Background: Lamin B receptor (LBR) is an integral nuclear envelope protein and contains a Tudor domain.
Results: The NMR structure of LBR-Tudor was determined and its interactions with nuclear proteins, histones, and nucleosomes were explored.
Conclusion: LBR-Tudor is not involved in recognition of methylated histones and binds free H3.
Significance: Tudor domains may act as histone chaperone-like platforms.
Keywords: Histone Methylation, NMR, Protein Domains, Protein Motifs, Protein Structure, Protein-Protein Interactions, Lamin B Receptor, Tudor Domain, Nuclear Envelope
Abstract
Lamin B receptor (LBR) is a polytopic protein of the nuclear envelope thought to connect the inner nuclear membrane with the underlying nuclear lamina and peripheral heterochromatin. To better understand the function of this protein, we have examined in detail its nucleoplasmic region, which is predicted to harbor a Tudor domain (LBR-TD). Structural analysis by multidimensional NMR spectroscopy establishes that LBR-TD indeed adopts a classical β-barrel Tudor fold in solution, which, however, features an incomplete aromatic cage. Removal of LBR-TD renders LBR more mobile at the plane of the nuclear envelope, but the isolated module does not bind to nuclear lamins, heterochromatin proteins (MeCP2), and nucleosomes, nor does it associate with methylated Arg/Lys residues through its aromatic cage. Instead, LBR-TD exhibits tight and stoichiometric binding to the “histone-fold” region of unassembled, free histone H3, suggesting an interesting role in histone assembly. Consistent with such a role, robust binding to native nucleosomes is observed when LBR-TD is extended toward its carboxyl terminus, to include an area rich in Ser-Arg residues. The Ser-Arg region, alone or in combination with LBR-TD, binds both unassembled and assembled H3/H4 histones, suggesting that the TD/RS interface may operate as a “histone chaperone-like platform.”
Introduction
Tudor domains are 50–70 amino acid modules, named after the synonymous Drosophila protein, which harbors 11 such copies in its molecule (1). Along with the chromodomain, PWWP, MBT, and Agenet, these modules comprise a structural superfamily, the so-called “Royal family.” The members of the Royal family occur in a variety of chromatin-associated proteins and are thought to originate from a common ancestor (2). Originally, Tudor domains were thought to be RNA-binding motifs, because they were first identified in RNA-binding proteins or ribonucleoprotein particles (3). However, subsequent structural and biochemical studies involving the survival motor neuron (SMN)2 protein suggested that Tudor domains may associate with symmetrically dimethylated Arg residues in spliceosomal Sm proteins (4–7). This hypothesis has received further support from more recent studies with a variety of proteins (8–13) and it is now clear that Tudor domains can bind either methylated Lys residues in H3 and H4 histone tails (14–18), or methylated arginines usually flanked by glycine residues (11–13, 19). These interactions involve the methylated side chains and a cluster of aromatic residues that constitute the so-called aromatic cage, present in many Tudor and chromodomains (20–22).
A putative Tudor domain has been recently identified by inspection of the amino acid sequence of the lamin B receptor (LBR). LBR is a ubiquitous integral protein of the nuclear envelope (NE) and was initially characterized by virtue of its ability to associate with nuclear lamin B (23). It is now thought to participate in a variety of nuclear functions, including tethering of the nuclear lamina to the inner nuclear membrane and “transient trapping” of nuclear components that are involved in chromatin remodeling and transcriptional inactivation (24–30).
The putative Tudor domain of LBR (hereon referred to as LBR-TD) is accommodated within the amino-terminal part of the protein (LBR-NT), which faces the nucleoplasm and has been shown to mediate binding to the nuclear lamina and peripheral heterochromatin. Sequence analyses suggest that LBR-NT contains three distinct regions (see scheme in Fig. 1A): (a) the LBR-TD, which spans the first 60 residues; (b) a highly charged 40-residue hinge region that is rich in Arg-Ser (RS) dipeptide motifs; and (c) a 110-amino acid segment (SGD) with no apparent sequence kinship to other proteins. The middle segment features of a “natively disordered” protein and exhibits multiple SRPK1 phosphorylation sites (29, 31, 32).
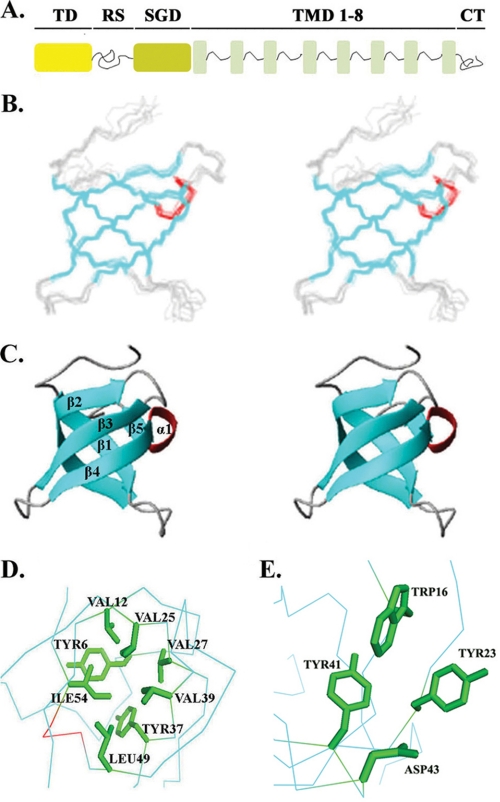
FIGURE 1.
Structure of LBR-TD. A, layout of LBR molecule. B, stereo view of the backbone atoms (N, Cα, C′) for residues of a NMR ensemble of 10 (out of 100 computed) lowest energy structures of LBR-TD. Secondary structure elements are colored cyan for β-strands and red for the 310-helix connecting β4 and β5. C, ribbon diagram of the closest to the mean LBR-TD structure. Secondary structure elements are colored as described above. D, rotated view of the Tudor domain structure showing the side chains of hydrophobic core residues in green. E, close view of the aromatic cage discussed in the text. The side chains of the three residues (Trp16, Tyr23, and Tyr41) that form the aromatic cluster are shown in green.
Biochemical studies have implicated LBR-NT in lamin B interactions (33). It is also likely that this part of the protein participates in other interactions, such as binding to histone H3/H4 oligomers (30), methyl-CpG-binding protein MeCP2 (34), and heterochromatin protein 1 (32). LBR-NT has also been implicated in binding linker DNA (35) and in LBR-LBR interactions (28), but the significance of these findings and the involvement of LBR-TD have not been precisely determined.
In an attempt to elucidate the interactions of LBR at the molecular level and further understand its in situ organization at the nuclear envelope, we have begun to dissect the LBR-NT into structurally/functionally relevant domains, starting from LBR-TD. Here, we report the solution structure of chicken LBR-TD and its interactions with other proteins and cellular components. Our biochemical data suggest that instead of binding to methylated arginine or lysine residues the LBR-Tudor-fold may have a histone chaperone-like activity, thus extending the range of functional roles of the Tudor family.
EXPERIMENTAL PROCEDURES
Expression and Purification of Recombinant Proteins
Chicken LBR cDNAs encoding the LBR subdomains Tudor (residues 1–62), RS (residues 63–100), SGD (residues 101–208), Tudor-RS (residues 1–100), and RSSGD (residues 63–208) were inserted into the NcoI/NotI sites of a modified pET24d (Novagen) expression vector encoding an amino-terminal His6-GST tag, followed by a TEV protease cleavage site, a generous gift from G. Stier (EMBL). The correct cDNA sequences of the expression clones were confirmed by DNA sequencing. Fusion proteins were expressed in BL21(DE3) cells according to standard procedures (36) and purified from bacterial lysates using Ni-NTA affinity chromatography. A uniformly 15N,13C-labeled polypeptide corresponding to LBR-TD was also prepared by growing Escherichia coli strain BL21(DE3) pLysS overexpressing LBR-TD in a minimal medium containing 15NH4Cl and [13C]glucose. The N-terminal tag was removed after TEV digestion and the pure Tudor domain was collected after passing through a second Ni-NTA column. NMR samples were dialyzed into 20 mm sodium phosphate buffer, pH 6.9, 100 mm NaCl and concentrated using an Amicon ultrafiltration device to a final concentration of 0.8 mm. The recombinant Tudor protein used for the NMR experiments in addition to residues 1–62 of chicken LBR carries four N-terminal residues (GAMG) from the TEV cleavage site.
Native calf thymus histones were purchased from Roche Diagnostics. A pET3a expression vector carrying the cDNAs encoding H3 and the histone H3 core region (amino acids 27–135) from Xenopus laevis was a kind gift from K. Luger, University of Colorado. Recombinant H3 and tail-less H3 were expressed in E. coli BL21(DE3) cells and purified under denaturing conditions using SP-Sepharose chromatography, as previously described (37). After removal of urea with extensive dialysis, the samples were lyophilized, dissolved in water, and their concentration was adjusted to 1 mg/ml. The yeast H3 tail region (1–46) was expressed in E. coli BL21(DE3) cells as a fusion protein with a N-terminal GST tag from a pGEX2T expression vector kindly provided by M. Grunstein, University of California, and was purified as previously described (38).
The recombinant human N-terminal region of LBR(1–201) was expressed from a pET15b expression vector encoding an amino-terminal His10 tag. The fusion protein was expressed in BL21(DE3) cells according to standard procedures (36) and purified from bacterial lysates under denaturing conditions using Ni-NTA affinity chromatography followed by protein refolding on the column (39).
Human MeCP2(1–486) was expressed as a fusion protein with an amino-terminal His6 tag from a pET30a expression vector, which was kindly provided by G. Badaracco, University of Insubria (34). The fusion protein was expressed in BL21(DE3) cells according to standard procedures (36) and purified under native conditions by using Ni-NTA-agarose beads.
Preparation of Nuclear Envelope Extracts
Turkey erythrocytes were obtained from whole blood and their nuclei were isolated with standard methods (28, 30, 40). Isolated nuclei were digested with MNase (100 units/ml of digestion buffer) for 10 min at 37 ºC (in 20 mm HEPES-KOH buffer, pH 7.4, 5 mm MgCl2, 0.025% Triton X-100, 1 mm CaCl2, 1 mm DTT, 1 mm PMSF, protease inhibitors: leupeptin, pepstatin, antipain, aprotinin at 2 μg/ml). The reaction was stopped with 2 mm EDTA and, after centrifugation at 10,000 × g, the resulting nuclear envelopes were washed with digestion buffer containing 2 mm EDTA. Nuclear extract was prepared with resuspension of the nuclear envelope pellet in 300 mm NaCl, 20 mm Tris-HCl, pH 7.5, 250 mm sucrose, 2 mm MgCl2, 1 mm EGTA, 1 mm DTT, 1 mm PMSF and protease inhibitors, followed by sonication. After ultracentrifugation for 30 min at 4 ºC and 300,000 × g, the soluble extract was collected and used in pull-down assays.
Isolation of Native Lamin B
Nuclei were isolated from rat liver according to standard methods (41). Isolated nuclei (5 ml) were resuspended in 0.1 mm MgCl2, 1 mm DTT, 0.5 mm PMSF (10 ml) and 15 ml of 10% sucrose, 20 mm Tris-HCl, pH 8.5, 0.1 mm MgCl2, 1 mm DTT, and 0.5 mm PMSF were added to the suspension. The preparation was digested with 500 μl of DNase I (2 mg/ml) for 15 min at ambient temperature under rotation and the suspension was centrifuged at 10,000 × g and 4 °C for 15 min. The pellet was resuspended in 10% sucrose, 20 mm Tris-HCl, pH 7.5, 1 mm MgCl2, 0.5 mm PMSF (10 ml) and 15 ml of 30% sucrose, 20 mm Tris-HCl, pH 7.5, 0.1 mm MgCl2, 1 mm DTT 0.5 mm PMSF were added. Another round of DNase I digestion was performed as above. The pellet was resuspended in 1 m KCl, 50 mm Tris-HCl, pH 7.5, 1 mm EDTA, 1 mm DTT, 0.5 mm PMSF (10 ml) and centrifuged as above. The final pellet was washed with ice-cold ddH2O and centrifuged at 10,000 × g and 4 °C for 45 min. The nuclear envelope pellet was extracted with 8 m urea, 10 mm Tris-HCl, pH 7.6, 4 mm EDTA, 1 mm DTT, 0.5 mm PMSF followed by sonication. The soluble material was collected after 35 min of ultracentrifugation at 300,000 × g and 18 °C. Soluble lamin B was isolated from the extract by using DE53 ion exchange chromatography. All buffers contained protease inhibitors, leupeptin, pepstatin, antipain, and aprotinin, to a final concentration of 2 μg/ml. Urea was removed from the desired elutions with extensive dialysis against 25 mm Tris-HCl, pH 8.5, 150 mm NaCl, 1 mm EGTA, 0.1 mm DTT, 1 mm PMSF. Antibodies against lamin B were prepared as previously described (42).
Isolation of Native H3-H4 Tetramers and Interaction Assays Using Sucrose Gradients
Native H3-H4 tetramers were isolated using hydroxylapatite chromatography (43). Briefly, nuclear extract from MNase-digested turkey erythrocyte nuclei was dialyzed against 10 mm phosphate buffer, pH 6.8, 1 mm DTT and subsequently passed through an equilibrated hydroxylapatite column (Bio-Rad). Different histone pairs were eluted from the column using increasing ionic strength buffers (the H3-H4 tetramer is eluted with 2 m NaCl).
For the sucrose gradient runs, GST-TD or Tudor protein samples were mixed with equimolar amounts of H3-H4 tetramer. The mixtures were dialyzed against 20 mm Tris, pH 7.5, 300 mm NaCl, 5% sucrose, 2 mm MgCl2, 0.1 mm EDTA, 1 mm DTT, 1 mm PMSF and subsequently concentrated to a total protein concentration of 1 mg/ml. 300-μl samples were coated on the top of a 5–20% sucrose gradient made in the same buffer in a total volume of 11 ml. Samples were spun at 40,000 × g in a SW40 Beckman rotor for 20 h at 4 ºC. Fractions of 500 μl were collected and run in a SDS-PAGE gel. Protein bands were visualized with Coomassie Brilliant Blue G-250.
Pull-down Assays
All reactions were carried out in Eppendorf tubes coated with 1% boiled/filtered fish skin gelatin. GST fusion proteins and GST alone as control (10–20 μg) were attached to glutathione-agarose beads. The beads were combined with nuclear extract or 15–30 μg of native/recombinant proteins in 300 mm NaCl buffer and incubated for 1 h at room temperature. The low-speed sediments were subsequently washed five times with the 300 mm NaCl buffer and once with isotonic buffer. Bound proteins were eluted with hot SDS sample buffer and run in a SDS-PAGE gel. Protein bands were visualized with Coomassie Brilliant Blue G-250 and by Western blot in the case of lamin B. Pull-down assays were repeated at high ionic strengths (up to 1 m).
The semi-quantitative experiments were performed in 400, 750, 1,000, 2,000, 5,000, and 10,000 μl of buffer with a constant amount of GST-TD and H3. Bands were quantified using Quantity-1 software (Bio-Rad). All experiments were repeated at least three times.
NMR Spectroscopy
NMR spectra of LBR-TD were recorded at 25 ºC on Bruker DRX 500 and DRX 600 NMR spectrometers equipped with triple resonance cryoprobes and pulsed field gradients. Multidimensional NMR spectra were processed with NMRPipe (44) and analyzed with NMRView (45). Backbone and side chain 1H, 15N, and 13C resonances were assigned using a set of triple resonance experiments (46). The 1H, 15N, and 13C chemical shifts of the backbone resonances were obtained from sensitivity-enhanced three-dimensional HNCA, CBCA(CO)NH, and CBCANH experiments. The side chain signals were assigned from three-dimensional HCCH-TOCSY and aromatic Cβ-Hδ/ϵ correlation experiments (47).
For NMR titrations, 1H-15N heteronuclear single quantum correlation (HSQC) experiments were recorded at 600 MHz proton frequency using a 50 μm 15N-labeled Tudor domain sample and an excess of the ligand in 20 mm sodium phosphate buffer (pH 6.9), 100 mm NaCl. Mono-, di-, and trimethylated Lys, as well as symmetrically and asymmetrically dimethylated Arg were added up to a molar ratio of 1:25, whereas native and recombinant histone H3 were added stepwise up to a molar ratio of 1:3 (LBR-TD:H3).
Structure Calculation
Distance restraints were derived from 15N- and 13C-edited three-dimensional NOESY experiments. Hydrogen bond restraints were also derived from identification of slow exchanging amide protons. Combined NOE cross-peak assignment and three-dimensional protein structure calculations were performed using the program CYANA (48). Torsion angle restraints were derived from TALOS (49). The final ensemble of structures was refined in a box of solvent molecules as described (50). The quality of the structure ensemble was evaluated using WHAT IF (51) and PROCHECK-NMR (52). Molecular images were generated with PyMol (53). Structure similarity searches were performed using the DALI (54) server. Coordinates have been deposited in the Protein Data Bank (accession code 2L8D). Chemical shift assignments and NOE peak lists have been deposited in the BioMagResBank (accession code 17402).
FRAP Experiments
Fluorescence recovery after photobleaching (FRAP) assays were performed on a Leica laser scanning confocal microscope (SP5) using suitable software and the 488-nm line of an argon laser. GFP-transfected cells were grown on special Petri dishes with coverslips attached and visualized in phenol-free culture medium buffered with HEPES-KOH. FRAP was performed with a bleach pulse of 6.5 s and initiated after 5 pre-bleach images. Post-bleach images (512 times 512 pixels) were collected for 307 s at low laser power (24%). Data were corrected for fluorescence quench and recovery observed in the entire cell and in the background.
RESULTS
Overview of Structure and Similarity to Royal Family Domains
The protein analyzed, LBR-TD, comprised residues 1–62 of chicken LBR (see schematic diagram in Fig. 1A) and was monomeric in solution, as assessed by analytical ultracentrifugation (data not shown). The solution structure of LBR-TD was determined by heteronuclear multidimensional NMR spectroscopy with distance restraints derived from three-dimensional 15N- and 13C-edited NOE spectra. Experimental restraints and structural statistics over the 10 lowest energy structures are summarized in supplemental Table S1. An ensemble of the 10 lowest-energy NMR structures and a ribbon representation of the average structure are presented in Fig. 1, B and C.
The NMR data show that residues 4–58 of LBR-TD form a well defined tertiary structure, with root mean square deviations of 0.53 Å for backbone atoms. All residues fall in the allowed regions of the Ramachandran plot. The amino-terminal residues 1–3 and carboxyl-terminal residues 59–62 are apparently disordered, as indicated by the paucity of inter-residue nuclear Overhauser effects (NOEs). The structured part adopts a β-barrel-like fold, consisting of five antiparallel β-strands. Strands β1–4 are connected by short turns, whereas strands β4 and β5 are linked by a short 310-helix. A β-bulge (residues 28–29) allows one strand (β2) to span both sides of the domain. The structure is stabilized by a well defined hydrophobic core that consists of residues Tyr6, Val12, Val25, Val27, Tyr37, Val39, Leu49, and Ile54 (Fig. 1D). These residues are conserved in most of the known TD sequences (2).
The side chains of the highly conserved amino acids Trp16 (loop 1), Tyr23 (strand β2), Tyr41 (strand β3), and Asp43 form a cluster on the surface of the domain (Fig. 1E). The location of these residues is highly reminiscent of the aromatic cages found in chromo and Tudor domains, which are thought to mediate recognition of methylated Lys or Arg side chains (4, 10, 12–15, 17–22) (Fig. 2A). The conformation of the side chains in the aromatic residues is well defined by a large number of NOE restraints in this area (supplemental Fig. S1).
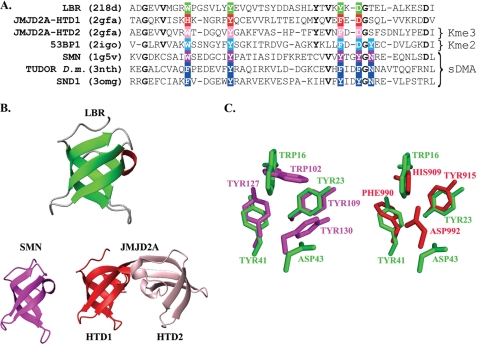
FIGURE 2.
Comparison of structures for closest structural homologues of LBR-TD. A, structure-based sequence alignment of the LBR-TD, the interdigitated Tudor domains of JMJD2A, and the Tudor domains of 53BP1, Drosophila TUDOR, SMN, and SND1 proteins. Residues that form the aromatic cage in each domain are boxed and in the colored background. Other invariant residues among the LBR and JMJD2A domains are bold. Notice that only the second domain of JMJD2A binds trimethyllysine (Kme3), 53BP1 binds dimethyllysine (Kme2), whereas the SMN, Drosophila TUDOR, and SND1 Tudor domains specifically recognize symmetrically dimethylated arginine (sDMA). Due to the interdigitated structure of JMJD2A the sequences for the superimposed structures are: HTD1, Thr900 … Glu925–Ile983 … Val1003; HTD2, Ala958 … Pro982–Thr926 … Ile946. B, ribbon diagrams of LBR-TD structural homologues. The structural homologues were identified by DALI and are displayed in green for LBR, magenta for SMN (PDB code 1g5v), red and pink for JMJD2A Tudor domains 1 (HTD1) and 2 (HTD2), respectively (PDB code 2gfa). C, close-up view of the superimposed aromatic cages of the three domains. Coloring scheme same as in B.
Structural homology searches using DALI (54) showed that the structure of the chicken LBR-TD is very similar to the TD of the SMN protein (PDB code 1g5v, Z score = 7.9, root mean square deviations = 1.8 Å) and the first of the two hybrid Tudor domains of the JMJD2A demethylase (PDB code 2gfa, Z score = 7.6, root mean square deviation = 1.7 Å), although the sequence identity with these two homologs did not exceed 19% (Fig. 2B). Extensive similarity (Z score = 8.0, root mean square deviation = 1.3) was also observed with a structure that corresponds to the Tudor domain of human LBR. This structure has been recently determined by NMR in the context of a Structural Genomics project by the RIKEN consortium and deposited in the data base (PDB code 2dig), but has not been further analyzed.
The amino acid composition of the LBR aromatic cage is very similar to that of the second hybrid Tudor domain of JMJD2A (HTD2), which is to date the only Tudor domain known to interact with trimethyllysine residues through a binding pocket that consists of only three aromatic residues and an aspartate (Fig. 2A). However, a more detailed comparison reveals that the aromatic cage of LBR-TD more strongly resembles the aromatic cage of the first hybrid Tudor domain of JMJD2A (HTD1) (Fig. 2, B and C), which is not involved in binding trimethylated lysine residues (15, 17). Given that methyllysine or methylarginine recognition by Tudor domains does not involve appreciable structural rearrangements of the aromatic cage upon binding (supplemental Fig. S2), it is therefore likely that LBR-TD is similar to HTD1 and does not recognize methylation marks.
Molecular Interactions and Functional Role of LBR-TD
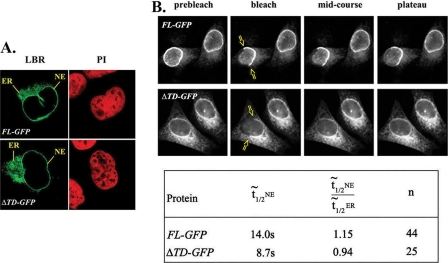
To explore the role of LBR-TD under in vivo conditions, we compared the properties of full-length LBR (FL-GFP) and a truncated LBR form lacking the entire LBR-TD module (ΔTD-GFP) in transiently transfected HeLa cells. As documented in Fig. 3A, FL-GFP and ΔTD-GFP exhibited a similar distribution, partitioning with the NE and the endoplasmic reticulum (ER). Furthermore, when a segment corresponding to half of the NE rim was photobleached (Fig. 3B, top), the two proteins recovered to a similar extent, yielding mobile fractions in the order of 0.6 and indicating that at steady-state a large fraction of the corresponding subunits does not exchange. However, assessing the relative recovery rates, we noticed that the median t[frax,1,2] calculated from plateau fluorescence was greater for FL-LBR (14.0 s) than for the mutant missing the LBR-TD (8.7 s). Moreover, when the rates of fluorescence recovery in the NE and ER were compared, the ratio of NE t[frax,1,2]/ER t[frax,1,2] was 1.15 for FL-LBR and 0.94 for the mutant (Fig. 3B and supplemental Table S1). From these data it can be inferred that absence of the Tudor domain renders NE-associated LBR relatively more mobile, presumably because the truncated protein fails to interact with some of the LBR partners.
FIGURE 3.
A, steady-state localization of full-length LBR (FL-GFP) and Tudor-truncated LBR (ΔTD-GFP) in transiently transfected HeLa cells. B, upper panel, FRAP data from transfected HeLa cells expressing FL-GFP and ΔTD-GFP LBR. Bottom panel, the table shows the median t[frax,1,2] when half of the nuclear envelope was bleached (t½NE), the ratio of median t[frax,1,2] between NE and ER and the number of independent experiments (n).
Interactions of LBR-TD in Vitro
As has been observed with other Tudor domains, no binding of LBR-TD to DNA and/or RNA could be detected by electrophoretic mobility shift assays (data not shown). Furthermore, when increasing amounts of mono-, di-, and trimethylated Lys, or symmetrically and asymmetrically dimethylated Arg were added to 15N-labeled LBR-TD (up to a molar ratio of 1:25), there was no appreciable chemical shift or intensity changes in the 1H-15N HSQC spectra (data not shown), suggesting that LBR-TD does not recognize free and methylated amino acids.
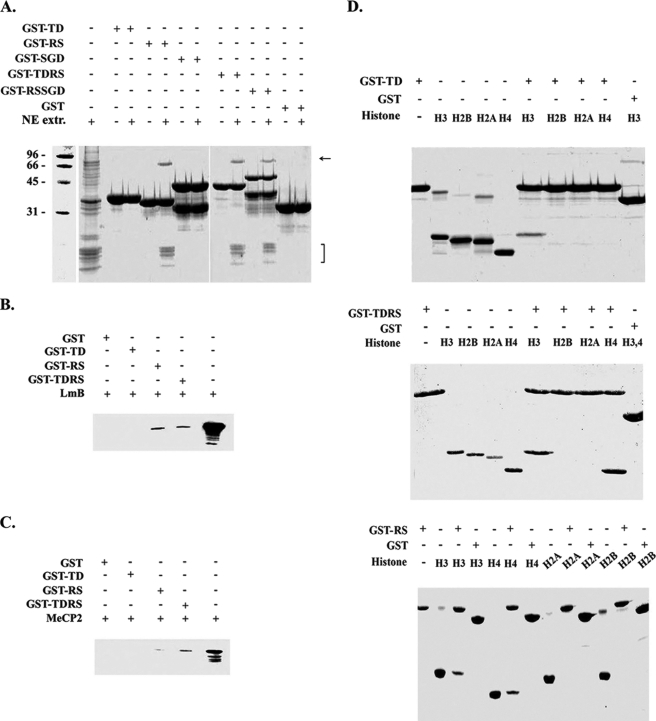
To better understand the role of LBR-TD at the molecular level, we studied its interactions with other nuclear proteins under in vitro conditions employing pull-down assays. For these purposes, we designed and expressed five recombinant proteins in bacteria, i.e. GST-TD, GST-RS, GST-SGD, GST-TDRS, and GST-RSSGD, covering the entire amino-terminal part of LBR (for a schematic diagram see Fig. 1A). When the various LBR derivatives were co-incubated with a NE-peripheral heterochromatin extract (for details and characterization, see Ref. 28), LBR-TD did not appear to bind any of the components present in the extract. However, GST-RS and all proteins containing the RS motif co-precipitated core histones and a 68-kDa polypeptide corresponding to nuclear lamin B (Fig. 4A). Lamin binding could be directly confirmed by co-incubating each recombinant protein with purified, rat liver lamin B and probing the corresponding precipitate with specific anti-lamin B antibodies (Fig. 4B). Although a band with appropriate Mr was not immediately obvious in the initial precipitates, specific binding was also detected when GST-RS and GST-TDRS were co-incubated with purified MeCP2, a chromatin protein shown to interact with LBR (Fig. 4C). Under the same conditions GST-TD failed to associate with this polypeptide.
FIGURE 4.
GST pull-down experiments with several N-terminal LBR regions. A, GST-fused LBR-TD, LBR-RS, LBR-SGD, LBR-TDRS, and LBR-RSSGD were incubated with nuclear envelope extracts, produced after MNase digestion of turkey erythrocyte nuclei. Bound proteins were eluted from GST beads with SDS sample buffer and visualized with Coomassie staining. B, GST-fused LBR-TD, LBR-RS, and LBR-TDRS were also incubated with: lamin B (B), recombinant MeCP2 (C), and purified native core histones (D).
LBR has also been shown to bind the isolated histones H3 and H4 (30). Based on this and taking into consideration the suggested role of the Tudor domains as potential “histone code readers,” we examined the in vitro interactions of LBR-TD, GST-RS, and GST-TDRS with native core histones isolated from calf thymus. As shown in Fig. 4D, LBR-TD bound efficiently to histone H3, but did not associate with histones H4, H2A, or H2B. On the other hand, both GST-RS and GST-TDRS bound specifically to histones H3 and H4.
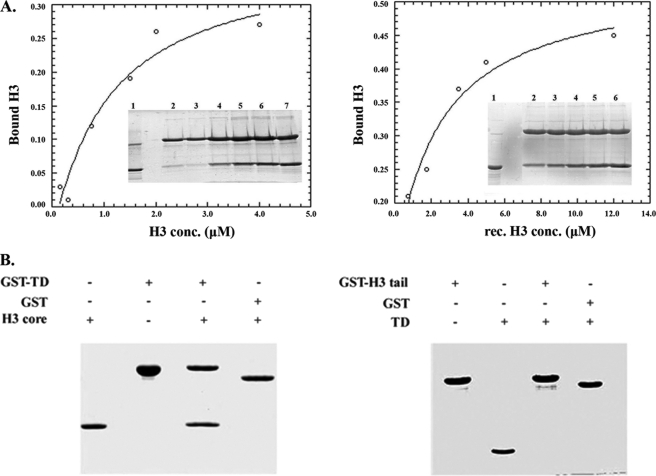
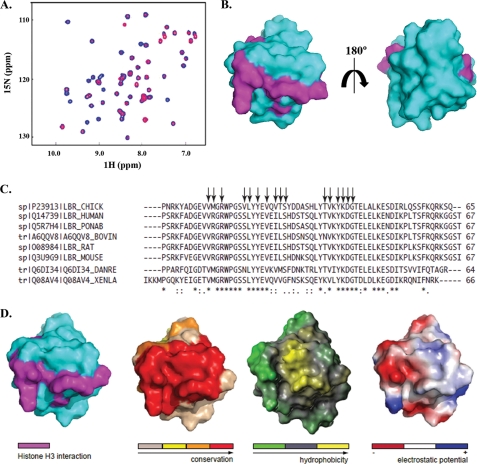
To ascertain that GST-TD binding to isolated H3 was specific, we repeated the assay in a quantitative fashion and under stringent ionic conditions, using both recombinant and native histones. As shown in Fig. 5A, binding was nearly stoichiometric, saturable, and could still occur in 1 m salt. Furthermore, these observations could be confirmed by NMR-based titration experiments, in which increasing amounts of calf thymus H3 were added to 15N-labeled LBR-TD and spectral changes were monitored by recording 1H-15N HSQC spectra. Extensive broadening and disappearance of several amide resonances upon H3 addition indicated that a LBR-TD·H3 complex was formed (Fig. 6A), presumably with an off-rate corresponding to an “intermediate exchange” binding regime (55). Histone interactions with their partners, such as chaperones or other chromatin-related proteins, are known to be transient and often accompanied by conformational changes upon binding (56). Therefore, their interface is highly dynamic and this is reflected in the NMR spectra of their complexes, although with the data at hand we cannot differentiate line broadening due to on/off binding exchange and conformational dynamics of the bound state. Residues perturbed by the interaction were identified based on the percentage of reduced signal intensity of amide resonances in LBR-TD upon addition of H3 at 1:3 molar ratio. These residues seemed to cover an extended area on one face of the LBR-TD surface, were not localized in the aromatic cluster or its vicinity (Fig. 6B), and are highly conserved in LBR sequences from different species (Fig. 6C). Notably, the same binding behavior was observed when LBR-TD was titrated with recombinant H3, reinforcing the idea that post-translational modifications of H3 were not required for this interaction (data not shown).
FIGURE 5.
Interaction of LBR-TD with histone H3. A, semiquantitative pull-down experiment using constant amounts of GST-Tudor and native H3 in serial dilutions with buffer, as detailed under ”Experimental Procedures.“ Experiments were repeated in the same manner with recombinant H3. Inset gels shows 20% of input native H3/recombinant H3. B, interaction of LBR-TD with core and tail parts of H3. GST pull-down assays with GST-fused LBR-TD and recombinant tail-less H3 (left panel) and with LBR-TD and GST-fused H3 N-terminal tail (right panel). Panels show Coomassie Blue-stained SDS-PAGE gels and 50% of input tailless H3 (left) or LBR-TD (right).
FIGURE 6.
Features of cLBR-TD-H3 interface. A, NMR-based titration experiment using 15N-labeled LBR-Tudor and increasing amounts of native or recombinant histone H3. A representative overlay of HSQC spectra for LBR-TD (blue) and for equimolar amounts of H3 and LBR-TD (red) is shown. B, mapping the interaction surface of histone H3 onto the LBR-TD structure of LBR. Surface representations of the LBR-TD structure that differ by a 180º rotation around the y axis. The side chains of residues with perturbed amide resonances upon titration with H3 are shown in magenta and the LBR-TD surface in light blue. C, sequence alignment of the LBR-TD from several species (from top to bottom: chicken, human, Pongo abelii, bovine, rat, mouse, zebrafis, and X. laevis). The conservation of residues across species is shown under the alignment. Symbols denote identical residues (*), highly similar residues (:) and similar residues (.). Arrows point to the amino acids comprising the interaction surface between cLBR-TD and H3 (identified by the NMR-chemical shift perturbation experiments). D, sequence conservation, hydrophobicity, and electrostatic potential of the H3 binding surface of cLBR-TD. Mapping on the surface of (from left to right): the residues of cLBR-TD comprising the interaction surface with H3, sequence conservation, hydrophobicity, and charge separation.
To get some insight on the type of contacts that could stabilize the LBR-TD/H3 interaction, we also examined the charge and the hydrophobic character of the binding surface (Fig. 6D). Based on this analysis, we can safely conclude that the binding is not solely dependent on electrostatic interactions, because there is a distinctly charged patch only in one part of the interaction surface, whereas the central part of the interface seems to be highly hydrophobic (as a result of the solvent exposed side chains of Val20, Leu21, and Tyr23). These observations imply that binding between LBR-TD and H3 is mediated by both electrostatic and hydrophobic interactions and is probably conserved in different organisms.
To identify the region of the histone H3 that was responsible for LBR-TD binding, we utilized different H3 fragments and assessed binding of LBR-TD to either H3 tails or tail-less H3. As shown in Fig. 5B, LBR-TD did not bind to the former peptide, but exhibited robust binding to the latter. Therefore, the association between the two proteins seems to be mediated by a region of the H3 molecule located in the so-called “histone-fold” domain.
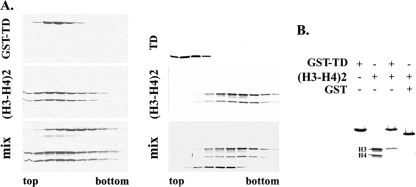
Puzzled by the fact that LBR-TD is able to associate with unassembled histone H3 but fails to bind core histone octamers present in NE-peripheral heterochromatin extracts, we repeated the experiments using an assortment of chromatin particles and subparticles. The results of these experiments were consistently negative (data not shown), except for H3-H4 tetramers isolated from salt-dissociated chromatin and hydroxylapatite column chromatography. As shown in Fig. 7A, both GST-TD and unfused TD obtained after cleavage with TEV protease exhibited mobility shifts when combined with H3-H4 tetramers and analyzed by rate zonal centrifugation in sucrose gradients. However, when an analogous experiment with H3-H4 tetramers was done in a column chromatography format (passing the histone oligomers through a glutathione-GST-TD column), we discovered that LBR-TD, instead of binding stably to H3-H4 tetramers, was apparently “stealing” H3 from the subparticles (Fig. 7B). From these results it would appear that binding of histone H3 to LBR-TD is antagonistic to binding to histone H4. This interpretation is graphically presented in the model shown in Fig. 8 and discussed in detail below.
FIGURE 7.
Interaction of LBR-TD with H3-H4 tetramers. LBR-TD either fused with GST (left panel) or unfused (right panel) was mixed with H3-H4 tetramers and run in 5–20% sucrose gradients. Fractions were collected and analyzed in SDS-PAGE gels stained with Coomassie Brilliant Blue. B, GST-fused LBR was incubated with isolated H3-H4 tetramer. Bound proteins were eluted from GST beads with SDS sample buffer and analyzed in a SDS-PAGE gel, stained with Coomassie Brilliant Blue.
FIGURE 8.
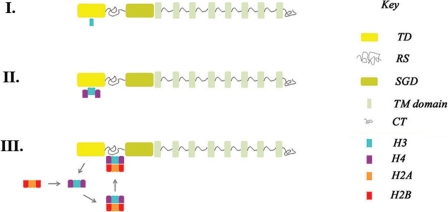
Hypothetical model for “chaperone-like” function of LBR-TD. According to this model, LBR-TD operates as a transient docking site for “free” H3 (I). Subsequent H3-H4 binding (II) triggers release of H3-H4 tetramers and deposition of nucleosome assembly intermediates to the adjacent RS region (III).
DISCUSSION
In this study we have determined the tertiary structure of chicken LBR-TD in solution and examined its interactions with a variety of substrates and potential binding partners. As expected, LBR-TD was found to adopt the characteristic β-barrel-fold of other Tudor domains and was apparently required for normal LBR dynamics in living cells. However, in contrast to a previously published report (33), our results showed that LBR-TD was not sufficient for lamin B binding in vitro, and that this interaction also required the adjacent Arg/Ser (RS) region.
Unlike other Tudor domains, LBR-TD did not associate with unmodified or modified Arg/Lys residues. Thus, no binding to free or methylated amino acids, the LBR-RS, Lys and Arg-rich histone tails, or to intact nucleosomal core particles was observed in vitro. This rules out the possibility that LBR-TD might operate as an “RS-trap” or as a “Lys/Arg modification reader,” in the fashion that other Tudor domains bind to Arg- and Lys-methylated proteins and peptides, Arg-Gly-rich sequences or free amino acids (4–19, 57). At a first glance this may seem paradoxical, because LBR-TD does contain an aromatic cage, similar to the second hybrid Tudor domain of JMJD2A, the chromodomain of heterochromatin protein 1 and other Royal family domains that can accommodate methylated Lys residues (2, 20–22). However, recognition of symmetrically dimethylated arginine by Tudor domains has been shown to require four aromatic residues in the binding pocket (Refs. 12 and 13 and Figs. 2A and supplemental Fig. S3), whereas the aromatic cage of LBR-TD comprises only three aromatic residues (Figs. 1E and 2B) and may therefore be incapable of accommodating a methylated guanidium group (supplemental Fig. S3). Furthermore, based on the structural data that are available, the same argument holds for dimethyllysine recognition, because 53BP1 utilizes four aromatic residues to line the binding pocket (14, 59). In a recent, comprehensive review article on LBR (26), the authors compared the structure and the aromatic cage of human LBR-TD with the ones from other Tudor domains and suggested that LBR-TD and 53BP1-TD, being structurally similar, might recognize the same ligand, i.e. H4-K20me2. However, based on the structure of the 53BP1·H4-K20me2 complex, the binding pocket of 53BP1 has been described before as a distinctly compact aromatic cage with four aromatic residues coming in direct contact with the methylated side chains and a fifth aromatic residue packing tightly against the ligand peptide backbone (14). The same mode of dimethyllysine recognition by 53BP1-TD has also been identified in the complex it forms with a p53-K382me2 peptide (58). Therefore, the aromatic cage of LBR-TD differs significantly from that of 53BP1 (see also supplemental Fig. S3). The only exception in the Tudor tendency to construct pockets with four aromatic residues for caging the methylated marks are the two hybrid Tudor domains of JMJD2A that form an interdigitated structure (15, 17). Although in both hybrid domains of JMJD2A the aromatic cages consist of three aromatic residues and an aspartate residue like the one found in LBR-TD, only the second of the two hybrid lobes (HTD2) is able to bind peptides containing a trimethyllysine residue (because the aromatic side chains are positioned orthogonally with respect to each other, which in turn generates sufficient space for accommodating the large trimethylammonium group). In the first hybrid lobe of JMJD2A (HTD1) the histidine side chain is oriented toward the inside of the cage-like enclosure and occupies a position that occludes caging of any methylated Lys. In LBR-TD, Trp16 has the same orientation as the histidine residue of HTD1-JMJD2A (supplemental Fig. S3) and (most likely) precludes LBR-TD from binding methylation marks. It should be noted that the relative disposition of the aromatic side chains of LBR-TD is well defined due to the large number of NOEs detected (supplemental Fig. S1). In addition, the arrangement of these residues in the chicken LBR-TD structure presented here and in the human LBR-TD structure previously determined is very similar (supplemental Fig. S4). In short, the aromatic pocket of LBR is too “open” to allow efficient caging of Arg or Lys dimethylated side chains and too restrictive for the trimethylated Lys side chain to fit.
Invariably, recognition of methylated ligands by the Royal family of domains depends largely on electrostatic stabilization mediated by cation-π interactions. Therefore, the surface of the binding pocket has a negatively charged electrostatic potential to attract the cationic moiety of the Lys or Arg residues. In LBR-TD the surface surrounding the cluster of aromatic residues is not as negatively charged as that of Tudor domains known to bind methylated marks (supplemental Fig. S5). Taken together, despite the overall fold similarity with other Tudor domains, these differences in the spatial arrangement of the aromatic rings that build the binding pocket in LBR-TD prohibit stable binding of the so-far characterized Tudor ligands.
Interestingly, a protein array approach for identifying novel methyllysine-dependent interactions of several Royal family domains, in agreement with our experimental observations and detailed structural analysis, also failed to detect any interaction between LBR-TD and variably methylated H3 or H4 tails (16, 59). This work was based on a protein-domain microarray screening and tested, among others, human LBR-TD as one of several chromatin-associated domains that might specifically recognize histone H3 and H4 tail peptides methylated to varying degrees on specific lysine or arginine residues. The results of this investigation revealed that LBR-TD failed to bind to any of the peptides used as baits, among them the H4-K20me2, but also H3-K79me2, H3-K4me1,2,3, H3-K9me1,2,3, and H4-K20me1,2,3. On the contrary, the Tudor domain of 53BP1 was found to recognize H3 and H4 peptides bearing dimethyl groups on H4-K20, H3-K4, and H3-K9. Nevertheless, we cannot exclude the possibility that LBR-TD could recognize a larger peptide ligand in the context of the full-length LBR protein as has been observed for methyllysine binding PHD-containing proteins (60).
Instead of mediating binding to methylated amino acids, we found that LBR-TD binds tightly and selectively to isolated histone H3 through a patch on its surface distinct from the aromatic cage and comprised of residues highly conserved in LBR sequences from different species. The binding is mediated by both electrostatic and hydrophobic interactions and is probably conserved in different organisms. In addition, our data indicate that H3-H4 oligomers and fully assembled nucleosomes interact with the RS region of LBR, which is physically contiguous with LBR-TD. Based on these findings, we propose a model (Fig. 8), whereby LBR-TD may in fact operate as a transient docking site or as a storage chaperone for newly imported histone H3 molecules, as, for example, CIA/ASF1 (56, 60, 61). In this scenario, H3-H4 binding may occur at a later phase, following chaperoning by LBR-TD. The assembly of H3-H4 tetramers might trigger their release from the Tudor domain and allow the deposition of nucleosome assembly intermediates (or complete histone octamers) to the adjacent RS region. Interestingly, an antiparallel β-sheet motif similar to that of LBR-TD has been recognized before as a recurring theme in histone chaperones and as a scaffold for histone binding elements (61, 62). Certainly, at this point and in the lack of sound experimental evidence, this is only a hypothesis and needs to be validated in a concrete biological context. However, this hypothesis highlights an alternative scenario for the functional role of LBR in the nuclear envelope, which remains elusive, despite its experimentally documented association with chromatin and nuclear lamina (26). This scenario is compatible with the presumed role of nuclear envelope as a platform for peripheral heterochromatin assembly and chromatin remodeling, based on a large body of experimental evidence (24–27).
Tudor domains are found in many proteins and serve distinct functions. To some extent, these diverse binding properties match the various different arrangements of the Tudor domains, which occur as single (SMN and LBR), or as tandem units (53BP1 and UHRF1). An interdigitated arrangement of tandem Tudor domains (JMJD2A), or an insertion of the Tudor domain into another fold (SND1), have also been observed. In addition, there are proteins that contain multiple repeats of such domains. For example, the Drosophila Tudor protein has 11 copies of the Tudor domain, whereas in the TDRD family the numbers range from 1 to 8. Based on their primary sequences and the general mechanisms used to recognize methylated ligands, it is not expected that all Tudor domains represent genuine methyl binders. Instead, it is likely that the stable Tudor-fold might serve architectural purposes and can be used as a platform for orienting other functional domains in the full-length protein. Such a role may apply to LBR-TD, because this domain adopts a stable globular structure and may thus assist in organizing the rest of the molecule and coordinating its interactions with chromatin particles.
Supplementary Material
Acknowledgments
We thank G. Stier (EMBL) for the generous gift of the pETM-30 plasmid, K. Luger (University of Colorado) for H3 plasmids, M. Grunstein (University of California) for H3-tail plasmids, G. Badaracco (University of Insubria) for MeCP2 plasmids and antibodies, E. Nikolakaki (Aristotle University of Thessaloniki) for help with cloning experiments at the early stages of this work, Y. Markaki and D. Makatsori (University of Ioannina) for providing advice with the nuclear envelope extract isolation and GST pull-down experiments, B. Simon (EMBL) for help with NMR titration experiments, A. Lustig (Biozentrum, University of Basel) for Analytical Ultracentrifugation experiments, and T. Papamarcaki (University of Ioannina) for materials. We thank the confocal laser microscope facility of the University of Ioannina for use of the Leica TCS-SP scanning confocal microscope.
Note Added in Proof
The results shown in Fig. 4, A, B, and C with GST-RS, GST-TDRS, GST-RSSGD are qualitatively the same when the LBR fusion proteins are either pretreated or treated during incubation with RNase to remove potential nucleic acid contamination originating from bacterial extracts.
This work was supported by the Greek Ministry of Education and the European Commission Program “Pythagoras II,” Greek General Secretariat for Research and Development Program “PENED2003,” and Grant IKYDAD 2004 Programme of Scientific Exchange and Cooperation between Greece and Germany.

This article contains supplemental Tables S1 and Figs. S1–S5.
The atomic coordinates and structure factors (code 2L8D) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SMN
- survival motor neuron protein
- LBR
- lamin B receptor
- LBR-TD
- LBR-Tudor domain
- FRAP
- fluorescence recovery after photobleaching
- RS
- Arg/Ser motifs
- NE
- nuclear envelope
- LBR-NT
- amino-terminal segment of LBR
- FL-GFP
- full-length LBR-GFP
- ΔTD
- truncated LBR form lacking the entire LBR-TD
- ER
- endoplasmic reticulum
- SGD
- second globular domain
- HSQC
- heteronuclear single quantum correlation
- PDB
- Protein Data Bank
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Talbot K., Miguel-Aliaga I., Mohaghegh P., Ponting C. P., Davies K. E. (1998) Hum. Mol. Genet. 7, 2149–2156 [DOI] [PubMed] [Google Scholar]
- 2. Maurer-Stroh S., Dickens N. J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C. P. (2003) Trends Biochem. Sci. 28, 69–74 [DOI] [PubMed] [Google Scholar]
- 3. Ponting C. P. (1997) Trends Biochem. Sci. 22, 51–52 [DOI] [PubMed] [Google Scholar]
- 4. Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M. (2001) Nat. Struct. Biol. 8, 27–31 [DOI] [PubMed] [Google Scholar]
- 5. Sprangers R., Groves M. R., Sinning I., Sattler M. (2003) J. Mol. Biol. 327, 507–520 [DOI] [PubMed] [Google Scholar]
- 6. Bühler D., Raker V., Lührmann R., Fischer U. (1999) Hum. Mol. Genet. 8, 2351–2357 [DOI] [PubMed] [Google Scholar]
- 7. Brahms H., Meheus L., de Brabandere V., Fischer U., Lührmann R. (2001) RNA 7, 1531–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Côté J., Richard S. (2005) J. Biol. Chem. 280, 28476–28483 [DOI] [PubMed] [Google Scholar]
- 9. Shaw N., Zhao M., Cheng C., Xu H., Saarikettu J., Li Y., Da Y., Yao Z., Silvennoinen O., Yang J., Liu Z. J., Wang B. C., Rao Z. (2007) Nat. Struct. Mol. Biol. 14, 779–784 [DOI] [PubMed] [Google Scholar]
- 10. Friberg A., Corsini L., Mourão A., Sattler M. (2009) J. Mol. Biol. 387, 921–934 [DOI] [PubMed] [Google Scholar]
- 11. Kirino Y., Vourekas A., Sayed N., de Lima Alves F., Thomson T., Lasko P., Rappsilber J., Jongens T. A., Mourelatos Z. (2010) RNA 16, 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu K., Chen C., Guo Y., Lam R., Bian C., Xu C., Zhao D. Y., Jin J., MacKenzie F., Pawson T., Min J. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 18398–18403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu H., Wang J. Y., Huang Y., Li Z., Gong W., Lehmann R., Xu R. M. (2010) Genes Dev. 24, 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Botuyan M. V., Lee J., Ward I. M., Kim J. E., Thompson J. R., Chen J., Mer G. (2006) Cell 127, 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Y., Fang J., Bedford M. T., Zhang Y., Xu R. M. (2006) Science 312, 748–751 [DOI] [PubMed] [Google Scholar]
- 16. Kim J., Daniel J., Espejo A., Lake A., Krishna M., Xia L., Zhang Y., Bedford M. T. (2006) EMBO Rep. 7, 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee J., Thompson J. R., Botuyan M. V., Mer G. (2008) Nat. Struct. Mol. Biol. 15, 109–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ramos A., Hollingworth D., Adinolfi S., Castets M., Kelly G., Frenkiel T. A., Bardoni B., Pastore A. (2006) Structure 14, 21–31 [DOI] [PubMed] [Google Scholar]
- 19. Charier G., Couprie J., Alpha-Bazin B., Meyer V., Quéméneur E., Guérois R., Callebaut I., Gilquin B., Zinn-Justin S., (2004) Structure 12, 1551–1562 [DOI] [PubMed] [Google Scholar]
- 20. Corsini L., Sattler M. (2007) Nat. Struct. Mol. Biol. 14, 98–99 [DOI] [PubMed] [Google Scholar]
- 21. Jacobs S. A., Khorasanizadeh S. (2002) Science 295, 2080–2083 [DOI] [PubMed] [Google Scholar]
- 22. Nielsen P. R., Nietlispach D., Mott H. R., Callaghan J., Bannister A., Kouzarides T., Murzin A. G., Murzina N. V., Laue E. D. (2002) Nature 416, 103–107 [DOI] [PubMed] [Google Scholar]
- 23. Worman H. J., Yuan J., Blobel G., Georgatos S. D. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 8531–8534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilson K. L., Foisner R. (2010) Cold Spring Harb. Perspect. Biol. 2, a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Georgatos S. D. (2001) EMBO J. 20, 2989–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Olins A. L., Rhodes G., Welch D. B., Zwerger M., Olins D. E. (2010) Nucleus 1, 53–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schirmer E. C., Foisner R. (2007) Exp. Cell Res. 313, 2167–2179 [DOI] [PubMed] [Google Scholar]
- 28. Makatsori D., Kourmouli N., Polioudaki H., Shultz L. D., McLean K., Theodoropoulos P. A., Singh P. B., Georgatos S. D. (2004) J. Biol. Chem. 279, 25567–25573 [DOI] [PubMed] [Google Scholar]
- 29. Nikolakaki E., Simos G., Georgatos S. D., Giannakouros T. (1996) J. Biol. Chem. 271, 8365–8372 [DOI] [PubMed] [Google Scholar]
- 30. Polioudaki H., Kourmouli N., Drosou V., Bakou A., Theodoropoulos P. A., Singh P. B., Giannakouros T., Georgatos S. D. (2001) EMBO Rep. 2, 920–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nikolakaki E., Drosou V., Sanidas I., Peidis P., Papamarcaki T., Iakoucheva L. M., Giannakouros T. (2008) Biochim. Biophys. Acta 1780, 214–225 [DOI] [PubMed] [Google Scholar]
- 32. Ye Q., Callebaut I., Pezhman A., Courvalin J. C., Worman H. J. (1997) J. Biol. Chem. 272, 14983–14989 [DOI] [PubMed] [Google Scholar]
- 33. Lin F., Noyer C. M., Ye Q., Courvalin J. C., Worman H. J. (1996) Hepatology 23, 57–61 [DOI] [PubMed] [Google Scholar]
- 34. Guarda A., Bolognese F., Bonapace I. M., Badaracco G. (2009) Exp. Cell Res. 315, 1895–1903 [DOI] [PubMed] [Google Scholar]
- 35. Ye Q., Worman H. J. (1994) J. Biol. Chem. 269, 11306–11311 [PubMed] [Google Scholar]
- 36. Sambrook J., Fritsch E. F., Maniatis T. M. (1989) in Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Luger K., Rechsteiner T. J., Flaus A. J., Waye M. M., Richmond T. J. (1997) J. Mol. Biol. 272, 301–311 [DOI] [PubMed] [Google Scholar]
- 38. Ling X., Harkness T. A., Schultz M. C., Fisher-Adams G., Grunstein M. (1996) Genes Dev. 10, 686–699 [DOI] [PubMed] [Google Scholar]
- 39. Oganesyan N., Kim S. H., Kim R. (2005) J. Struct. Funct. Genomics 6, 177–182 [DOI] [PubMed] [Google Scholar]
- 40. Georgatos S. D., Blobel G. (1987) J. Cell Biol. 105, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Blobel G., Potter V. R. (1966) Science 154, 1662–1665 [DOI] [PubMed] [Google Scholar]
- 42. Maison C., Horstmann H., Georgatos S. D. (1993) J. Cell Biol. 123, 1491–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Simon R. H., Felsenfeld G. (1979) Nucleic Acids Res. 6, 689–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 45. Johnson B. A. (2004) Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 46. Sattler M., Schleucher J., Griesinger C. (1999) Prog. NMR Spectrosc. 34, 93–158 [Google Scholar]
- 47. Yamazaki T., Forman-Kay J. D., Kay L. E. (1993) J. Am. Chem. Soc. 115, 11054–11055 [Google Scholar]
- 48. Herrmann T., Güntert P., Wüthrich K. (2002) J. Biomol. NMR 24, 171–189 [DOI] [PubMed] [Google Scholar]
- 49. Cornilescu G., Delaglio F., Bax A. (1999) J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 50. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 51. Vriend G. (1990) J. Mol. Graph. 8, 52–56 [DOI] [PubMed] [Google Scholar]
- 52. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 53. Delano W. L. (2002) The Pymol Molecular Graphics System, Schrödinger, LLC, New York [Google Scholar]
- 54. Holm L., Sander C. (1995) Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 55. Matsuo H., Walters K. J., Teruya K., Tanaka T., Gassner G. T., Lippard S. J., Kyogoku Y., Wagner G. (1999) J. Am. Chem. Soc. 121, 9903–9904 [Google Scholar]
- 56. Mousson F., Lautrette A., Thuret J. Y., Agez M., Courbeyrette R., Amigues B., Becker E., Neumann J. M., Guerois R., Mann C., Ochsenbein F. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5975–5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grenon M., Costelloe T., Jimeno S., O'Shaughnessy A., Fitzgerald J., Zgheib O., Degerth L., Lowndes N. F. (2007) Yeast 24, 105–119 [DOI] [PubMed] [Google Scholar]
- 58. Roy S., Musselman C. A., Kachirskaia I., Hayashi R., Glass K. C., Nix J. C., Gozani O., Appella E., Kutateladze T. G. (2010) J. Mol. Biol. 398, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang Y., Lu Y., Espejo A., Wu J., Xu W., Liang S., Bedford M. T. (2010) Mol. Cell 40, 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Horton J. R., Upadhyay A. K., Qi H. H., Zhang X., Shi Y., Cheng X. (2010) Nat. Struct. Mol. Biol. 17, 38–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Natsume R., Eitoku M., Akai Y., Sano N., Horikoshi M., Senda T. (2007) Nature 446, 338–341 [DOI] [PubMed] [Google Scholar]
- 62. Park Y. J., Luger K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1248–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.