrial iron acquisition under iron-limiting conditions. J. Biol. Chem. 278, 40612–40620 [DOI] [PubMed] [Google Scholar]
- 59. Carman G. M., Han G. S. (2009) Regulation of phospholipid synthesis in yeast. J. Lipid Res. 50, S69-S73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carman G. M. (1997) Phosphatidate phosphatases and diacylglycerol pyrophosphate phosphatases in Saccharomyces cerevisiae and Escherichia coli. Biochim. Biophys. Acta 1348, 45–55 [DOI] [PubMed] [Google Scholar]
- 61. Oshiro J., Han G. S., Carman G. M. (2003) Diacylglycerol pyrophosphate phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1635, 1–9 [DOI] [PubMed] [Google Scholar]
- 62. Kumar A., Agarwal S., Heyman J. A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Cheung K. H., Miller P., Gerstein M., Roeder G. S., Snyder M. (2002) Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Habeler G., Natter K., Thallinger G. G., Crawford M. E., Kohlwein S. D., Trajanoski Z. (2002) YPL.db: the Yeast Protein Localization database. Nucleic Acids Res. 30, 80–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Han G. S., Johnston C. N., Carman G. M. (2004) Vacuole membrane topography of the DPP1-encoded diacylglycerol pyrophosphate phosphatase catalytic site from Saccharomyces cerevisiae. J. Biol. Chem. 279, 5338–5345 [DOI] [PubMed] [Google Scholar]
- 65. Karanasios E., Han G. S., Xu Z., Carman G. M., Siniossoglou S. (2010) A phosphorylation-regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc. Natl. Acad. Sci. U.S.A. 107, 17539–17544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Choi H. S., Su W. M., Morgan J. M., Han G. S., Xu Z., Karanasios E., Siniossoglou S., Carman G. M. (2011) Phosphorylation of phosphatidate phosphatase regulates its membrane association and physiological functions in Saccharomyces cerevisiae: identification of Ser(602), Thr(723), and Ser(744) as the sites phosphorylated by CDC28 (CDK1)-encoded cyclin-dependent kinase. J. Biol. Chem. 286, 1486–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lyons T. J., Gasch A. P., Gaither L. A., Botstein D., Brown P. O., Eide D. J. (2000) Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. U.S.A. 97, 7957–7962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. O'Hara L., Han G. S., Peak-Chew S., Grimsey N., Carman G. M., Siniossoglou S. (2006) Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281, 34537–34548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Guruprasad K., Reddy B. V., Pandit M. W. (1990) Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng. 4, 155–161 [DOI] [PubMed] [Google Scholar]
- 70. Irie K., Takase M., Araki H., Oshima Y. (1993) A gene, SMP2, involved in plasmid maintenance and respiration in Saccharomyces cerevisiae encodes a highly charged protein. Mol. Gen. Genet. 236, 283–288 [DOI] [PubMed] [Google Scholar]
- 71. Thomas B. J., Rothstein R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 72. Lamping E., Lückl J., Paltauf F., Henry S. A., Kohlwein S. D. (1994) Isolation and characterization of a mutant of Saccharomyces cerevisiae with pleiotropic deficiencies in transcriptional activation and repression. Genetics 137, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nikawa J., Hosaka K., Yamashita S. (1993) Differential regulation of two myo-inositol transporter genes of Saccharomyces cerevisiae. Mol. Microbiol. 10, 955–961 [DOI] [PubMed] [Google Scholar]
- 74. Gaspar M. L., Aregullin M. A., Jesch S. A., Nunez L. R., Villa-García M., Henry S. A. (2007) The emergence of yeast lipidomics. Biochim. Biophys. Acta 1771, 241–254 [DOI] [PubMed] [Google Scholar]