Background: How does a limited pool of <108 T cell receptors (TCRs) provide immunity to >1015 antigens?
Results: A single TCR can respond to >one million different decamer peptides.
Conclusion: This unprecedented level of receptor promiscuity explains how the naïve TCR repertoire achieves effective immunity.
Significance: TCR degeneracy has enormous potential to be the root cause of autoimmune disease.
Keywords: Antigen Presentation, Major Histocompatibility Complex (MHC), T Cell, T Cell Biology, T Cell Receptor, Autoimmunity, Cellular Immune Response, Diabetes, Insulin, Pancreatic Islets
Abstract
The T cell receptor (TCR) orchestrates immune responses by binding to foreign peptides presented at the cell surface in the context of major histocompatibility complex (MHC) molecules. Effective immunity requires that all possible foreign peptide-MHC molecules are recognized or risks leaving holes in immune coverage that pathogens could quickly evolve to exploit. It is unclear how a limited pool of <108 human TCRs can successfully provide immunity to the vast array of possible different peptides that could be produced from 20 proteogenic amino acids and presented by self-MHC molecules (>1015 distinct peptide-MHCs). One possibility is that T cell immunity incorporates an extremely high level of receptor degeneracy, enabling each TCR to recognize multiple peptides. However, the extent of such TCR degeneracy has never been fully quantified. Here, we perform a comprehensive experimental and mathematical analysis to reveal that a single patient-derived autoimmune CD8+ T cell clone of pathogenic relevance in human type I diabetes recognizes >one million distinct decamer peptides in the context of a single MHC class I molecule. A large number of peptides that acted as substantially better agonists than the wild-type “index” preproinsulin-derived peptide (ALWGPDPAAA) were identified. The RQFGPDFPTI peptide (sampled from >108 peptides) was >100-fold more potent than the index peptide despite differing from this sequence at 7 of 10 positions. Quantification of this previously unappreciated high level of CD8+ T cell cross-reactivity represents an important step toward understanding the system requirements for adaptive immunity and highlights the enormous potential of TCR degeneracy to be the causative factor in autoimmune disease.
Introduction
The mammalian T cell receptor (TCR)6 orchestrates immune responses by binding to foreign peptides presented at the cell surface in the context of major histocompatibility complex (MHC) molecules. Recognition is mediated by the highly variable complementarity-determining regions of the αβ TCR (1, 2). A priori, the TCR repertoire must be broad enough to respond to all foreign peptides that can bind to self-MHC molecules (3). If this were not the case, then pathogens could rapidly evolve to exploit such deficiencies in immune coverage. Current estimates of human αβ TCR diversity suggest that there are <108 different antigen receptors in the naïve T cell pool (4), a number that is dwarfed by the potential number of antigenic peptide-MHC molecules that could be encountered. Although next generation sequencing technologies may lead to an increased estimate of TCR diversity, such future revisions are unlikely to alter the fact that a relatively small number of TCRs must, and do, provide effective immune recognition of all peptides that can be generated from 20 proteogenic amino acids and that also bind self-MHC molecules (>1015 distinct peptide-MHCs). This represents a particular biochemical challenge to the immune system because the TCR, unlike the B cell receptor, cannot undergo affinity maturation in the form of somatic hypermutation.
It is unclear how the limited naïve T cell pool responds to a multitude of ligands that it has never encountered before and cannot adapt to at the protein sequence level. One possibility is that T cell immunity inherently features an extremely high level of receptor degeneracy, enabling each TCR to recognize multiple peptides. However, clonal selection theory suggests that individual T cells are specific for a single peptide-MHC molecule with recognition of alternative ligands unlikely. In contrast, studies published in the 1990s demonstrated that T cells can recognize several peptides (5–11). Since then, observations of TCR degeneracy have continued to accumulate in the literature (12–19). In addition, other studies have shown that TCRs can recognize distinct peptides in the context of non-self-MHC, a phenomenon known as alloreactivity (20–25). The majority of previous studies of TCR degeneracy have examined sets of between 2 and 200 peptides, with one recent study examining ∼4,000 peptides (19). Given that the entire universe of decamer peptides alone comprises >1013 distinct amino acid sequences, the proportion of the peptide universe at this length that has been examined in the most comprehensive study to date (19) remains extremely small (<0.000000045%).
The aim of this study was to probe the entire decamer peptide universe systematically to quantify how many peptides a single TCR can recognize in the context of a single MHC molecule. We demonstrate an unprecedented level of receptor degeneracy that allows a single monoclonal T cell to respond to >one million distinct peptides. As such, the TCR represents one of the most remarkable biological receptors and by far the most promiscuous known.
EXPERIMENTAL PROCEDURES
Generation and Maintenance of an Autoimmune CD8+ T Cell Clone
The 1E6 CD8+ T cell clone specific for the human leukocyte antigen (HLA) A*0201-restricted autoantigen preproinsulin peptide ALWGPDPAAA (PPI15–24) was generated as described previously (26).
Decamer Combinatorial Peptide Library (CPL) Scan
The decamer combinatorial peptide library (Pepscan) contains a total of 9.36 × 1012 (= (10 + 19) ×199) different decamer peptides and is divided into 200 different peptide mixtures (see Fig. 1). In every peptide mixture, one position has a fixed l-amino acid residue and all other positions are degenerate, with the possibility of any one of 19 natural l-amino acids being incorporated in each individual position (cysteine is excluded). Each library mixture consists of 3.2 × 1011 (199) different decamer peptides in approximately equimolar concentrations. For CPL screening, 1E6 CD8+ T cells were washed and rested overnight in RPMI 1640 medium containing 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mm l-glutamine, and 2% heat-inactivated fetal calf serum (all Invitrogen). In 96-well U-bottom plates, 6 × 104 C1R-A2 cells were incubated with various peptide library mixtures (at 100 μg/ml) in duplicate for 2 h at 37 °C. Following peptide pulsing, 3 × 104 1E6 CD8+ T cells were added, and the assay was incubated overnight at 37 °C. Subsequently, the supernatant was harvested and assayed for MIP1β by ELISA according to the manufacturer's instructions (R&D Systems).
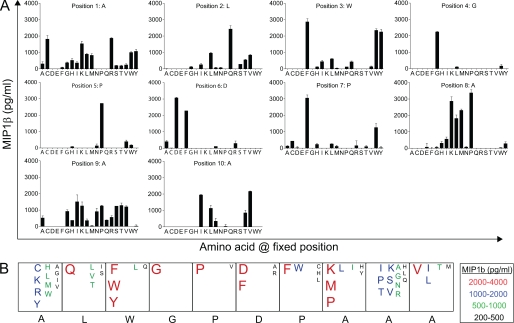
FIGURE 1.
Decamer CPL scan of the 1E6 CD8+ T cell clone. A, 6 × 104 C1R-A2 cells were pulsed in duplicate with each mixture from a decamer CPL (100 μg/ml) at 37 °C. After 2 h, 3 × 104 1E6 CD8+ T cells were added and incubated overnight. Supernatant was harvested and assayed for MIP1β. B, data from A are displayed as a box plot summary. The index insulin peptide sequence is shown below the boxes in black.
CD8+ T Cell Effector Function Assays: MIP1β ELISA
Individual peptides were assayed for agonist activity by MIP1β ELISA as described above. Functional sensitivity is expressed by the pEC50 of each peptide with respect to the TCR. This is defined as −1 × the base 10 logarithm (p) of the 50% efficacy concentration (EC50); a greater functional sensitivity is indicated by a larger pEC50 value, which was estimated as described in supplemental Equations.
Overview of Sampling Approaches Used to Quantify TCR Degeneracy
Although the TCR has an appreciable degeneracy, it is still specific enough that sampling peptides at random will most likely result in less than ∼10 strong agonists for every ∼10,000 peptides sampled. For this reason, we employed conditioned sampling. In one approach, we sampled from motif-restricted peptide sets. This results in lower bound estimates of the actual number of agonist ligands, in that any agonist not fitting the prescribed motif is excluded from the sample. In general, more stringent motifs exclude more agonists but provide better resolution at the high pEC50 end of the curve, and vice versa; for this reason, a range of motifs of varying stringency was used. In a second approach (CPL-based importance sampling), we sampled from the entire peptide universe with bias toward peptides that were likely to elicit a response, then estimated a true distribution by applying a correction weighting to the observations (i.e. dividing back by the bias).
Sampling Equations
Motif-restricted Sampling
A sampling motif specifies, for each of the m positions in an m-mer peptide, one or more amino acid residues that may occur at that position. If np alternative residues have been specified at position p, the probability that a sampled peptide has a given residue at position p equals 1/np if the given residue is one of the np given alternatives, and zero otherwise. Consider a sample of n peptides, consisting of peptides P[1], P[2], …, P[n], with measured functional sensitivities pEC50[1], pEC50[2], …, pEC50[n]. The cross-reactivity of the TCR is expressed by the number of peptides whose pEC50 exceeds a given value ω. The degeneracy of the TCR is then represented by determining this number for a range of ω values, estimated as follows.
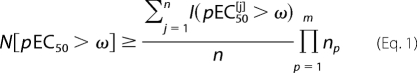 |
Here, N[pEC50 > ω] is the number of peptides that have a pEC50 for the TCR that exceeds ω, and
if sample peptide j has a pEC50 that exceeds ω;
otherwise. The sampling motif typically excludes a number of agonists (unless np = 20 at every position, i.e. the “universal motif”, which was not used). This exclusion means that the quantity on the right in Equation 1 always underestimates the true N[pEC50 > ω]. This means that the motif-based method provides a lower boundary to TCR degeneracy, i.e. a conservative estimate.
CPL-based Importance Sampling
The idea of sampling motifs can be generalized to the well known strategy of importance sampling by specifying, for all positions and amino acids, the probability that a given amino acid occurs at a given position. Then, the probability of drawing a peptide P that has amino acid residue P(p) at position p is given by
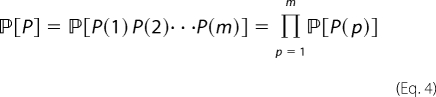 |
where ℙ[P(p)] is the probability of drawing amino acid P at position p. Standard rules of probability stipulate that for each fixed p, the ℙ[P(p)] sum to unity over the 20 amino acids. The CPL was used to generate distributions that could be expected to bias the sample toward good agonists. However, to correct for the bias in the estimate, observations must be weighted by the reciprocal of the bias. Accordingly, degeneracy was estimated as follows:
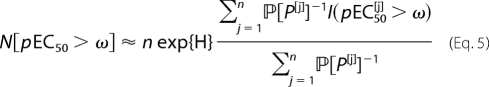 |
where H is the entropy of the sampling distribution, defined by
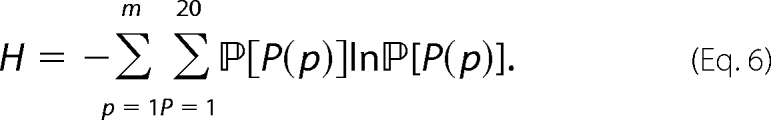 |
The factor n exp{H} in Equation 5 is an estimate of the effective sample size. This estimate is based on a standard result concerning the probability of sampling a fixed set of n items. A well known theorem (27) states that this probability is close to exp{−n H}. On the other hand, in unbiased sampling from a set of size N, the probability of obtaining any given n-element sample is exactly equal to N−n. Together, these observations indicate that exp{H} can be interpreted as the “diameter” of the population from which the peptide is drawn. Consequently, an n-element sample of m-mer peptides probes an effective set size of
the approximation is accurate when n is tiny compared with exp{−H}, as is the case in the experiments reported here.
RESULTS
CPL Screening Reveals the Potential for TCR Degeneracy
To determine the extent of T cell cross-reactivity, we probed the peptide recognition degeneracy of the autoimmune CD8+ T cell clone IE6 using a CPL comprising 9.36 × 1012 different decamer peptides (Fig. 1). Using this approach, we were able to scan every amino acid at every position of the peptide within a random residue “backbone” and build a detailed picture of the molecular landscape preferred by the 1E6 TCR. The 1E6 clone was generated from a patient with type 1 diabetes and is the only documented example of an autoreactive CD8+ T cell that can kill human pancreatic islet β-cells (26). Killing is mediated via recognition of residues 15–24 (ALWGPDPAAA) of the autoantigen preproinsulin bound to HLA A*0201 on the β-cell surface. The majority of HLA A*0201+ patients with type I diabetes recognize the preproinsulin 15–24 epitope (26), and HLA A*0201 is known to confer an increased risk of disease (28, 29). The number of amino acids that were recognized by the 1E6 clone was restricted in the central region of the peptide (residues 4–6), suggesting that this TCR makes the majority of its peptide contacts with these residues. In contrast, recognition was highly degenerate at the remaining positions. This degeneracy was confirmed by the ability of 1E6 T cells to recognize a panel of peptides robustly with any of the 20 natural proteogenic l-amino acids at peptide position 8 (supplemental Fig. S1), with half of the substitutions leading to increased levels of functional sensitivity. Similar results were obtained with corresponding scans at other degenerate positions (data not shown). The CPL scan results also revealed that the index peptide is suboptimal in all positions outside the central region (residues 4–6). Thus, positional peptide degeneracy is extreme at 7 of 10 positions, hinting at the potential for a single TCR to recognize a multitude of different amino acid combinations.
Quantifying the Number of Decamer Peptides That Can Be Recognized by a Single TCR
We next sought to quantify the number of decamer peptides that can be recognized by the 1E6 clone. The total number of decamers that can be made by combinations of the 20 natural proteogenic l-amino acids is 2010 (1.02 × 1013). Only a small proportion (∼1–3%) of all peptides are predicted to bind any given MHC (3, 19), although our own experiments predict that this percentage could be far greater for HLA A*0201 (data not shown). Even with the most conservative estimates of MHC binding (19), the number of potential antigenic HLA A*0201-restricted decamer peptides is still extremely large (1.02 × 1011) and precludes screening all possibilities in T cell recognition assays. To overcome this problem, we screened sets of 30 peptides sampled (Mathematica®; supplemental Fig. S5) from larger motif-restricted or CPL-based importance-sampled sets differing in total size from 225 to 1.66 × 108 individual peptides as described below.
Quantifying T Cell Cross-reactivity Using Motif-restricted Sampling
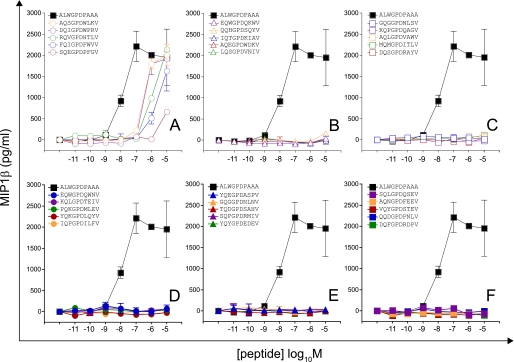
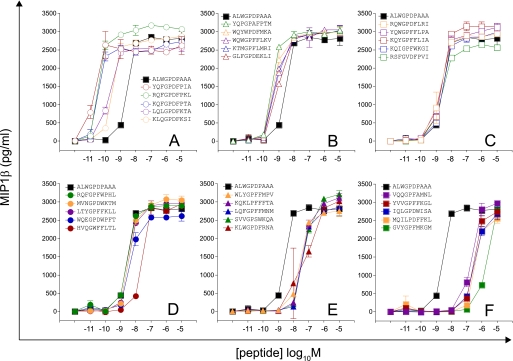
Motif-restricted peptide sets were designed based on CPL evidence for amino acid preference at each position of the peptide. First, we screened 30 peptides sampled at random from a total set size of 225 (Motif I, RQWGPDP{A/C/D/F/H/I/K/L/M/N/P/R/S/V/Y}{A/C/G/H/I/K/L/M/N/P/Q/R/S/T/V}A; Fig. 2 and supplemental Fig. S2). Values of pEC50 (−1 × the base 10 logarithm of the EC50) as a measure of functional sensitivity were estimated for all peptides using simultaneous curve fitting (as described in supplemental Equation S1, Fig. S6, and Table S3). Accordingly, increases in functional sensitivity translate into increases in the pEC50 value. The 1E6 clone recognized all peptides within this subset efficiently, with 24 of 30 peptides eliciting greater levels of functional sensitivity than the index peptide. A further 30 peptides were sampled at random from a total set size of 5,776 (Motif II, RQWGP{D/F}{P/F}XX{A/I/L/V}, where X denotes any one of the amino acids excluding cysteine (Fig. 3 and supplemental Fig. S2 and Table S3). One peptide from this subset was recognized poorly; 16 were recognized with pEC50 >7, with a total of 8 peptides recognized more efficiently than the index peptide (Fig. 3 and supplemental Fig. S2). A further two motif-restricted sets of increasing degeneracy were screened (Motif III, RQXGPDXXXA, total set size 194; and Motif IV, XQXGPDXXXV, total set size 195; X denotes any one of the amino acids excluding cysteine; Figs. 4 and 5 and supplemental Fig. S3 and Table S3). The extent of peptide recognition when peptides are sampled at random from large subsets demonstrates the considerable degree of degeneracy exhibited by the clonal 1E6 TCR.
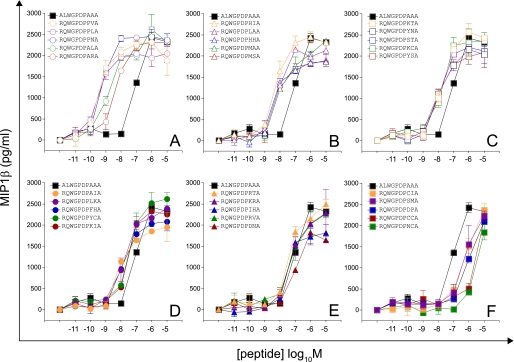
FIGURE 2.
Recognition of 30 peptides sampled at random from a large peptide set (motif, RQWGPDP{A/C/D/F/H/I/K/L/M/N/P/R/S/V/Y}{A/C/G/H/I/K/L/M/N/P/Q/R/S/T/V}A; total set size = 225). 6 × 104 C1R-A2 cells were pulsed with peptides at various concentrations. After 2 h, 3 × 104 1E6 CD8+ T cells were added and incubated overnight. Supernatant was harvested and assayed for MIP1β. Each panel displays titrations of five different peptides relative to index. A, titrations of peptides with the highest functional sensitivities. F, titrations of peptides with the lowest functional sensitivities. Error bars, S.D. from the mean of two replicates. pEC50 values for each peptide are displayed in supplemental Table S3.
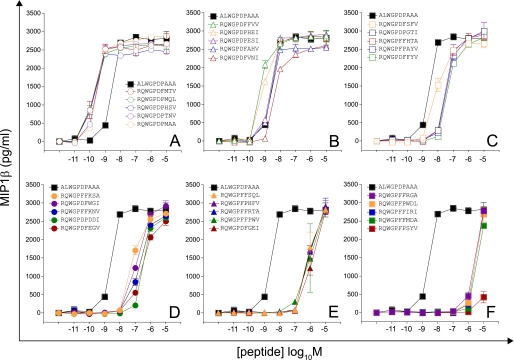
FIGURE 3.
Recognition of 30 peptides sampled at random from a large peptide set (motif,RQWGP{D/F}{P/F}XX{A/I/L/V}; total set size = 5,776). Details are as described for Fig. 2.
FIGURE 4.
Recognition of 30 peptides sampled at random from a large peptide set (motif, RQXGPDXXXA; total set size = 194). Details are as described for Fig. 2.
FIGURE 5.
Recognition of 30 peptides sampled at random from a large peptide set (motif, XQXGPDXXXV; total set size = 195). Details are as described for Fig. 2.
Quantifying T Cell Cross-reactivity Using CPL-based Importance Sampling
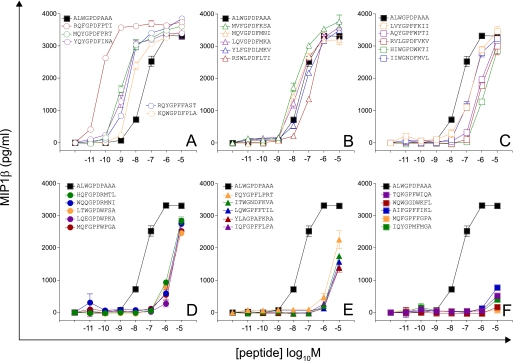
A limitation of sampling from motif-restricted sets is that strong ligands will invariably be excluded, resulting in an underestimate of the true number of agonists (i.e. a lower bound estimate of the number of different peptides that a single TCR can recognize). To obtain a more accurate estimate of true T cell cross-reactivity, we employed CPL-based importance sampling, which makes no assumptions about TCR contact or MHC class I-binding residues. Importance sampling ensures that every peptide has a chance of being sampled (although cysteine was excluded to avoid the potential for oxidation), but incorporates bias toward strong agonists predicted using CPL scan data. This bias is adjusted for to yield unbiased estimates of agonist numbers (see under “Sampling Equations”). Raw data from the primary CPL scan were modified as described in supplemental Table S1 and subsequently normalized (supplemental Table S2) to provide a peptide sampling distribution biasing the sample toward good agonists. The chance of picking a peptide is the product of the normalized weights assigned to each amino acid residue at each given peptide position. Two sets of 30 peptides were drawn from an effective set size of 1.66 × 108 (calculated from the sampling entropy; see under “Sampling Equations,” Equation 7). Of a total of 60 peptides, 34 were recognized efficiently with a pEC50 > 7, and 22 were better agonists than the index peptide (Figs. 6 and 7 and supplemental Fig. S4 and Table S3). Interestingly, just this set of 60 peptides identified four peptides with functional sensitivities 100-fold better than the index peptide, despite differing from the index peptide sequence (ALWGPDPAAA) at 6 (YQFGPDFPIA, KQFGPDFPTA) or 7 (RQFGPDFPKL, RQFGPDFPTI) positions (Figs. 6 and 7 and supplemental Fig. S4 and Table S3). Thus, CPL-based importance sampling demonstrates that a large proportion of peptides from a biased set of 1.66 × 108 peptides would be recognized and that these recognized peptides can differ considerably from the index peptide.
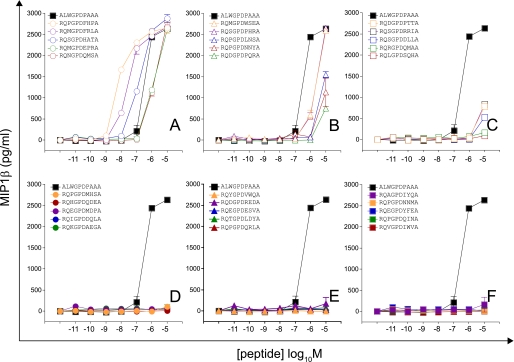
FIGURE 6.
Recognition of 30 peptides drawn from a CPL-based importance sampling set with effective size = 1. 66 × 108 (calculated from the sampling entropy) (first set). Details are as described for Fig. 2.
FIGURE 7.
Recognition of 30 peptides drawn from a CPL-based importance sampling set with effective size = 1. 66 × 108 (calculated from the sampling entropy) (second set). Details are as described for Fig. 2.
A Single TCR Can Recognize More Than One Million Different Peptides
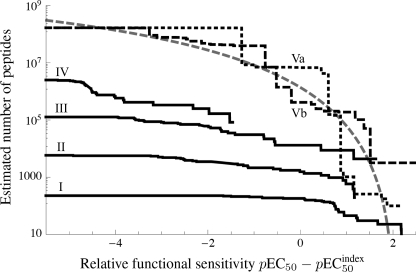
The pEC50 expresses the potency of a ligand, often referred to as “functional avidity” (supplemental Fig. S6). Cross-reactivity can be quantified precisely by specifying the number of ligands that the TCR recognizes with a pEC50 of at least a given value ω. This number decreases as ω increases; an insight into the nature of TCR degeneracy is afforded by plotting this agonist number as a function of ω. Estimation of this number was performed using Equations 1 and 5, resulting in Fig. 8. The motif-restricted estimate (solid lines in Fig. 8) is a lower boundary, which becomes tighter as the degeneracy of the motif increases; however, this advantage is offset by the decreasing chances of finding good agonists in a sample of 30 peptides. The estimates based on the biased sampling (dashed lines in Fig. 8) indicate that in the order of one million agonists exist for 1E6 that are as least as good as the index peptide. For comparison, the curve derived from TCR activation theory (30, 31) is also shown (gray dashed line in Fig. 8; supplemental Equation S2).
FIGURE 8.
1E6 CD8+ T cells can recognize more than one million different decamer peptides. I, RQWGPDP{A/C/D/F/H/I/K/L/M/N/P/R/S/V/Y}{A/C/G/H/I/K/L/M/N/P/Q/R/S/T/V}A (set size 225; 30 peptides sampled at random). II, RQWGP{D/F}{P/F}XX{A/I/L/V} (set size 5,776; 30 peptides sampled at random). III, RQXGPDXXXA (set size 194; 30 peptides sampled at random). IV, XQXGPDXXXV (set size 195; 30 peptides sampled at random). In the motifs, X denotes any one of the 19 amino acids excluding cysteine. Va and Vb, two replicates of a biased sampling set (effective set size 1.66 × 108, calculated from the sampling entropy); each set of 30 peptides was sampled with bias toward strong agonists, where the bias weights were based on the primary CPL scan. Relative functional sensitivities (pEC50 − pEC50 index) are plotted as survivor curves. Gray dashed line, theoretical curve (supplemental Equation S2, with α = 4.5, β = 10, γ = 2, N0 = 208 anchorable peptides). The biased samples (black dashed and dotted lines) estimate the TCR degeneracy spectrum, whereas the motif-based samples (black solid lines) provide a lower boundary to the TCR degeneracy spectrum.
DISCUSSION
Despite the huge potential importance of TCR degeneracy to human health, there has never been a comprehensive attempt to quantify the number of peptides that can be recognized by a single TCR. To address this issue, we examined the extent of cross-reactivity exhibited by a single autoimmune T cell clone with pathogenic relevance in human type I diabetes (1E6). Our analysis demonstrates that the 1E6 TCR can recognize ∼500 peptides within a factor 2 of the optimal agonist (i.e. peptides that have a functional sensitivity that is at least 50% of the functional sensitivity of the optimal agonist). An estimated 60,000 peptides have a functional sensitivity within a factor 10 of the optimal agonist, and ∼1.3 × 106 peptides are within a factor 100 of the optimal agonist. These considerations are especially significant given that the functional sensitivity of 1E6 CD8+ T cells for the index peptide is at least 100-fold lower than the optimal agonist; this is illustrated by RQFGPDFPTI, the functional sensitivity of which is 100-fold better than the index. Almost 10 million peptides are within a factor 1,000 of the optimal agonist, but such weak agonists will not generally be physiologically significant unless presented at very high copy numbers. Taken together, these results indicate that the 1E6 TCR has >one million significant peptide agonists at concentrations with the potential to be physiologically relevant.
When putting TCR degeneracy into perspective, it is important to realize that individual TCRs capable of recognizing 106 decamer peptides still only have a less than 1 in 107 chance of cross-reacting with any peptide chosen at random from the entire decamer peptide universe (∼1013). A high level of cross-reactivity is therefore amply compatible with the degree of specificity required for self/non-self determination. Furthermore, the number of decamer peptides that it is possible to make from the entire human proteome (excluding post-translational amino acid modifications) is only one millionth of the possible peptide universe at this length. Functional recognition of ∼106 decamer peptides by a single TCR translates into a frequency of cross-reactivity of 1:100,000 (assuming that 1% of peptides bind to MHC), which is likely to be the most accurate estimate of this parameter to date due to the comprehensive nature of our approach. The probability of cross-reactivity with any individual peptide is an important consideration in terms of viral escape, bystander activation and autoimmune side effects, and the results presented here fit well with theoretical considerations of T cell immunity (3). It should be noted, however, that the degeneracy curves are estimates based on samples that constitute a small fraction of the number of possible peptide ligands and, hence, should be considered as depicting an order-of-magnitude estimate that is supported by the lower bounds inferred from motif-based samples.
The 1E6 CD8+ T cell clone was chosen for these studies to highlight the huge potential for T cell cross-reactivity as a possible cause of autoimmunity. In support of the generality of our findings, we have also observed high levels of degeneracy at some positions in a 9-amino acid residue nonautoimmune epitope (32). Furthermore, the recognition of longer, MHC class II-restricted peptides by CD4+ T cells with a “TCR footprint” of similar size could ensure that these cells incorporate the capacity to recognize tens, or possibly even hundreds, of millions of peptides at physiologically relevant surface densities (33). The reality of T cell cross-reactivity, as quantified here, has far reaching implications. It provides an explanation for how a limited pool of TCRs can provide the broad antigenic coverage that is required for effective immunity. In addition, the extent of TCR degeneracy suggests that almost any peptide antigen can be improved for any given cognate TCR, in the sense of there being at least one stronger agonist than the original index peptide, thereby providing scope for rational therapeutic interventions based on the directed manipulation of T cell immunity.
Supplementary Material
Acknowledgments
We thank Don Mason and Bent Jakobsen for critical reading of the manuscript and David Rand for careful scrutiny of the mathematics.
This work was supported by the Biotechnology and Biological Sciences Research Council Grant BB/H001085/1 and the United Kingdom Department of Health via the National Institute for Health Research comprehensive Biomedical Research Centre award to Guy's and St. Thomas' NHS Foundation Trust in partnership with King's College London (to A. S. and M. P.).
- TCR
- T cell receptor
- CPL
- combinatorial peptide library
- HLA
- human leukocyte antigen
- MIP
- macrophage inflammatory protein.
REFERENCES
- 1. Rudolph M. G., Wilson I. A. (2002) The specificity of TCR/pMHC interaction. Curr. Opin. Immunol. 14, 52–65 [DOI] [PubMed] [Google Scholar]
- 2. Rudolph M. G., Stanfield R. L., Wilson I. A. (2006) How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 [DOI] [PubMed] [Google Scholar]
- 3. Mason D. (1998) A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunol. Today 19, 395–404 [DOI] [PubMed] [Google Scholar]
- 4. Arstila T. P., Casrouge A., Baron V., Even J., Kanellopoulos J., Kourilsky P. (1999) A direct estimate of the human αβ T cell receptor diversity. Science 286, 958–961 [DOI] [PubMed] [Google Scholar]
- 5. Wraith D. C., Bruun B., Fairchild P. J. (1992) Cross-reactive antigen recognition by an encephalitogenic T cell receptor. Implications for T cell biology and autoimmunity. J. Immunol. 149, 3765–3770 [PubMed] [Google Scholar]
- 6. Bhardwaj V., Kumar V., Geysen H. M., Sercarz E. E. (1993) Degenerate recognition of a dissimilar antigenic peptide by myelin basic protein-reactive T cells. Implications for thymic education and autoimmunity. J. Immunol. 151, 5000–5010 [PubMed] [Google Scholar]
- 7. Reay P. A., Kantor R. M., Davis M. M. (1994) Use of global amino acid replacements to define the requirements for MHC binding and T cell recognition of moth cytochrome c (93–103). J. Immunol. 152, 3946–3957 [PubMed] [Google Scholar]
- 8. Evavold B. D., Sloan-Lancaster J., Wilson K. J., Rothbard J. B., Allen P. M. (1995) Specific T cell recognition of minimally homologous peptides: evidence for multiple endogenous ligands. Immunity 2, 655–663 [DOI] [PubMed] [Google Scholar]
- 9. Wucherpfennig K. W., Strominger J. L. (1995) Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell 80, 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hemmer B., Fleckenstein B. T., Vergelli M., Jung G., McFarland H., Martin R., Wiesmüller K. H. (1997) Identification of high potency microbial and self ligands for a human autoreactive class II-restricted T cell clone. J. Exp. Med. 185, 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kersh E. N., Shaw A. S., Allen P. M. (1998) Fidelity of T cell activation through multistep T cell receptor ζ phosphorylation. Science 281, 572–575 [DOI] [PubMed] [Google Scholar]
- 12. Kissler S., Anderton S. M., Wraith D. C. (2002) Cross-reactivity and T-cell receptor antagonism of myelin basic protein-reactive T cells is modulated by the activation state of the antigen presenting cell. J. Autoimmun. 19, 183–193 [DOI] [PubMed] [Google Scholar]
- 13. Nino-Vasquez J. J., Allicotti G., Borras E., Wilson D. B., Valmori D., Simon R., Martin R., Pinilla C. (2004) A powerful combination: the use of positional scanning libraries and biometrical analysis to identify cross-reactive T cell epitopes. Mol. Immunol. 40, 1063–1074 [DOI] [PubMed] [Google Scholar]
- 14. Crawford F., Huseby E., White J., Marrack P., Kappler J. W. (2004) Mimotopes for alloreactive and conventional T cells in a peptide-MHC display library. PLoS Biol. 2, E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee J. K., Stewart-Jones G., Dong T., Harlos K., Di Gleria K., Dorrell L., Douek D. C., van der Merwe P. A., Jones E. Y., McMichael A. J. (2004) T cell cross-reactivity and conformational changes during TCR engagement. J. Exp. Med. 200, 1455–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kan-Mitchell J., Bajcz M., Schaubert K. L., Price D. A., Brenchley J. M., Asher T. E., Douek D. C., Ng H. L., Yang O. O., Rinaldo C. R., Jr., Benito J. M., Bisikirska B., Hegde R., Marincola F. M., Boggiano C., Wilson D., Abrams J., Blondelle S. E., Wilson D. B. (2006) Degeneracy and repertoire of the human HIV-1 Gag p17(77–85) CTL response. J. Immunol. 176, 6690–6701 [DOI] [PubMed] [Google Scholar]
- 17. Wilson D. B., Wilson D. H., Schroder K., Pinilla C., Blondelle S., Houghten R. A., Garcia K. C. (2004) Specificity and degeneracy of T cells. Mol. Immunol. 40, 1047–1055 [DOI] [PubMed] [Google Scholar]
- 18. Dai S., Huseby E. S., Rubtsova K., Scott-Browne J., Crawford F., Macdonald W. A., Marrack P., Kappler J. W. (2008) Crossreactive T cells spotlight the germ line rules for αβ T cell-receptor interactions with MHC molecules. Immunity 28, 324–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishizuka J., Grebe K., Shenderov E., Peters B., Chen Q., Peng Y., Wang L., Dong T., Pasquetto V., Oseroff C., Sidney J., Hickman H., Cerundolo V., Sette A., Bennink J. R., McMichael A., Yewdell J. W. (2009) Quantitating T cell cross-reactivity for unrelated peptide antigens. J. Immunol. 183, 4337–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Udaka K., Tsomides T. J., Eisen H. N. (1992) A naturally occurring peptide recognized by alloreactive CD8+ cytotoxic T lymphocytes in association with a class I MHC protein. Cell 69, 989–998 [DOI] [PubMed] [Google Scholar]
- 21. Reiser J. B., Darnault C., Guimezanes A., Grégoire C., Mosser T., Schmitt-Verhulst A. M., Fontecilla-Camps J. C., Malissen B., Housset D., Mazza G. (2000) Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat. Immunol. 1, 291–297 [DOI] [PubMed] [Google Scholar]
- 22. Archbold J. K., Macdonald W. A., Miles J. J., Brennan R. M., Kjer-Nielsen L., McCluskey J., Burrows S. R., Rossjohn J. (2006) Alloreactivity between disparate cognate and allogeneic pMHC-I complexes is the result of highly focused, peptide-dependent structural mimicry. J. Biol. Chem. 281, 34324–34332 [DOI] [PubMed] [Google Scholar]
- 23. Felix N. J., Allen P. M. (2007) Specificity of T-cell alloreactivity. Nat. Rev. Immunol. 7, 942–953 [DOI] [PubMed] [Google Scholar]
- 24. Archbold J. K., Ely L. K., Kjer-Nielsen L., Burrows S. R., Rossjohn J., McCluskey J., Macdonald W. A. (2008) T cell allorecognition and MHC restriction: a case of Jekyll and Hyde? Mol. Immunol. 45, 583–598 [DOI] [PubMed] [Google Scholar]
- 25. Macdonald W. A., Chen Z., Gras S., Archbold J. K., Tynan F. E., Clements C. S., Bharadwaj M., Kjer-Nielsen L., Saunders P. M., Wilce M. C., Crawford F., Stadinsky B., Jackson D., Brooks A. G., Purcell A. W., Kappler J. W., Burrows S. R., Rossjohn J., McCluskey J. (2009) T cell allorecognition via molecular mimicry. Immunity 31, 897–908 [DOI] [PubMed] [Google Scholar]
- 26. Skowera A., Ellis R. J., Varela-Calviño R., Arif S., Huang G. C., Van-Krinks C., Zaremba A., Rackham C., Allen J. S., Tree T. I., Zhao M., Dayan C. M., Sewell A. K., Unger W. W., Drijfhout J. W., Ossendorp F., Roep B. O., Peakman M. (2008) CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope. J. Clin. Invest. 118, 3390–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karlin S., Taylor H. M. (1974) A First Course in Stochastic Processes, 2nd Ed., pp. 495–502, Academic Press, New York [Google Scholar]
- 28. Nejentsev S., Howson J. M., Walker N. M., Szeszko J., Field S. F., Stevens H. E., Reynolds P., Hardy M., King E., Masters J., Hulme J., Maier L. M., Smyth D., Bailey R., Cooper J. D., Ribas G., Campbell R. D., Clayton D. G., Todd J. A. (2007) Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450, 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Todd J. A., Walker N. M., Cooper J. D., Smyth D. J., Downes K., Plagnol V., Bailey R., Nejentsev S., Field S. F., Payne F., Lowe C. E., Szeszko J. S., Hafler J. P., Zeitels L., Yang J. H., Vella A., Nutland S., Stevens H. E., Schuilenburg H., Coleman G., Maisuria M., Meadows W., Smink L. J., Healy B., Burren O. S., Lam A. A., Ovington N. R., Allen J., Adlem E., Leung H. T., Wallace C., Howson J. M., Guja C., Ionescu-Tîrgoviste C., Simmonds M. J., Heward J. M., Gough S. C., Dunger D. B., Wicker L. S., Clayton D. G. (2007) Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat. Genet. 39, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. van den Berg H. A., Rand D. A., Burroughs N. J. (2001) A reliable and safe T cell repertoire based on low-affinity T cell receptors. J. Theor. Biol. 209, 465–486 [DOI] [PubMed] [Google Scholar]
- 31. van den Berg H. A., Rand D. A. (2007) Quantitative theories of T-cell responsiveness. Immunol. Rev. 216, 81–92 [DOI] [PubMed] [Google Scholar]
- 32. Wooldridge L., Laugel B., Ekeruche J., Clement M., van den Berg H. A., Price D. A., Sewell A. K. (2010) CD8 controls T cell cross-reactivity. J. Immunol. 185, 4625–4632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hiemstra H. S., Drijfhout J. W., Roep B. O. (2000) Antigen arrays in T cell immunology. Curr. Opin. Immunol. 12, 80–84 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.