Background: HAMP domains accept signals from membrane receptors and propagate them possibly via rotation.
Results: Using targeted mutations based on bioinformatics, sequence comparisons, and structures, we characterize the role of particular amino acids in signaling.
Conclusion: Results are compatible with a gearbox model of HAMP rotation.
Significance: Various models of HAMP domain-mediated signaling are testable.
Keywords: Adenylate Cyclase (Adenylyl Cyclase), Chemotaxis, Escherichia coli, Mycobacterium tuberculosis, Receptors, HAMP Domain, Two Component System
Abstract
HAMP domains, ∼55 amino acid motifs first identified in histidine kinases, adenylyl cyclases, methyl-accepting chemotaxis proteins, and phosphatases, operate as signal mediators in two-component signal transduction proteins. A bioinformatics study identified a coevolving signal-accepting network of 10 amino acids in membrane-delimited HAMP proteins. To probe the functionality of this network we used a HAMP containing mycobacterial adenylyl cyclase, Rv3645, as a reporter enzyme in which the membrane anchor was substituted by the Escherichia coli chemotaxis receptor for serine (Tsr receptor) and the HAMP domain alternately with that from the protein Af1503 of the archaeon Archaeoglobus fulgidus or the Tsr receptor. In a construct with the Tsr-HAMP, cyclase activity was inhibited by serine, whereas in a construct with the HAMP domain from A. fulgidus, enzyme activity was not responsive to serine. Amino acids of the signal-accepting network were mutually swapped between both HAMP domains, and serine signaling was examined. The data biochemically tentatively established the functionality of the signal-accepting network. Based on a two-state gearbox model of rotation in HAMP domain-mediated signal propagation, we characterized the interaction between permanent and transient core residues in a coiled coil HAMP structure. The data are compatible with HAMP rotation in signal propagation but do not exclude alternative models for HAMP signaling. Finally, we present data indicating that the connector, which links the α-helices of HAMP domains, plays an important structural role in HAMP function.
Introduction
The ability to sense extracellular stimuli and trigger appropriate responses is important for all living cells. Therefore, a molecular understanding of the mechanisms of transmembrane signaling is essential. A feature of many signaling proteins is their modular organization (1). Bacterial sensory histidine kinases and chemoreceptors typically show an extracytoplasmic sensor domain, a transmembrane segment, and a cytoplasmic output or transmitter domain. Such ensembles are also found in bacterial membrane-anchored adenylyl cyclases, Ser/Thr protein kinases, and phosphatases (2). These sensor proteins often possess distinct regions between extra- and intracellular domains that presumably undergo conformational changes during signal propagation. One of these regions is the HAMP domain, first found in histidine kinases, adenylyl cyclases (ACs),2 methyl-accepting chemotaxis proteins, and phosphatases (3) but also present in many other proteins (4). A co-evolution between individual families of HAMP domains and output domains is evident (4) and several examples in which HAMP domains were swapped without loss of function suggest a common mechanism for signal propagation (5–8).
HAMP domains are dimeric, parallel four-helix bundles with coiled coil properties to which each monomer contributes two amphipathic α-helices (AS1 and AS2) joined in a right-handed way by a connector of at least 12 amino acids (9–12). The first structure of a HAMP domain was determined from the Af1503 protein of the archaeon Archaeoglobus fulgidus (Ref. 6) (see Fig. 1A) and has since been confirmed in HAMP domains from other proteins (13–15). The Af1503 HAMP domain adopts an unusual complementary x-da conformation that is related to the canonical coiled coil form by a 26° axial rotation of all four helices (Fig. 2). A mechanistic model for signal transduction was proposed; the so-called “gearbox” model, in which HAMP alternates between these two states (a detailed description is in Ref. 6). The model predicted testable positional movements of individual amino acids in the coiled-coil structure of the HAMP domain but did not predict receptor features such as type of ligand, serine responsiveness, or the degree of enzyme inhibition. Because the archaeal Af1503 HAMP domain lacks an intrinsic output domain, we employed the catalytic domain of the mycobacterial AC Rv3645 (cyclase homology domain (CHD)) as a reporter to biochemically test rotation as a potential mechanism of signaling through HAMP domains. Consistent with a transition between different packing modes, when the volume of a conserved small side chain at a core position (Ala-291 in Af1503 HAMP) was increased by point mutations, the maximal catalytic velocity of the HAMP-CHD chimera decreased (6). Structurally, we recently demonstrated that such substitutions induce a gradual transition in packing of the HAMP domain involving both changes in helix rotation toward a knobs-into-holes packing and changes in bundle shape, consistent with the proposed two-state model (16).
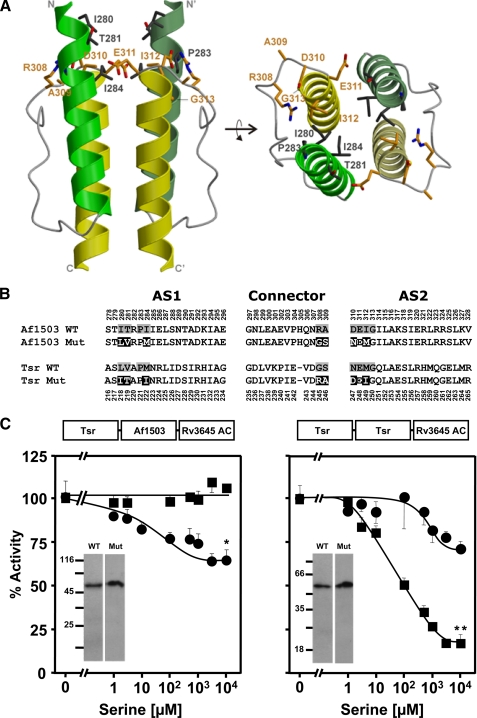
FIGURE 1.
The signal-accepting network. A, shown is the location of the signal-accepting network of Af1503 HAMP in side (left) and top (right) view. The residues forming the network are shown in stick representation and Corey-Pauling-Koltun coloring. In the homodimer only a single set of residues is identified by numbering (HAMP structure from A. fulgidus from Ref. 6). B, shown is the amino acid sequence of Tsr and Af1503 HAMP. The residues comprising the signal-accepting network are shaded gray, and mutations in Af1503 Mut and Tsr Mut are inverted. C, shown is in vitro AC-activity (basal activity was set as 100%) of the mutants compared with the unmutated chimeras. Above each plot is the scheme of the chimera. The left panel shows serine response of the Af1503 HAMP wild-type chimera and the mutated chimera with the network of Tsr (circles). 100% activity was 19 ± 1.6 and 14.6 ± 1.5 nmol of cAMP·mg−1min−1, respectively. Maximal serine inhibition of the mutant was 34 ± 9%. The right panel shows Tsr HAMP wild-type chimera and its mutant (circles). 100% activity was 18 ± 0.6 for the wild-type and 0.7 ± 0.02 nmol of cAMP·mg−1min−1 for the mutant. Inset, Western blots of 0.5-μg membrane preparation of each construct show no degraded proteins. **, p < 0.05 compared with respective concentration point of unmutated construct. **, p < 0.001. p value was calculated using a two-tailed Student's t tests (see “Experimental Procedures”).
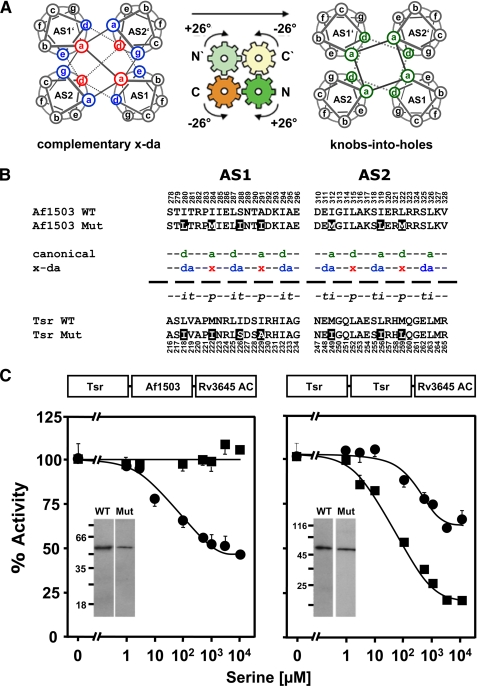
FIGURE 2.
Exchange of x-da exchange residues between Af1503 and Tsr HAMP domains. A, shown is a schematic view of the gearbox model. Helical wheel diagrams show the interconversion from complementary x-da packing (left) to knobs-into-holes packing (right) by a concerted 26° axial rotation of all four helices (indicated by arrows). Complementary x-da packing uses three positions to form the hydrophobic core, one in x-geometry (red) and two in da-geometry (blue; left). Canonical knobs-into-holes packing uses two core residues, both with the same geometry (green; right). B, shown are amino acid sequences of AS1 and AS2 from Tsr and Af1503 HAMP and its mutants. The x-da and canonical (a–g) nomenclature of the differing packing modes is indicated in the middle, with core positions colored and labeled according to their geometries in each packing mode (see above). In addition, the transient, permanent, and intermediate residues are labeled t, p, and i. Residues that were exchanged are inverted. C, serine response of the x-da mutants (circles) compared with the respective unmutated chimeras is shown. Constructs with HAMP from Af1530 are shown on the left and Tsr on the right. 100% activity was 18 ± 0.6 and 19 ± 1.6 nmol of cAMP·mg−1min−1 for Tsr and Af1503 HAMP wild-type chimeras, respectively, and 30 ± 2.3 and 21 ± 1.5 nmol of cAMP·mg−1min−1 for the corresponding mutated chimeras. Western blots of 0.5-μg membrane preparation of each construct are shown as insets. No proteolytic fragments were observed.
Formerly, functional chimeras were generated in which the sensor parts of the serine (Tsr) or the aspartate chemotaxis receptors (Tar) were connected via different HAMP domains (from Tsr, Tar, and Rv3645) to the CHD of Rv3645 (17). These chimeras were regulated in vitro as well as in vivo by their respective ligands l-serine and l-aspartate (17). The biochemical properties of these chemotaxis receptors such as ligand specificity and sensitivity, responses to previously characterized targeted receptor mutations, and requirement for dimerization were identical to those determined in chemotaxis experiments (17). In addition, functionality of the chimeric constructs was demonstrated in vivo (17).
Although it was an attractive idea to join the Tsr receptor to the Rv3645 CHD via the structurally defined Af1503 HAMP, such a tripartite chimera was unresponsive to serine (16). Serine regulation was achieved by substitution of Ala-291 with Phe, supporting the notion of rotation as a potentially common mechanism in HAMP-mediated signal propagation (16). Here we used this Tsr-AC reporter system with the Tsr or Af1503 HAMP domains and carried out a targeted mutational analysis based on sequence and structural features to investigate the role of particular amino acids in signal propagation.
EXPERIMENTAL PROCEDURES
Materials
The mycobacterial AC Rv3645 (331–549) Af1503 HAMP (278–331), Tsr (1–215), Tar (1–213), and Tsr-HAMP (216–268) were available in the laboratory. Radiochemicals were from Hartmann Analytic and GE Healthcare. Enzymes were purchased from either Roche Diagnostics or New England Biolabs. Other chemicals were from Fluka, Merck, Roche Diagnostics, Roth, and Sigma.
Plasmid Construction
Standard molecular biology methods were used. The chimeras Tsr1–215-Tsr HAMP216–268-Rv3645 CHD331–549 (17) and Tsr1–215-Af1503 HAMP278–331- Rv3645 CHD331–549 (16) were used as templates. Point mutations were introduced by site-directed mutagenesis using restriction sites located nearby or by fusion PCR using corresponding sense and antisense primers (from MWG Biotech and Biomers). Primer sequences are available on request. For Tar1–213- Af1503 HAMP278–331-Rv3645 CHD331–549 constructs the coding sequence of Tar (amino acids 1–213) replaced the Tsr coding sequence in the respective Tsr- Af1503 HAMP-Rv3645 CHD chimera. For the constructs Tsr HAMP216–268-Rv3645 CHD331–549, the coding sequence of Tsr sensor (amino acids 1–215) was removed from Tsr-Tsr HAMP-Rv3645 CHD. All constructs were cloned into BamHI and HindIII restriction sites of either pQE30 or pQE80L plasmids (Qiagen). This added an N-terminal MRGS-His6 affinity tag. The fidelity of all constructs was confirmed by sequencing.
Expression and Preparation of Recombinant Proteins
Plasmids of sensor-HAMP-CHD constructs (Tsr-Tsr HAMP-Rv3645 CHD, Tsr-Af1503 HAMP-Rv3645 CHD, and Tar-Af1503 HAMP-Rv3645 CHD) were transformed into Escherichia coli BL21(DE3)[pRep4]. Cultures were grown at 30 °C in Luria-Bertani broth containing 100 μg/μl of ampicillin and 50 μg/ml kanamycin. Expression was induced with 100 μm isopropyl 1-thio-β-d-galactopyranoside for 4–5 h at 22 °C. Bacteria were washed with 50 mm Tris/HCl, 1 mm EDTA, pH 8, and stored at −80 °C. Membrane fractions were prepared as previously described (17) and assayed for AC activity. The integrity of all recombinant proteins was examined by Western blotting using a purified mouse monoclonal RGS-His4 antibody (Qiagen). Proteolytic degradation was not observed.
Adenylyl Cyclase Assay
AC activity was determined by measurement of cAMP produced at 37 °C for 10 min in 100 μl (18). Standard reactions contained 20 μg of membrane protein, 50 mm Tris/HCl, pH 7.5, 22% glycerol, 3 mm MnCl2, 200 μm [α-32P] ATP, and 2 mm [2,8-3H]cAMP to monitor yield during product isolation. Creatine kinase (0.23 mg/ml) and creatine phosphate (3 mm) were used as an ATP regenerating system. At least two independent protein preparations were carried out. Data are the means ± S.E. of four or more measurements. To more easily compare data, concentration-response curves for serine or aspartate were normalized setting basal AC activity (no ligand addition) as 100%. As a control, membrane fractions from E. coli cells containing the empty expression vector were assayed for AC activity. The activities were in the range of the column background and were neglected. Student's t tests were used for statistical evaluation when necessary. A p value of ≤0.05 was considered significant.
RESULTS AND DISCUSSION
A Coevolving Network within HAMP as Signal Receiver
A bioinformatics study showed that in HAMP domains, key positions evolved in three defined networks (4) within which residues are thought to act in a concerted way to yield a specific functionality. One of these networks is found mainly in membrane-bound HAMP domains and comprises 10 residues located directly beneath the cytosolic membrane (Fig. 1A). Because of its location it was proposed to receive the signal emanating from the last transmembrane helix and translate it into a structural change of the HAMP domain (4). Here we designate it as the signal-accepting network. The network is formed by a ring of residues encompassing the hydrophobic core of AS1 and AS2 as well as solvent-exposed positions from the C-terminal part of the connector (Fig. 1A).
To probe the functionality of this network, we exchanged the respective residues between the Tsr and Af1503 HAMP domains in the reporter chimeras (Fig. 1B). As demonstrated earlier, the reporter chimera with the Af1503 HAMP was not regulated by serine (16). Conversion of the signal-accepting network in Af1503 HAMP to that found in Tsr HAMP resulted in a modest, yet significant serine regulation (p < 0.05; Fig. 1C, left, and Table 1), indicating that one or more of the Tsr-specific residues enabled signal transmission. This notion was confirmed by an equivalent construct in which signaling through Tsr HAMP was significantly impaired when its signal-accepting network was substituted by that of Af1503 HAMP (Fig. 1C, right). The serine concentration-response curve was shifted far to the right, and maximal serine inhibition, although significant (p < 0.001), was reduced from 82 to 28%, whereas the apparent IC50 for serine was increased from 66 to 840 μm (Fig. 1C, right, and Table 1). Inspection of individual substitutions in Tsr HAMP indicated that most of them were conservative conversions, e.g. Leu → Ile or Met → Ile, which have an identical side-chain volume (163–166 Å3). Although differences in flexibility and shape exist between Leu, Ile, and Met, the respective substitutions were considered to be irrelevant. Mutations Val → Thr and Asn → Asp were not considered relevant because their presence in another Tsr HAMP mutant was not detrimental (supplemental Fig. S1).
TABLE 1.
Role of the signal accepting network
In vitro adenylyl cyclase activities, maximal inhibition by serine, and IC50 serine concentrations of mutants in the signal-accepting network of Af1503 HAMP and Tsr are shown. ND, not determined.
| Mutation | AC activity | % of inhibition | IC50 |
|---|---|---|---|
| nmol·mg−1min−1 | by 10 mm serine | μm | |
| Tsr receptor1–215 Af1503 HAMP278–331 Rv3645 CHD331–549 | |||
| WT | 19.1 ± 1.6 | No inhibition | |
| Network from Tsr (Fig. 1C) | 14.6 ± 1.5 | 34 ± 8.8 | 15 ± 40 |
| R308G | 13.1 ± 4.0 | No inhibition | |
| A309S | 9.5 ± 0.6 | No inhibition | |
| R308G/A309S | 9.4 ± 1.2 | No inhibition | |
| Tsr receptor + HAMP1–268 Rv3645 CHD331–549 | |||
| WT | 18.4 ± 0.6 | 82 ± 2.7 | 66 ± 18 |
| Network from Af1503 (Fig. 1C) | 0.7 ± 0.02 | 28 ± 6.6 | 840 ± 247 |
| G245R | 0.4 ± 0.05 | 57 ± 10 | 108 ± 42 |
| S246A | 1.5 ± 0.10 | 84 ± 2.5 | ND |
| G245R/S246A | 0.7 ± 0.03 | 58 ± 2.0 | 111 ± 66 |
| E248K | 0.3 ± 0.02 | No inhibition | |
We, therefore, tested whether the effects may have been due to two conversions in the connector (Gly → Arg and Ser → Ala; Fig. 1B). We generated single and double mutants at these positions in both Af1503 and Tsr HAMP chimeras. The three mutants in Af1503 (R308G, A309S, and R308G/A309S) were not regulated by serine as the parent chimera (Table 1). In the three corresponding mutants in Tsr (G245R, A246S, and G245R/A246S) serine regulation was unaffected (Table 1).
One residue of the network, the glutamate at the beginning of AS2 (Glu-248 in Tsr), is the most highly conserved residue in >14,000 HAMP domains. It was identified as a critical residue in previous studies (19). The mutation E248K in Tsr HAMP completely abrogated serine regulation (Table 1), i.e. this particular glutamate is indispensible for transduction of the serine signal through the Tsr HAMP domain. Notably, without the sensory input and transmembrane domains, the E248K mutation had no effect on AC activity in a corresponding chimera (supplemental Table S1). Taken together, the data supported the notion that a network which was proposed based on bioinformatics (4) may operate as a receiver and transducer system for the signal coming down from the upstream Tsr sensory domain.
Importance of the Core Positions; Interplay between Permanent and Transient Core Residues
In the complementary x-da packing of the Af1503 HAMP structure, the core of the coiled-coil packing consists of one residue in x-geometry and two adjacent residues in da geometry (6). In a canonical packing these residues correspond to positions a, d, and e of the heptad repeat in AS1 and a, d, and g, in AS2 (Fig. 2A). For clarity, only the latter nomenclature is used in Fig. 2A as the x-da nomenclature strictly refers to the geometry (cf. legend of Fig. 2A). The residues are further categorized into those that are permanently central to the core in both states (permanent), those that rotate into the center of the core (intermediate), and those that rotate out of the core (transient) from x-da to canonical packing (Fig. 2, A and B, and Ref. 4). These residues should be central to a signal-induced rearrangement of the core. Therefore, we swapped the seven x-da residues which differed between the Tsr and Af1503 HAMP domains and examined serine regulation (Fig. 2B). In the Af1503 HAMP chimera with the Tsr residues, we observed a gain in serine regulation (Fig. 2C, left). Maximal inhibition was 54%, and the IC50 was at 28 μm serine. In the parallel construct in which the seven differing x-da residues of Af1503 substituted the corresponding residues in Tsr HAMP, the opposite was observed. Inhibition by serine was decreased to 36 from 82% in the parent construct, and the IC50 for serine increased 4.5-fold from 66 to 295 μm (Fig. 2C, right).
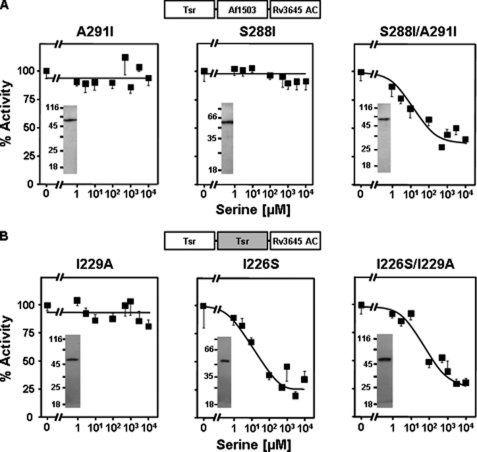
We had previously identified the x residue Ala-291 in the Af1503 HAMP domains as essential for signal propagation. Increasing the side-chain size at this core position in Af1503 HAMP caused a graded decrease in basal activity of a soluble HAMP-AC chimera and led us to propose that this residue is involved in tuning the conformational output of the HAMP domain (6). When we introduced the largest structurally acceptable side chain at this position (A291F) in the Tsr-Af1503 HAMP-AC chimera, we gained serine regulation in the previously unresponsive protein (16). Because in the core swapping experiments between Af1503 and Tsr this position was the site of one of the substitutions (A291I in Af1503), we analyzed whether the responsive phenotype might be due to mutation of A291I alone. This was not the case (Fig. 3A, left). Obviously, the isoleucine at position Ala-291 did not by itself allow the adjustment of the equilibrium between the proposed x-da and knobs-into-holes conformations toward a signaling state, whereas the larger phenyl-side chain in A291F did (16). We also examined the non-conservative substitution S288I. Again the single mutation did not result in serine regulation (Fig. 3A, middle). In the S288I/A291I double mutant responsiveness was accomplished. Serine inhibited with an IC50 of 6.9 μm (Fig. 3A, left, and Table 2). These data indicated a concerted interaction between Ser-288 and Ala-291.
FIGURE 3.
The functional coupling of permanent and transient core residues Ser-288 and Ala-291 in Af1503 and Ile-226 and IIe-229 in Tsr. A, shown are serine responses of the A291I, S288I, and S288I/A291I mutants in the Af1503 HAMP. S288I and A291I were not regulated. With the double mutant S288I/A291I, regulation by serine is gained. Maximal inhibition was 59 ± 1.6%. Basal activities of the S288I, A291I, and S288I/A291I mutants were 3.4 ± 0.3, 21.5 ± 2.4, and 17.7 ± 1.5 nmol of cAMP·mg−1min−1, respectively. B, shown are serine responses of Tsr HAMP mutants I226S, I229A, and I226S/I229A. Basal activities were, respectively, 20 ± 1, 3.4 ± 0.5, and 5.1 ± 0.4 nmol of cAMP·mg−1min−1. Maximal serine inhibition was 70 ± 6% for the double mutant and 65 ± 11% for the I226S mutant. The mutation S229I disrupted serine regulation. Western blots (0.5 μg of membrane preparation) indicated similar expression levels for all mutants and no proteolysis of the recombinant proteins.
TABLE 2.
Importance of the core positions; interplay between permanent and transient core residues in Af1503 and Tsr HAMP domains
In vitro adenylyl cyclase activities, maximal inhibition by 10 mm serine, and IC50 serine concentrations of mutants at transient positions of Af1503 HAMP and Tsr are shown.
| Mutation | AC activity | % of inhibition | IC50 |
|---|---|---|---|
| nmol·mg−1min−1 | by 10 mm serine | μm | |
| Tsr receptor1–215 Af1503 HAMP278–331 Rv3645 CHD331–549 | |||
| WT | 19.1 ± 1.6 | No inhibition | |
| x-da from Tsr | 21.2 ± 1.5 | 54 ± 3.5 | 28 ± 12.5 |
| A291I | 3.4 ± 0.3 | No inhibition | |
| S288I | 21.5 ± 2.4 | No inhibition | |
| S288I/A291I | 17.7 ± 1.5 | 59 ± 1.6 | 6.9 ± 5.7 |
| T281I | 13.3 ± 1.0 | No inhibition | — |
| T281I/A291I | 3.1 ± 0.2 | 34 ± 7.8 | 284 ± 383 |
| A295I | 5.6 ± 1.2 | No inhibition | |
| A291I/A295I | 3.8 ± 0.1 | No inhibition | |
| E311I | 4.9 ± 0.4 | No inhibition | |
| A291I/E311I | 0.9 ± 0.1 | No inhibition | |
| S318I | 23.6 ± 2.6 | No inhibition | |
| A291I/S318I | 5.4 ± 0.3 | 52 ± 5.4 | 6.2 ± 1.8 |
| S325I | 13.1 ± 0.4 | No inhibition | |
| A291I/S325I | 4.6 ± 0.4 | 38 ± 4.7 | 7 ± 2.7 |
| Tsr receptor + HAMP1–268 Rv3645 CHD331–549 | |||
| WT | 18.4 ± 0.6 | 82 ± 2.7 | 66 ± 18 |
| x-da from Af1503 | 30.0 ± 2.3 | 36 ± 6.7 | 295 ± 122 |
| I226S | 3.4 ± 0.5 | 65 ± 11 | 123 ± 221 |
| I229A | 20 ± 1 | No inhibition | |
| WT | 18.4 ± 0.6 | 82 ± 2.7 | 66 ± 18 |
Analysis of the equivalent residues in the Tsr HAMP chimera confirmed the functional coupling between these positions, although their individual effects showed differences to those in Af1503 HAMP (Fig. 3B). Mutation I229A in the Tsr HAMP, corresponding to A291I in Af1503, abrogated regulation (Fig. 3B, left). This result bolsters the prominent role of this particular permanent core position in affecting the conformational switch during signaling, as discussed previously (6). Conversely, the mutation I226S in the Tsr HAMP had only a minor effect; i.e. it increased the IC50 for serine. However, in the double mutant I226S/I229A, serine regulation was restored (Fig. 3B, right, and Table 2). This emphasized the critical interaction of these positions in the Tsr as well as Af1503 HAMP domains.
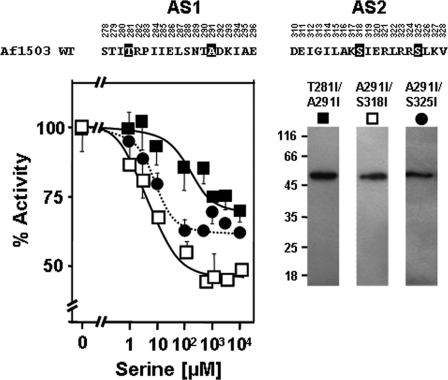
In the gearbox model, Ser-288 in Af1503 and Ile-226 in Tsr are transient core residues, i.e. residues that rotate out of the core during the transition from complementary x-da to knobs-into-holes packing (Fig. 2, A and B). An exhaustive analysis of HAMP sequences showed that residues at these positions are usually small, favoring complementary x-da packing, whereas their low hydrophobicity supports the canonical knobs-into-holes packing, indicating a role for the transient core residues in maintaining a balance between the two conformational states (4). In light of this hypothesis, in the S288I/A291I double mutant the increase in side-chain size at Ala-291, which favors knobs-into-holes packing (16), would have been balanced by an increase in hydrophobicity at the transient core position Ser-288, which favors complementary x-da packing (4). This prompted us to systematically explore whether the other transient core positions of Af1503 HAMP (Thr-281, Ala-295, Glu-311, Ser-318, and Ser-325) would also couple with the A291I mutation and result in serine responsiveness when converted to large, hydrophobic residues. We mutated them individually to isoleucine and investigated their effect alone and in conjunction with A291I (Table 2). None of the individual mutants was controlled by serine, but four of six double mutants (including S288I/A291I; see above) were serine-regulated (Fig. 4). Although they differed in minor, in our opinion insignificant aspects (Table 2), they were considered as equivalent in modulating HAMP signaling. In Af1503 HAMP, the majority of transient core residues thus appeared to interact and couple with the permanent core residue Ala-291 for switching irrespective of the layer in which they are positioned in the four helix bundle.
FIGURE 4.
Interactions of Ala-291 in Af1503 with transient amino acids in serine signaling. Combination of A291I with Ile mutants at transient positions Thr-281, Ser-318, and Ser-325 results in serine regulation of chimeras (the mutated amino acids are inverted in the sequence above). Basal activities were set at 100%: filled squares, T281I/A291I; open squares, A291I/S318I; circles, A291I/S325I. Western blots of 0.5 μg membrane preparation of each construct are shown at the right with the respective symbol on top.
There were two exceptions; A295I and E311I failed to couple with A291I and did not gain serine responsiveness (Table 2). We note that the equivalent positions in Tsr (Ala-233 and Glu-248) are sensitive to mutation as most point mutations abolished the serine response in physiological experiments (19). As shown above for Tsr HAMP, the glutamate Glu-248 has an indispensible function within the signal accepting network in transducing the signal from the last transmembrane span (Table 1 and supplemental Table S1). We surmise that Ala-295 may be important for the correct formation of the hinge region, which positions the connector with respect to the HAMP helices (see below).
In parallel to the Tsr-Af1503 HAMP-AC chimera, we also generated a chimera containing the sensory domain of Tar instead of Tsr. We attempted to gain aspartate regulation in conjunction with the double mutant S288I/A291I of the Af1503 HAMP domain. However, this mutant was not aspartate-regulated (supplemental Table S2). Only with the mutation of a further transient core residue in Af1503 HAMP, S318I, generating the triple mutant S288I/A291I/S318I, did we observe inhibition by aspartate, albeit at rather high concentrations (IC50 = 510 μm) and only to a maximal inhibition of 28% (supplemental Table S2). This result confirmed the functional coupling of transient core residues with the permanent core residue Ala-291 and also suggested that their effect is cumulative, lending support to their role in balancing HAMP between different conformational states.
The Connector Affects HAMP Conformation and Plays Role in Signaling
To span the distance of 26 Å between the C terminus of AS1 and the N terminus of AS2, 12 amino acids are minimally required. The connector in HAMP domains does not have a conserved length and occasionally carries insertions of considerable size (e.g. 130 residues in yeast SLN1). The extent of sequence conservation in the connector is also rather moderate (4). A glycine (Gly-297 in Af1503) is positioned at the hairpin turn at the N terminus of AS1 and, hence, is highly conserved (4, 6). Furthermore, two hydrophobic positions (HR), Leu-299 and Val-303 in Af1503, are often conserved (20). Thus, in many HAMP domains the sequence GXHR1XXXHR2 is maintained. Possibly this pattern stabilizes HAMP conformations in ways that are variable among HAMPs, suggesting individual domain conformations (6, 13–14, 19). Although canonical and divergent HAMP domains may use slightly different mechanisms in signal transmission, the connector contributes in both to the rotational rearrangements of the α-helices by formation of several new contacts, in addition to physically holding AS1 and AS2 together. To examine the effect of an extended length of the linker and the need for a physical linkage, we engineered a tobacco etch virus protease restriction site of seven additional amino acids (ENLYFQG) into the Tsr HAMP connector between Asp-244 and Gly-245. Serine inhibition was fully retained in the resulting chimera, in agreement with insertions made at this position in previous analysis (20). However, proteolytic cleavage of the connector with the tobacco etch virus protease abolished regulation, attesting to the fact that the connector is needed for signaling and that the α-helices in the HAMP domain do not per se possess high affinity for each other and must be physically connected (see supplemental Fig. S2 and supplemental Table S3).
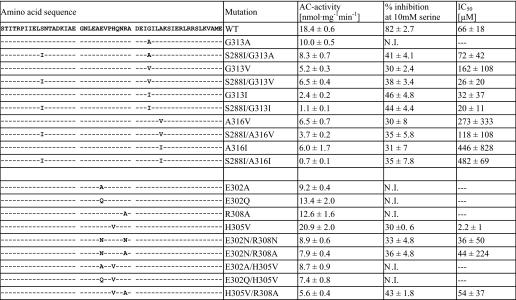
In the Af1503 HAMP structure, several contacts between the connector and AS1 and AS2 are observed. Two residues in AS2 (Gly-313, Ala-316) are located in a region of close contact between the connector and AS2 and presumably have small side chains due to steric constraints. Based on our model of the knobs-into-holes conformation (6), which predicts space for larger side chains at these positions, we hypothesized that mutations to large hydrophobic side chains at Gly-313 and Ala-316 should have similar effects as mutations at the permanent core position Ala-291. This was experimentally probed by introduction of respective point mutations (Table 3). The G313V and G313I mutants were significantly inhibited by serine (Table 3), an effect observed at position Ala-291 only for the largest side chain (A291F). IC50 concentrations for serine were significantly lower, with the bulkier isoleucine substitution (32 μm) than with the valine substitution (162 μm; Table 3). In combination with the mutation S288I, which was shown to enable signaling in conjunction with A291I (see above), the sensitivity toward serine was enhanced for both mutants, whereas the G313A mutant, individually unaffected by serine, now also was regulated (Table 3). Results obtained for position Ala-316 were only partly in agreement with our hypothesis (Table 3). The single mutants A316V and A316I were both inhibited by serine, but the concentrations required for inhibition were high (IC50 concentrations >100 μm) and the combination with S288I did not enhance serine sensitivity, indicating that in contrast to Ala-291 and Gly-313, mutations at this position might not be functionally coupled to mutations at transient core positions. Taken together, these results nevertheless show that interactions between the HAMP helices and the connector may play a critical role in signal propagation. These results also appear to support the notion of a balance between two conformational states in which mutations favoring knobs-into-holes can be compensated for by mutations favoring complementary x-da packing.
TABLE 3.
In vitro adenylyl cyclase activities, maximal inhibition by 10 mm serine, and IC50 concentrations for serine of the mutants made at positions Gly-313 and Ala-316 of Af1503 HAMP and at salt bridge residues in the connector of Af1503 HAMP. N.I., no inhibition
Furthermore, we investigated the effects of the disruption of three salt bridges between connector and helices (supplemental Fig. S3). Mutations predicted to interfere in formation of salt bridges were introduced at respective connector residues (Glu-302, His-305, and Arg-308). The results were inconclusive, as no general pattern was observed (Table 3). The observation that single mutations in the connector directly affected serine response of the chimera implicated the connector in signaling. Although our data do not allow us to establish a general mechanism of how connector sequences affect signaling in different proteins, they distinctly demonstrate a participation of the connector in signal propagation.
Conclusion
Compared with scoring swimming behavior in physiological experiments in vivo, the chimeras generated in this study provide a biochemical system to in vitro examine signal transmission through the HAMP domain (17, 19). Our chimeric in vitro system is suited to quantitatively study the effect of single or multiple HAMP mutations on Tsr-initiated monodirectional transmembrane signaling as class III ACs are unknown to be the subject of sensory adaptation processes. Because the membrane-bound reporter AC itself has a HAMP domain, these chimeras constitute a system in which related components were linked. Because of the lack of a ligand for the AC membrane domain, we connected a biologically characterized chemotaxis receptor with a biochemically characterized output domain. The activities of the membrane-bound Rv3645 AC, the soluble HAMP-CHD complex, and the soluble cyclase catalytic domain are similar to those of the chimeras used here. Actually Rv3645 AC activity is affected by structural modifications of its own HAMP and the Rv3645 HAMP itself propagates the serine signal of Tsr in respective hybrid constructs (17, 21–22). Our chimeras are similar to functional hybrid histidine kinases, e.g. Taz, in which the sensor module of the E. coli aspartate chemoreceptor Tar is fused via a HAMP domain to the transmitter domain of the histidine kinase EnvZ and signaling is assayed in vivo via induction of β-galactosidase (8, 23–24). All those hybrids share an identical domain composition and allow investigation of various aspects of transmembrane signaling with different output assays.
Here we investigated (i) the signal-accepting network of HAMP domains that was predicted by a bioinformatics study (4). In general, the phenotypes of the mutants generated along those lines biochemically confirmed the functionality of such a network as an interacting unit. (ii) We established that the connector which bridges the distance between the C-terminal end of AS1 and the N-terminal beginning of AS2 is required to obtain serine control. Proteolytic clipping of the connector abrogated signaling. (iii) We classified individual amino acids as permanent, intermediate, and transient core residues based on the proposed gearbox model of rotation and established a rationale for their interactions in HAMP-mediated signaling. In general, our biochemical data are fully compatible with the gearbox model of HAMP rotation but cannot yet exclude other mechanisms of HAMP signaling, such as the dynamic bundle model (19). The few instances that did not fall in line with our predictions and expectations probably indicate that beyond rotation, additional conformational processes take place in signal transmission, possibly either upstream (cable) and/or downstream (linker) of the HAMP domain (25). We note that HAMP rotation as a potential mechanism of signal propagation does not necessarily clash with a piston movement of the second transmembrane helix (for review, see Ref. 26) because our experiments provide no information about upstream conformational movements. This would require structural knowledge of the entire signaling complex including its transmembrane helices. Our future experiments aim in this direction.
Supplementary Material
Acknowledgments
We thank U. Kurz for technical assistance and Dr. S. Parkinson, University of Utah for many discussions and valuable suggestions.
This work was supported by the Deutsche Forschungsgemeinschaft (SFB 766, TP B8) and by institutional funds of the Max Planck Society.

This article contains supplemental Table S1–S3 and Figs. S1–S3.
- AC
- adenylyl cyclase
- CHD
- cyclase homology domain.
REFERENCES
- 1. Parkinson J. S., Kofoid E. C. (1992) Annu. Rev. Genet. 26, 71–112 [DOI] [PubMed] [Google Scholar]
- 2. Galperin M. Y. (2004) Environ. Microbiol. 6, 552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aravind L., Ponting C. P. (1999) FEMS Microbiol. Lett. 176, 111–116 [DOI] [PubMed] [Google Scholar]
- 4. Dunin-Horkawicz S., Lupas A. N. (2010) J. Mol. Biol. 397, 1156–1174 [DOI] [PubMed] [Google Scholar]
- 5. Appleman J. A., Chen L. L., Stewart V. (2003) J. Bacteriol 185, 4872–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hulko M., Berndt F., Gruber M., Linder J. U., Truffault V., Schultz A., Martin J., Schultz J. E., Lupas A. N., Coles M. (2006) Cell 126, 929–940 [DOI] [PubMed] [Google Scholar]
- 7. Zhu Y., Inouye M. (2003) J. Biol. Chem. 278, 22812–22819 [DOI] [PubMed] [Google Scholar]
- 8. Utsumi R., Brissette R. E., Rampersaud A., Forst S. A., Oosawa K., Inouye M. (1989) Science 245, 1246–1249 [DOI] [PubMed] [Google Scholar]
- 9. Appleman J. A., Stewart V. (2003) J. Bacteriol. 185, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kishii R., Falzon L., Yoshida T., Kobayashi H., Inouye M. (2007) J. Biol. Chem. 282, 26401–26408 [DOI] [PubMed] [Google Scholar]
- 11. Williams S. B., Stewart V. (1999) Mol. Microbiol. 33, 1093-1102 [DOI] [PubMed] [Google Scholar]
- 12. Butler S. L., Falke J. J. (1998) Biochemistry 37, 10746–10756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swain K. E., Falke J. J. (2007) Biochemistry 46, 13684–13695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Airola M. V., Watts K. J., Bilwes A. M., Crane B. R. (2010) Structure 18, 436–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Watts K. J., Johnson M. S., Taylor B. L. (2008) J. Bacteriol 190, 2118–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferris H. U., Dunin-Horkawicz S., Mondéjar L. G., Hulko M., Hantke K., Martin J., Schultz J. E., Zeth K., Lupas A. N., Coles M. (2011) Structure 19, 378–385 [DOI] [PubMed] [Google Scholar]
- 17. Kanchan K., Linder J., Winkler K., Hantke K., Schultz A., Schultz J. E. (2010) J. Biol. Chem. 285, 2090–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salomon Y., Londos C., Rodbell M. (1974) Anal. Biochem. 58, 541–548 [DOI] [PubMed] [Google Scholar]
- 19. Zhou Q., Ames P., Parkinson J. S. (2009) Mol. Microbiol. 73, 801–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ames P., Zhou Q., Parkinson J. S. (2008) J. Bacteriol. 190, 6676–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hammer A. (2004) Klonierung und Charakterisierung von fünf Adenylatcyclasen aus Stigmatella aurantiaca und Mycobakterium tuberculosis. Doctoral dissertation, Eberhard-Karls-Universität, Tübingen, Germany [Google Scholar]
- 22. Linder J. U., Hammer A., Schultz J. E. (2004) Eur. J. Biochem. 271, 2446–2451 [DOI] [PubMed] [Google Scholar]
- 23. Jin T., Inouye M. (1994) J. Mol. Biol. 244, 477–481 [DOI] [PubMed] [Google Scholar]
- 24. Baumgartner J. W., Kim C., Brissette R. E., Inouye M., Park C., Hazelbauer G. L. (1994) J. Bacteriol. 176, 1157–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kitanovic S., Ames P., Parkinson J. S. (2011) J. Bacteriol. 193, 5062–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hazelbauer G. L., Falke J. J., Parkinson J. S. (2008) Trends Biochem. Sci. 33, 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.