Background: Molecular chaperones assist in the folding of metastable proteins implicated in neurodegenerative diseases.
Results: Huntingtin is a heat shock protein 90 (Hsp90) client protein.
Conclusion: Hsp90 inhibition-mediated degradation of soluble mutant huntingtin is independent of a cellular heat shock response.
Significance: Mechanisms targeting Hsp90 chaperone function may provide new treatments for Huntington disease.
Keywords: Heat Shock Protein, Huntington Disease, Neurodegeneration, Neurodegenerative Diseases, Polyglutamine Disease, Ubiquitin, Hsp70, Hsp90, Heat Shock Factor 1
Abstract
Mechanisms to reduce the cellular levels of mutant huntingtin (mHtt) provide promising strategies for treating Huntington disease (HD). To identify compounds enhancing the degradation of mHtt, we performed a high throughput screen using a hippocampal HN10 cell line expressing a 573-amino acid mHtt fragment. Several hit structures were identified as heat shock protein 90 (Hsp90) inhibitors. Cell treatment with these compounds reduced levels of mHtt without overt toxic effects as measured by time-resolved Förster resonance energy transfer assays and Western blots. To characterize the mechanism of mHtt degradation, we used the potent and selective Hsp90 inhibitor NVP-AUY922. In HdhQ150 embryonic stem (ES) cells and in ES cell-derived neurons, NVP-AUY922 treatment substantially reduced soluble full-length mHtt levels. In HN10 cells, Hsp90 inhibition by NVP-AUY922 enhanced mHtt clearance in the absence of any detectable Hsp70 induction. Furthermore, inhibition of protein synthesis with cycloheximide or overexpression of dominant negative heat shock factor 1 (Hsf1) in HdhQ150 ES cells attenuated Hsp70 induction but did not affect NVP-AUY922-mediated mHtt clearance. Together, these data provided evidence that direct inhibition of Hsp90 chaperone function was crucial for mHtt degradation rather than heat shock response induction and Hsp70 up-regulation. Co-immunoprecipitation experiments revealed a physical interaction of mutant and wild-type Htt with the Hsp90 chaperone. Hsp90 inhibition disrupted the interaction and induced clearance of Htt through the ubiquitin-proteasome system. Our data suggest that Htt is an Hsp90 client protein and that Hsp90 inhibition may provide a means to reduce mHtt in HD.
Introduction
Huntington disease (HD)3 is a progressive neurodegenerative disease characterized by brain atrophy and motor, cognitive, and psychiatric symptoms. Patients also suffer from muscle atrophy, weight loss, and metabolic disturbances. HD is caused by the expansion of a trinucleotide repeat resulting in an elongated glutamine stretch close to the N terminus of the huntingtin (Htt) protein (1). The length of the polyglutamine expansion correlates with disease onset (2). The dominant mode of inheritance supports the hypothesis that the extended polyglutamine stretch confers a toxic gain of function to huntingtin, possibly due to structural changes of the mutant protein, and aberrant interactions with different cellular pathways. Multiple potential pathogenic mechanisms of mutant huntingtin (mHtt) have been proposed, including proteasome impairment, mitochondrial dysfunction, transcriptional dysregulation, impaired intracellular transport, and cell death induced by the formation of toxic aggregates containing N-terminal mHtt fragments (2–4). Reversal of inducible mHtt overexpression, RNA interference, and antisense oligonucleotide studies have all demonstrated amelioration of HD-like symptoms upon reduction of mHtt expression levels (5–8). Therefore, mechanisms to reduce the cellular load of the disease-causing mHtt protein, such as via enhancement of its clearance and degradation, represent promising therapeutic strategies.
Heat shock proteins play an important role in protein folding and quality control. In the context of polyglutamine diseases, such as HD, heat shock protein 70 (Hsp70; Hspa1a/b), Hsp40 (Dnajb1), and Hsp90 (Hsp90aa1 and Hsp90ab1) have been the subject of several studies. Elevation of Hsp70 levels has been found to be neuroprotective in several animal models (9). For instance, Hsp70 overexpression suppressed neuropathology and improved motor function in a spinocerebellar ataxia mouse model (10). Furthermore, Hsp70 and Hsp40 attenuated assembly of polyglutamine proteins into amyloid-like fibrils (11, 12).
Hsp90 comprises about 1–2% of total cellular protein (13). It uses ATP hydrolysis to fold and maturate client proteins and interacts with more than 20 co-chaperones (13, 14). Notably, Hsp90 is described to preferentially bind to partially folded intermediates, suggesting a role in the maturation of metastable proteins late in their folding pathway (13, 15, 16). Currently, more than 200 Hsp90 clients have been identified, including a range of oncogenic proteins (17). Hsp90 inhibitors, such as the geldanamycin derivatives 17-allylamino-17-demethoxygeldanamycin and 17-dimethylaminoethylamino-17-demethoxygeldanamycin as well as other structural classes, including NVP-AUY922, are in clinical development as cancer therapies (17, 18). Pharmacological inhibition of Hsp90 induces a heat shock response (HSR) after release of the transcription factor heat shock factor 1 (Hsf1) from the Hsp90 chaperone complex. Hsf1 in turn induces the expression of other heat shock proteins, such as Hsp70 (9, 13). Induction of Hsp70 after Hsp90 inhibition using geldanamycin inhibited mHtt exon 1 protein aggregation (19), whereas loss of Hsp70 exacerbated pathogenesis in a mouse model of HD (20).
We conducted a high throughput screen aimed at identifying molecules that enhance degradation or clearance of soluble mHtt. Among the hits identified that reduced mHtt levels without overt toxic and nonspecific effects, we discovered a single class of compounds possessing a known mechanism of action, Hsp90 inhibition. Unexpectedly, an HSR induced after Hsp90 inhibition was not required for the degradation of soluble mHtt. We provide evidence that mutant and wild-type Htt are client proteins of Hsp90 and that the chaperoning function of Hsp90 is critical for maintaining the stability of mHtt in different cellular systems. Pharmacological inhibition of Hsp90 destabilizes mHtt and facilitates its clearance through the ubiquitin-proteasome system (UPS).
EXPERIMENTAL PROCEDURES
High Throughput Screening
The screen was conducted in a 1536-well format as described previously (21). PicoGreen (Invitrogen, P11495; 1:1200) and caspase 3/7 (Promega, G8091) assays to exclude putative toxic compounds were performed according to instructions from the manufacturers.
Cell Lines
HN10-Htt cell lines were cultured as described (22). mHtt expression was induced by the addition of 750 nm RSL1 to the growth medium. Mouse embryonic stem (ES) cells expressing a 1000-amino acid fragment or full-length mHtt (Q145) from the ROSA26 locus and HdhQ150 ES cells (23) were cultured in 3i-medium (24). Neurons were derived from ES cells as described (25). HEK293T cells were cultured in DMEM supplemented with Glutamax (Invitrogen, 32430) and 10% fetal calf serum.
Transient Transfections
Transfections of HEK293T and HN10 cells with plasmids/siRNAs were done using Lipofectamine 2000 (Invitrogen, 11668-019). The plasmids used were human Htt573Q72/pER (CMV promoter), hemagglutinin (HA)-tagged ubiquitin/pSG5 (SV40 promoter), HSF-1 dominant negative/pcDNA5 (26), and GFP/pLL3.7 (CMV promoter). For siRNA transfections, HN10–573Q72 cells (105 cells in 500 μl) were seeded into 24-well dishes containing 50 μl of siRNA stock solutions in Opti-MEM (Invitrogen) and 50 μl of transfection reagent (Lipofectamine diluted 1:50 in Opti-MEM), and the cells were cultured for 48 h. Mixtures of siRNAs targeting mouse Hsp90aa1 and Hsp90ab1 (Qiagen; final concentration of each siRNA, 12.5 nm) or negative control siRNAs (Ambion AM4611 and AM4613; 25 nm) were used (Hsp90si1: Hsp90aa1, 5′-cccgtgaaatgctgcaacaaa; Hsp90ab1, 5′-aacaaagaagatcaaagagaa; Hsp90si2: Hsp90aa1, 5′-cagaaagaatttggtcaagaa; Hsp90ab1, 5′-ccgcaagaacatcgtcaagaa).
Western Blot and Antibodies
Cells were harvested and lysed in ice-cooled lysis buffer (phosphate-buffered saline (Invitrogen, 14190), 1% Triton X-100), containing protease inhibitors (Roche Applied Science Complete, 11836145001). Lysates were kept on ice for 15 min before centrifugation for 10 min at 13,000 rpm at 4 °C. The BCA assay (Thermo Scientific, 23227) was used for protein quantification. Samples were diluted in NuPAGE loading buffer (Invitrogen, NP0007 and NP0009) and heated for 10 min at 95 °C. The samples (ES cells, 15 μg; HN10 cells, 10 μg) were loaded onto 4–12% NuPAGE Bis-Tris gels or 3–8% Tris acetate gels (Invitrogen, NP0335 and EA03752). Semidry protein transfer (Ancos, LV8428062) to PVDF membranes (Millipore, Immobilon-P PVH00010) was performed in NuPAGE transfer buffer (Invitrogen, NP0006-1) for 1 h at 15 V, and the membrane was then incubated for 1 h in 20 mm Tris-Cl, 137 mm NaCl, pH 7.6, 0.1% Tween 20, 5% (w/v) dried milk. Incubation with primary antibodies was done overnight (o/N) at 4 °C before secondary horseradish peroxidase-conjugated goat anti-mouse (Chemicon, AP127P) or goat anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, 111-035-144) were applied for 1 h. The ECL reagent (GE Healthcare) was used for protein visualization. Densitometric quantification of Western blots was done from x-rays using the ImageJ software (signals were normalized to tubulin). The following primary antibodies were used: 2B7 (anti-Htt; custom production by NanoTools, Freiburg, Germany), MW1 (anti-poly(Q); Developmental Studies Hybridoma Bank; Ref. 27), Hsp90 (Stressgen, SPS-771 and SPA-830), Hsp25 (Stressgen, SPA-801), Hsp40 (Stressgen, SPA-400), Hsp70 (Stressgen, SPA-810), Hsf1 (Stressgen, SPA-901), α-tubulin (Abcam, ab28037), Akt (Cell Signaling Technology, 9272), phospho-Akt (Cell Signaling Technology, 9275), ubiquitin (Millipore, MAB1510), p23 (Alexis, ALX-804-023), and HA (Roche Applied Science, 12CA5).
Co-immunoprecipitation
HN10 and ES cells were lysed (Dounce homogenizer) in ice-cold IP lysis buffer (20 mm Tris-HCl, pH 7.4, 50 mm NaCl, 5 mm MgCl2, 1 mm EDTA, 20 mm sodium molybdate, 4 mm sodium orthovanadate, 0.02% Nonidet-P40, Roche Applied Science Complete protease inhibitors). The samples were left on ice for 15 min and centrifuged for 10 min at 13,000 rpm at 4 °C. 150 μl of lysate (∼150 μg of protein) were incubated o/N with 2 μg of Hsp90 or p23 antibody. Afterward, the samples were incubated for 30 min with 15 μl of protein G-Sepharose 4 fast flow beads (GE Healthcare, 17-0618-01) and subsequently washed three times with lysis buffer and once in lysis buffer containing 20 mm Hepes, pH 8.0. NuPAGE loading buffer was added to the dried beads, and the samples were heated for 10 min at 95 °C.
Compounds
NVP-AUY922 was synthesized at Novartis (10 mm stock solution in dimethyl sulfoxide (DMSO)). Proteasome inhibitors epoxomicin and MG132 (Calbiochem, 324800 and 474790), cycloheximide (Sigma-Aldrich), and RSL1 (New England Biolabs) were used.
Ubiquitination Assay
HN10 and HEK293T cells were transiently transfected with HA-ubiquitin and Htt573Q72 plasmids. After 24 h, the medium was replaced, and the cells were treated o/N with Hsp90 and/or proteasome inhibitors. Two days after transfection, the cells were lysed in 50 mm Hepes, pH 8.0, 250 mm NaCl, 5 mm EDTA, 0.1% Nonidet-P40, Roche Applied Science Complete protease inhibitors), and aliquots (∼500 μg of total protein) were incubated o/N with 5 μg of anti-Htt 2B7 antibody and subsequently for 30 min with 15 μl of protein G-Sepharose 4 beads. HA-ubiquitin immunoprecipitation was done using an anti-HA magnetic bead kit (μMACS HA, Miltenyi Biotec).
Time-resolved Förster Resonance Energy Transfer (TR-FRET)
Assays were performed as described previously (21, 28, 29). Briefly, cells were lysed in PBS, 1% Triton X-100 and incubated at room temperature for 30 min with shaking. Then, 5 μl of lysate (HN10 cells, 2 μg; ES cells, 5 μg) were transferred to a low volume plate (Greiner Bio-One, 784080), and 1 μl of antibody mixture added. For detection of Htt573Q72 expressed from the HN10 cell line, the antibodies 2B7-terbium (Tb)/β1-d2 (intact Htt573Q72) or 2B7-Tb/MW1-d2 (total Htt573Q72) were used. mHtt expressed from HdhQ150 cells was measured with 2B7-Tb/MW1-d2. Fluorophore labeling of antibodies was performed by CisBio Bioassays (Parc Marcel Boiteux, France). TR-FRET measurements were done after 1-h incubation at room temperature using an EnVision reader (PerkinElmer Life Sciences; excitation, 320 nm, time delay, 100 ms; integration time, 400 ms). Values (means from at least three experiments ±S.D.; duplicate or triplicate determinations) are expressed as the ratio between the emissions at 665 and 620 nm; background signals (antibodies in lysis buffer) were subtracted. The t test was used to assess significance.
RESULTS
High Throughput Screen Identifies Hsp90 Inhibitors as Enhancers of Mutant Huntingtin Degradation
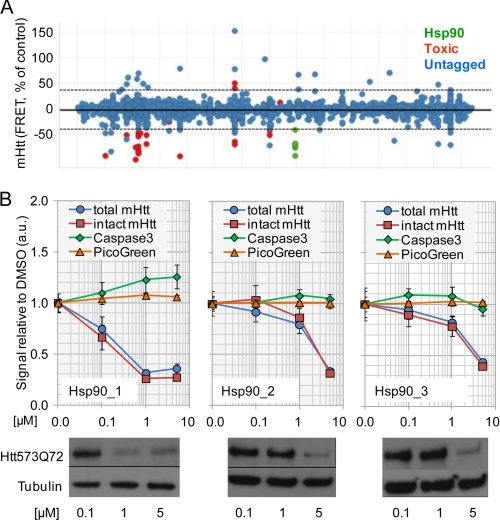
Our aim was to identify mechanisms to reduce cellular levels of soluble mHtt. To this end, screening of the Novartis compound library (∼2 × 106 compounds) was performed using a mouse hippocampal HN10 cell line expressing an inducible β1 epitope-tagged, 573-amino acid N-terminal fragment of human Htt with 72 glutamine residues (Htt573Q72) as described (22). This cell line does not produce readily detectable mHtt aggregates. Soluble mHtt levels were measured using a sensitive, homogeneous TR-FRET assay (21, 29). Toxic compounds and structures that interfered with the TR-FRET assay readout were excluded as described (21). Compounds affecting mHtt levels by inhibition of the inducible expression system were identified in HN10 cells expressing Gaussia luciferase from the expression vector as used for mHtt (not shown). The remaining hits were then selected for further validation (Fig. 1A). Compounds reducing mHtt protein levels were confirmed in concentration-response curves, and effects were compared with readouts for cytotoxicity, such as caspase 3 activation and DNA fragmentation (Fig. 1B). After review of the chemical structures, it became apparent that several hits, comprising different structural classes, had previously been characterized to act by a common mechanism of action, ATP competitive inhibition of Hsp90. Hsp90 inhibitory activity of compounds identified from the screen was confirmed in radicicol binding assays as described (not shown) (30). The compounds caused a concentration-dependent reduction of the Htt573Q72 protein expressed in the HN10 cell line without overt cytotoxic effects as measured by TR-FRET and Western blot readouts (Fig. 1B).
FIGURE 1.
High throughput screening identifies Hsp90 inhibitors as modulators of mHtt degradation. A, visualization of primary screen data for compounds with known mode of action. The Hsp90 inhibitor compounds are labeled. The dotted line denotes the cutoff used for hit selection (3 × S.D.). B, examples of profiles of three different Hsp90 inhibitor structures identified from the screen by biochemical (upper panel) and Western blot analyses (lower panel). TR-FRET detection of the entire Htt573Q72 fragment was done with the antibody combination (2B7-Tb and β1-d2) binding to the N-terminal 17 amino acids of Htt and to the C-terminal β1 tag, respectively (intact Htt). MW1-d2 binds to the poly(Q) region of mHtt and in combination with 2B7-Tb detects N-terminal mHtt fragments (total Htt). DNA fragmentation (PicoGreen) and caspase 3 activation assays were done to assess cytotoxicity. a.u., arbitrary units. Error bars denote S.D.
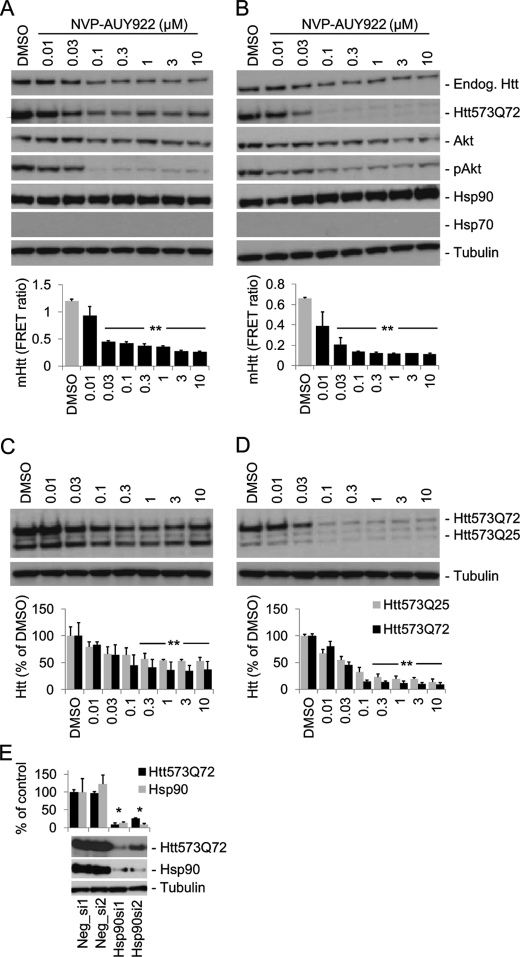
For further characterization of the mechanism of mHtt clearance after Hsp90 inhibition, we used a potent and selective Hsp90 inhibitor compound, NVP-AUY922, that had been described previously (31, 32). In a manner similar to the Hsp90 inhibitors shown in Fig. 1B, NVP-AUY922 concentration-dependently reduced Htt573Q72 protein levels. Application of 30 nm or higher concentrations to HN10-Htt573Q72 cells significantly reduced mHtt protein as evidenced by Western blots and quantified using TR-FRET (p < 0.001; Fig. 2, A and B). The expression of endogenous wild-type Htt in HN10 cells was also reduced, however with lower efficacy (Fig. 2A). NVP-AUY922 treatment reduced the levels of phosphorylated Akt and to a lesser extent of the non-phosphorylated form as expected for an Hsp90 client protein (14). Interestingly, in HN10 cells, NVP-AUY922 did not induce expression of Hsp70 protein, an established marker for the HSR after Hsp90 inhibition (Figs. 2, A and B, and 4A) (9, 13). The observed potency of NVP-AUY922 correlated well with IC50 values of 8 and 21 nm at Hsp90α and Hsp90β, respectively, from binding experiments (32). Destabilization of soluble mHtt could induce the formation of aggregates. However, we did not observe aggregates in the gel slots after NVP-AUY922 treatment (supplemental Fig. S1).
FIGURE 2.
Pharmacological and genetic Hsp90 inhibition induces mHtt degradation. HN10-Htt cell lines were cultured for 3 days in medium containing 750 nm RSL1 to induce Htt expression. The cells were then treated with NVP-AUY922 as indicated in RSL1-containing medium (A and C; “steady state”; harvested after 24 h) or in medium without RSL1 to turn off inducible Htt expression (B and D; “washout”; harvested after 16 h). A and B, Western blots (upper panels) and TR-FRET quantifications (lower panels) of NVP-AUY922 effects in HN10-Htt573Q72 cells. C and D, NVP-AUY922 effects on HN10 cells co-expressing mutant (Htt573Q72) and wild-type (Htt572Q25) fragments. Because the TR-FRET assay lacks the ability to detect only wild-type Htt, quantification was done by densitometry (lower panels). E, siRNA-mediated down-modulation of Hsp90 decreases Htt573Q72 in HN10 cells. Induced cells were transfected with different siRNA mixtures targeting Hsp90aa1 and Hsp90ab1 (Hsp90si1/2) and control siRNAs (Neg_si1/2) as described under “Experimental “Procedures.” The signals were normalized to Neg_si1; mHtt and Hsp90 were quantified by TR-FRET (2B7-Tb and MW1-d2) and densitometry, respectively. **, p < 0.001; *, p < 0.01; n = 3. Error bars denote S.D. Endog., endogenous; pAkt, phosphorylated Akt.
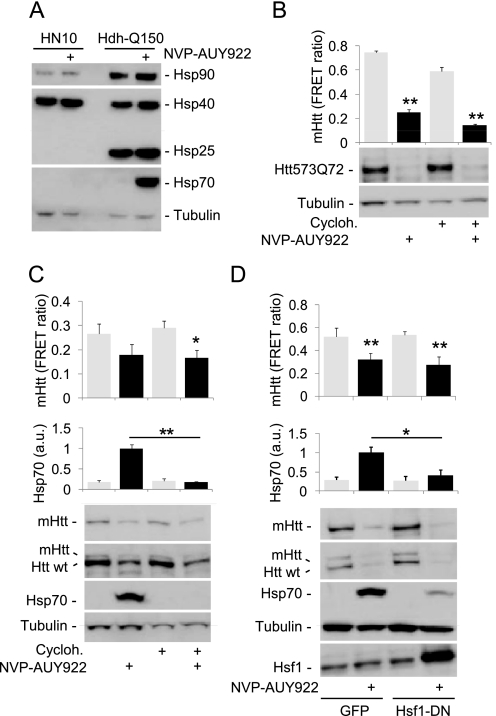
FIGURE 4.
Degradation of mHtt after Hsp90 inhibition does not require induction of Hsp70. A, Western blot analysis of the expression of heat shock proteins in lysates from HN10-Htt573Q72 (HN10) and HdhQ150 cells. In contrast to HdhQ150 cells, 0.3 μm NVP-AUY922 does not induce Hsp70 and Hsp25 in HN10 cells. B and C, application of 0.3 μm cycloheximide for 24 h to HN10 (B) or HdhQ150 cells (C) does not abolish mHtt degradation induced by 0.3 μm NVP-AUY922 (24-h application; control, DMSO; n = 4). HN10-Htt573Q72 cells were cultured in medium without expression inducer ligand RSL1 from the time of compound addition onward (washout). In HdhQ150 cells, 0.3 μm cycloheximide abolishes Hsp70 induction. D, transient overexpression (36 h) of Hsf1-DN in HdhQ150 cells attenuates NVP-AUY922-induced Hsp70 induction (*, p < 0.01 versus GFP; n = 3) but does not affect mHtt degradation (**, p < 0.001 versus DMSO; n = 6). B–D, upper panels, mHtt TR-FRET and Hsp70 quantifications; lower panels, Western blots. Error bars denote S.D. a.u., arbitrary units; Cycloh., cycloheximide.
To assess the selectivity of the effect over wild-type Htt, the Hsp90 inhibitor was applied to an HN10 cell line expressing both the mutant (Htt573Q72) and the wild-type (Htt573Q25) Htt fragments. NVP-AUY922 treatment affected both mutant and wild-type forms; however, the decline of mHtt appeared somewhat more pronounced compared with the wild-type fragment (Fig. 2, C and D).
To rule out the possibility that the observed effects of NVP-AUY922 on Htt degradation were mediated by interference with the inducible expression system, cells were cultured in medium without the inducer ligand RSL1 from the time of Hsp90 inhibitor application onward (Fig. 2, B and D). Removal of RSL1 turns off inducible Htt transcription. Also under these conditions, Htt573Q72 protein clearance was significantly enhanced compared with DMSO vehicle treatment (p < 0.001), suggesting that NVP-AUY922 acts at the Htt protein but not at the RNA level.
Transfection of HN10-Htt573Q72 cells with mixtures of siRNAs targeting both cytoplasmic Hsp90 isoforms (Hsp90aa1 and Hsp90ab1) substantially reduced Hsp90 protein levels compared with control siRNAs (Fig. 1E). This led to a concomitant decrease of Htt573Q72 protein, thus demonstrating that non-pharmacological inactivation of Hsp90 by siRNAs also reduced cellular levels of mHtt (Fig. 1E).
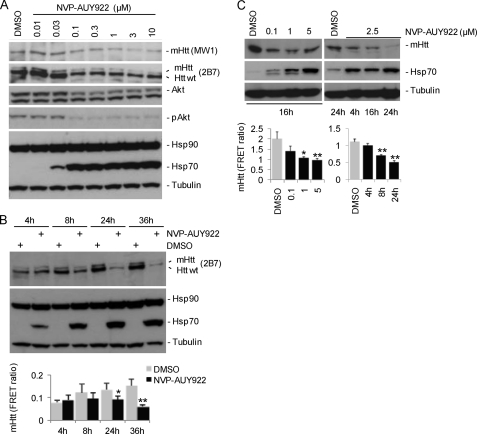
We extended our studies to knock-in HdhQ150 ES cells expressing full-length mHtt (Fig. 3). As observed for the HN10 cell line, the presence of NVP-AUY922 reduced both mutant as well as wild-type Htt as shown by Western blots with mHtt- (MW1) and pan-selective Htt antibodies (2B7; Fig. 3A). The MW1 antibody binds to expanded poly(Q) only and does not detect wild-type mouse Htt (29). NVP-AUY922 induced a strong up-regulation of Hsp70 in ES cells (Figs. 2 and 3) in contrast to HN10 cells. Lysates were analyzed at different time points after initiating treatment (Fig. 3B). A concentration of 0.3 μm NVP-AUY922 caused a significant reduction of mHtt compared with control-treated cells after 36 h (p < 0.01; Fig. 3B). To investigate the effect of Hsp90 inhibition on full-length human mHtt in a neuronal context, mouse ES cells expressing mHtt were differentiated into neurons as described (25). NVP-AUY922 concentration- and time-dependently reduced mHtt protein levels as measured by Western blot and TR-FRET (Fig. 3C). As observed with HdhQ150 ES cells, NVP-AUY922 caused a concentration-dependent induction of Hsp70 protein expression (Fig. 3C).
FIGURE 3.
NVP-AUY922 application reduces levels of full-length mHtt in HdhQ150 ES cells and in ES cell-derived neurons. HdhQ150 ES cell (heterozygous) Western blot data for mHtt and selected marker proteins after o/N application (A) or application of 0.3 μm NVP-AUY922 for different time frames (B) are shown. B, lower panel, quantification of mHtt by TR-FRET (2B7-Tb and MW1-d2; *, p = 0.012; **, p = 0.006 versus DMSO at time points; n = 3). C, ES cells expressing full-length mHtt with 145 glutamine residues were differentiated into neurons and cultured for 14 days. NVP-AUY922 at the different concentrations indicated was applied o/N or for the indicated time points. Upper panel, Western blots; lower panel, quantification of mHtt by TR-FRET (2B7-Tb/MW1-d2). *, p = 0.002; **, p < 0.001 (versus DMSO; n = 3). Error bars denote S.D. pAkt, phosphorylated Akt.
Inhibition of Hsp90 Chaperone Function Rather than Induction of Heat Shock Response Is Critical for NVP-AUY922-induced Degradation of Soluble mHtt
Pharmacological Hsp90 inhibition can facilitate protein degradation either through disruption of the Hsp90 chaperone-client protein complex or indirectly via Hsf1-mediated induction of other heat shock proteins, such as Hsp70 and Hsp40. We noted a lack of Hsp70 and Hsp25 (Hspb1) induction after NVP-AUY922 treatment in the HN10 cell line used in the primary screen (Fig. 4A), suggesting that the HSR is not essential for the degradation of soluble mHtt after Hsp90 inhibition. Application of a 0.3 μm concentration of the protein synthesis inhibitor cycloheximide to HN10 cells for 24 h did not block NVP-AUY922-induced mHtt degradation (Fig. 4B). Similarly, cycloheximide did not affect mHtt degradation in HdhQ150 cells but completely abolished Hsp70 induction (Fig. 4C). Furthermore, overexpression of dominant negative Hsf1 (Hsf1-DN) (26) in HdhQ150 cells attenuated Hsp70 induction but did not block mHtt degradation induced by NVP-AUY922 (Fig. 4D). The data suggested that Hsp90 inhibition-mediated degradation of soluble mHtt was independent of Hsp70 induction.
Mutant and Wild-type Htt Are Client Proteins of Hsp90
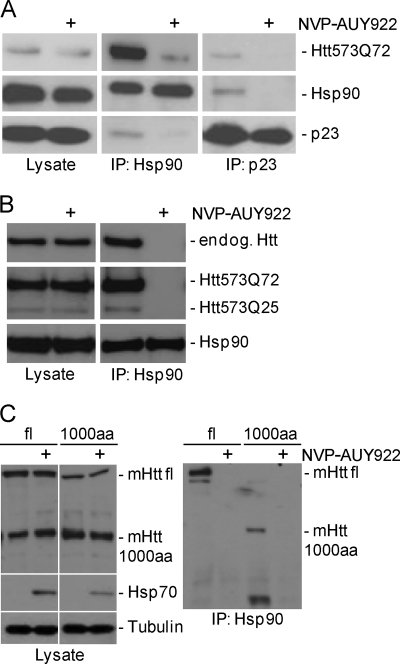
Co-immunoprecipitation experiments were performed on Htt-expressing HN10 cells treated for 4 h with 0.3 μm NVP-AUY922 or DMSO (Fig. 5, A and B). An antibody directed against Hsp90 co-immunoprecipitated Htt573Q72 as well as the Hsp90 co-chaperone p23 (33) from lysates of DMSO but not from NVP-AUY922-treated cells (Fig. 5A). Likewise, Htt573Q72 and Hsp90 were co-immunoprecipitated by p23 antibodies from lysates of DMSO but not from NVP-AUY922-treated cells (Fig. 5A). Pharmacological Hsp90 inhibition promotes dissociation of p23 from the Hsp90-client protein complex (33), and thus the absence of co-immunoprecipitation after compound treatment provided a control for Hsp90 inhibition by NVP-AUY922. Hsp90 antibody also co-immunoprecipitated the wild-type Htt (Htt573Q25) fragment and endogenous full-length wild-type Htt from HN10 cells (Fig. 5B). Hsp90/mHtt interaction was also observed in lysates from ES cells expressing either an N-terminal 1000-amino acid fragment or full-length mHtt (Fig. 5C). Again, the molecular interaction was disrupted after Hsp90 inhibitor treatment. In summary, the co-immunoprecipitation data suggested that mutant and wild-type Htt are client proteins of Hsp90 and that inhibition of Hsp90 chaperoning activity led to destabilization and subsequent degradation of Htt.
FIGURE 5.
Mutant and wild-type Htt interact with Hsp90 chaperone complex. Induced Htt-expressing HN10 cell lines (A and B) or ES cells (C) were incubated for 4 h with 0.3 μm NVP-AUY922 or DMSO in medium without expression inducer ligand RSL1 (washout). Immunoprecipitation (IP) was done using the antibodies as indicated. Whole cell lysates (Lysate) were loaded as a control. A, Hsp90 and p23 antibodies co-immunoprecipitate Htt573Q72 from HN10 cells; the interaction is abolished by NVP-AUY922. B, Hsp90 antibody co-immunoprecipitates Htt573Q72 and Htt573Q25 fragments as well as endogenous wild-type Htt from HN10 cells co-expressing mutant and wild-type fragments. C, Hsp90 antibody co-immunoprecipitates mHtt from ES cell lines expressing either a full-length (fl) or a 1000-amino acid N-terminal fragment transgene (1000aa) with 145 glutamine residues. NVP-AUY922 or DMSO application was for 4 h. Note that full-length transgenic mHtt (fl) migrates close to wild-type Htt endogenously (endog.) expressed from the cells and that there is a nonspecific band at the size of the 1000-amino acid fragment detected with Htt antibody 2B7.
Hsp90 Inhibition Facilitates mHtt Degradation through Proteasome
To further investigate the mechanisms of mHtt clearance after Hsp90 inhibition, HN10-Htt573Q72 and Htt573Q25 cells were treated with NVP-AUY922 at different concentrations in the presence or absence of the proteasome inhibitor epoxomicin. The clearance of the wild-type Htt573Q25 was attenuated by epoxomicin (Fig. 6A). In contrast, Htt573Q72 degradation was not significantly affected by epoxomicin when applied alone, suggesting that mHtt normally is not effectively cleared through the proteasome (Fig. 6A). However, in the presence of NVP-AUY922, epoxomicin treatment partially attenuated Htt573Q72 degradation. We postulate that mHtt, in contrast to wild-type Htt, became a substrate for the UPS only when released from the Hsp90 complex.
FIGURE 6.
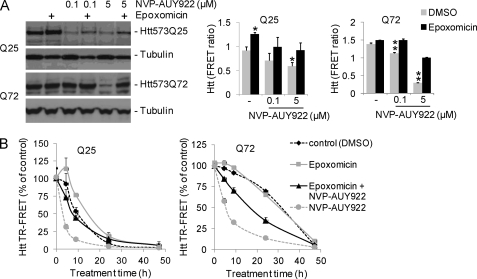
Hsp90 inhibition induces proteasome-dependent mHtt degradation. A and B, induced HN10–573Q25 (Q25) and HN10-Htt573Q72 (Q72) cells were treated o/N with 200 nm epoxomicin and/or NVP-AUY922 at the different concentrations indicated. Western blots (left panel) and Htt quantifications by TR-FRET are shown (right panels; n = 3). A, application of NVP-AUY922 reduces mHtt protein in Htt573Q25 and Htt573Q72 cells, and this effect is partly attenuated by epoxomicin. **, p < 0.001 versus DMSO and epoxomicin; *, p < 0.01 versus DMSO. B, Hsp90 inhibition accelerates Htt degradation kinetics. HN10–573Q25 and HN10-Htt573Q72 cells were cultured in medium without expression inducer ligand RSL1 from the time of 0.3 μm NVP-AUY922/50 nm epoxomicin treatment onward. Cells were harvested at the different time points indicated. NVP-AUY922-induced mHtt degradation is partly reversed by epoxomicin. Error bars denote S.D.
To further investigate degradation kinetics, HN10-Htt573Q72 and Htt573Q25 cells were cultured for 3 days in medium containing a 750 nm concentration of the mHtt expression-inducing ligand RSL1. Subsequently, the cells were cultured in non-inducing medium (washout) in the presence or absence of 5 μm NVP-AUY922 and/or 50 nm epoxomicin. Again, Hsp90 inhibition by NVP-AUY922 facilitated both mutant and wild-type Htt degradation, which was partially attenuated by proteasome inhibition (Fig. 6B).
Ubiquitination of mHtt Is Increased upon Hsp90 Inhibition
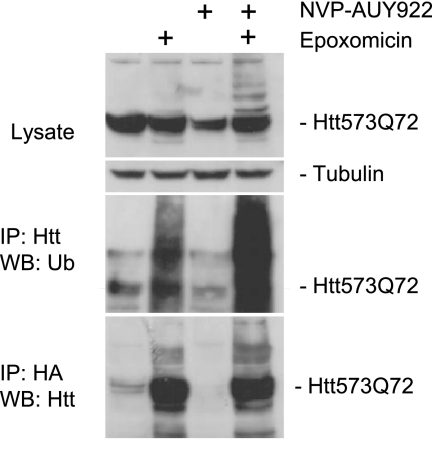
Proteasomal degradation requires protein polyubiquitination. We failed to conclusively demonstrate ubiquitination of the Htt573Q72 fragment expressed from the Htt-HN10 cell lines probably due to limited detection sensitivity (not shown). Therefore, we switched to a transient expression system. Plasmid constructs expressing HA-tagged ubiquitin and Htt573Q72 were transiently transfected into HN10 cells. The cells were then treated overnight with NVP-AUY922 in the presence or absence of the proteasome inhibitor epoxomicin. Immunoprecipitation was conducted with antibodies directed against HA and Htt (Fig. 7). Proteasome inhibition revealed basal levels of mHtt ubiquitination that increased in the presence of Hsp90 inhibition, supporting the conclusion that Hsp90 inhibition triggered Htt ubiquitination (Fig. 7). Qualitatively similar data were obtained with Htt573Q72 expressed in HEK293 cells using the proteasome inhibitor MG132 (supplemental Fig. S2).
FIGURE 7.
mHtt is ubiquitinated upon Hsp90 inhibition. HN10 cells were transiently transfected with expression constructs for Htt573Q72 and HA-tagged ubiquitin (Ub). 24 h later, 1 μm NVP-AUY922, 200 nm proteasome inhibitor epoxomicin, or a combination of the two was applied o/N. Immunoprecipitation (IP) was performed using anti-HA (Roche Applied Science, 12CA5) and anti-Htt antibodies (2B7). WB, Western blot.
DISCUSSION
We identified Hsp90 inhibitors as enhancers of mHtt degradation from a high throughput compound screen. The screen was aimed at exploring mechanisms that reduce cellular levels of soluble mHtt as this would be expected to lead to a reduction of the total cellular pool of mHtt, including aggregates and other, potentially toxic intermediate forms of mHtt. Treatment with NVP-AUY922 reduced mHtt levels in different cell types, including ES cell-derived neurons (Figs. 2 and 3).
To investigate a possible selective effect on mHtt, an HN10 cell line co-expressing both mutant (Htt573Q72) and wild-type (Htt573Q25) forms was analyzed. NVP-AUY922-induced reduction of both mutant and wild-type Htt was somewhat more efficacious at mHtt (Fig. 2, C and D). Similarly, in HdhQ150 cells, both mutant and wild-type full-length Htt were affected by Hsp90 inhibitor treatment (Fig. 3). However, substantially lower protein expression levels of mutant compared with wild-type Htt prohibited a meaningful quantification of NVP-AUY922 effects in this cell line. Co-immunoprecipitation experiments did not provide evidence of Hsp90 binding preference for mutant versus wild-type Htt (Fig. 5B). However, it is tempting to speculate that the difference observed in terms of proteasome sensitivity for wild-type and mutant Htt in the absence of Hsp90 inhibitors (Fig. 6) may be caused by a difference in affinity for Hsp90. In summary, it is evident that Hsp90 inhibition by NVP-AUY922 enhanced the clearance of both mutant and wild-type Htt (Figs. 2, 3, 5, and 6). Genetic inactivation of both Htt alleles is lethal during development, demonstrating essential functions of wild-type Htt (34). Therefore, the value of mechanisms targeting both mutant and wild-type Htt remains questionable. However, siRNA and antisense oligonucleotide approaches targeting both mutant and wild-type Htt ameliorated HD-like symptoms and did not produce overt side effects in animal models (7), suggesting that a concomitant decrease of both mutant and wild-type Htt may be beneficial in HD.
Disruption of the Hsp90-client complex facilitates proteasomal degradation of client proteins (13). Hsp90 inhibitors also induce a stress response via Hsf1, leading to induction of other heat shock proteins, such as Hsp70 and Hsp40. This can mediate an indirect clearance of protein aggregates especially via Hsp70-dependent E3 ubiquitin ligases (9, 13). We identified Hsp90 inhibitors in a screen using an Htt573Q72-overexpressing hippocampal HN10 cell line. In this cell line, mHtt, however, was reduced after Hsp90 inhibitor treatment in the absence of any detectable Hsp70 induction (Figs. 2 and 4). In contrast, the Hsp70 antibody used readily detected a strong induction of Hsp70 after NVP-AUY922 treatment of mouse HdhQ150 cells and in mouse ES cell-derived neurons (Figs. 3 and 4). In support of the HN10 cell data, a previous study on rat primary hippocampal neurons documented a lack of Hsf1 and Hsp70 induction after heat shock in contrast to several other neuronal cell types investigated (35). Inhibition of protein synthesis with cycloheximide or overexpression of dominant negative Hsf1 attenuated Hsp70 induction in HdhQ150 cells but did not inhibit NVP-AUY922-induced mHtt degradation (Fig. 4), demonstrating that disruption of the Hsp90-client protein complex, rather than Hsp70 induction, facilitated mHtt clearance. Studies on the androgen receptor mutated in spinobulbar muscular atrophy further support the notion that destabilizing the Hsp90-client protein complex can induce clearance of polyglutamine proteins independently of an HSR. 17-Allylamino-17-demethoxygeldanamycin-induced clearance of mutant androgen receptor appeared to be uncoupled from Hsp70 as only limited amounts of Hsp70 and Hsp40 were induced in vivo (36). Furthermore, the Hsp90 inhibitor 17-dimethylaminoethylamino-17-demethoxygeldanamycin enhanced proteasomal clearance of mutant androgen receptor even when Hsp70 induction was blocked by siRNAs (37). Moreover, Hsp90 inhibition blocked the formation of mutant androgen receptor aggregates in Hsf1 knock-out mouse embryonic fibroblasts that cannot induce Hsp70 and Hsp40 (38). In summary, the data provide strong evidence that the mechanism of Hsp90 inhibitor-mediated degradation of soluble mHtt is the disruption of the Hsp90-mHtt client protein complex. Of note, a recent study has revealed an impairment of the HSR in HD mouse models (39). Our data suggest that the HSR is not essential for Hsp90-mediated degradation of soluble mHtt.
Co-immunoprecipitation revealed a physical interaction of mutant and wild-type Htt with the Hsp90 chaperone complex (Fig. 5), and pharmacological inhibition of Hsp90 induced Htt degradation (Figs. 1–4 and 6). Thus, considering established criteria for Hsp90 clients (13), our data support the conclusion that mutant and wild-type Htt are client proteins of Hsp90. In the absence of Hsp90 inhibitor, clearance of wild-type Htt573Q25 but not of mutant Htt573Q72 was dependent on the activity of the proteasome, suggesting that association to the Hsp90 complex may protect mHtt from proteasome-dependent degradation (Fig. 6A). However, proteasome inhibition partially attenuated the clearance of mHtt after NVP-AUY922 treatment (Fig. 6), demonstrating that degradation through the UPS is facilitated after release of mHtt from the Hsp90 chaperone complex. In support of this degradation pathway, mHtt ubiquitination was increased after Hsp90 inhibition (Fig. 7). Possibly, Hsp90-associated mHtt remains protected from the intervention of ubiquitin ligases, thereby explaining why mHtt becomes a substrate for UPS degradation only when dissociated from the Hsp90 complex. Nevertheless, in the presence of NVP-AUY922, mHtt degradation was only partially attenuated by proteasome inhibition. This may provide evidence for additional, yet to be elucidated, epoxomicin-independent cellular degradation pathways of mHtt. The cellular mechanisms of mHtt degradation and a possible contribution of altered UPS functionality to disease pathology are still under debate. Although some studies reported an impairment of the UPS, others concluded that mHtt does not lead to proteasomal dysfunction (40–43). In the transgenic R6/2 model of HD, which is characterized by rapid disease progression, proteasome activity was not altered compared with wild-type mice (44).
In conclusion, our data show that clearance of soluble mHtt in different cellular systems, including ES cell-derived neurons, can be induced via Hsp90 inhibition. mHtt is stabilized by the Hsp90 chaperone complex, and pharmacological inhibition facilitates mHtt release and proteasomal degradation. This effect is independent of a general HSR and Hsp70 induction. Because Hsp90 inhibition is expected to influence a variety of client proteins, it is currently uncertain whether targeting Hsp90 is selective enough to provide a means for therapeutic intervention in HD. However, further investigation of Hsp90 inhibitors and of mechanisms targeting Hsp90 co-chaperone functions in HD are warranted.
Supplementary Material
Acknowledgments
We thank Ursula Müller, Stephan Grüninger, and Gabi Schutzius for expert technical assistance; Jens Richter and Audrey Marcel for culturing ES cell lines; Harm H. Kampinga for providing the Hsf1-DN construct; Joachim Nozulak for chemistry support; and Gregor Lotz and Susan Ide for discussions and comments on the manuscript. We thank the CHDI Foundation and TaconicArtemis for providing the Rosa26 ES cell lines. The MW1 antibody was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa.
This work was supported by the European Community's Seventh Framework Programme FP7/2008 under Grant Agreement 215618.

This article contains supplemental Figs. S1 and S2.
- HD
- Huntington disease
- DN
- dominant negative
- Hsf1
- heat shock factor 1
- Hsp
- heat shock protein
- HSR
- heat shock response
- Htt
- huntingtin
- RSL1
- N′-tert-butyl-N′-(3,5-dimethylbenzoyl)-2-ethyl-3-methoxybenzohydrazide
- TR-FRET
- time-resolved FRET
- mHtt
- mutant huntingtin
- o/N
- overnight
- UPS
- ubiquitin-proteasome system
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. The Huntington's Disease Collaborative Research Group (1993) A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Ross C. A., Tabrizi S. J. (2011) Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 10, 83–98 [DOI] [PubMed] [Google Scholar]
- 3. Fecke W., Gianfriddo M., Gaviraghi G., Terstappen G. C., Heitz F. (2009) Small molecule drug discovery for Huntington's Disease. Drug Discov. Today 14, 453–464 [DOI] [PubMed] [Google Scholar]
- 4. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 5. DiFiglia M., Sena-Esteves M., Chase K., Sapp E., Pfister E., Sass M., Yoder J., Reeves P., Pandey R. K., Rajeev K. G., Manoharan M., Sah D. W., Zamore P. D., Aronin N. (2007) Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl. Acad. Sci. U.S.A. 104, 17204–17209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McBride J. L., Boudreau R. L., Harper S. Q., Staber P. D., Monteys A. M., Martins I., Gilmore B. L., Burstein H., Peluso R. W., Polisky B., Carter B. J., Davidson B. L. (2008) Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. U.S.A. 105, 5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sah D. W., Aronin N. (2011) Oligonucleotide therapeutic approaches for Huntington disease. J. Clin. Investig. 121, 500–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yamamoto A., Lucas J. J., Hen R. (2000) Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101, 57–66 [DOI] [PubMed] [Google Scholar]
- 9. Turturici G., Sconzo G., Geraci F. (2011) Hsp70 and its molecular role in nervous system diseases. Biochem. Res. Int. 2011, 618127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cummings C. J., Sun Y., Opal P., Antalffy B., Mestril R., Orr H. T., Dillmann W. H., Zoghbi H. Y. (2001) Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum. Mol. Genet. 10, 1511–1518 [DOI] [PubMed] [Google Scholar]
- 11. Lotz G. P., Legleiter J., Aron R., Mitchell E. J., Huang S. Y., Ng C., Glabe C., Thompson L. M., Muchowski P. J. (2010) Hsp70 and Hsp40 functionally interact with soluble mutant huntingtin oligomers in a classic ATP-dependent reaction cycle. J. Biol. Chem. 285, 38183–38193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muchowski P. J., Schaffar G., Sittler A., Wanker E. E., Hayer-Hartl M. K., Hartl F. U. (2000) Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A. 97, 7841–7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taipale M., Jarosz D. F., Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 [DOI] [PubMed] [Google Scholar]
- 14. Pratt W. B., Toft D. O. (2003) Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp. Biol. Med. 228, 111–133 [DOI] [PubMed] [Google Scholar]
- 15. Rutherford S. L., Lindquist S. (1998) Hsp90 as a capacitor for morphological evolution. Nature 396, 336–342 [DOI] [PubMed] [Google Scholar]
- 16. Luo W., Sun W., Taldone T., Rodina A., Chiosis G. (2010) Heat shock protein 90 in neurodegenerative diseases. Mol. Neurodegener. 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kamal A., Burrows F. J. (2004) Hsp90 inhibitors as selective anticancer drugs. Discov. Med. 4, 277–280 [PubMed] [Google Scholar]
- 18. Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M. F., Fritz L. C., Burrows F. J. (2003) A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 425, 407–410 [DOI] [PubMed] [Google Scholar]
- 19. Sittler A., Lurz R., Lueder G., Priller J., Lehrach H., Hayer-Hartl M. K., Hartl F. U., Wanker E. E. (2001) Geldanamycin activates a heat shock response and inhibits huntingtin aggregation in a cell culture model of Huntington's disease. Hum. Mol. Genet. 10, 1307–1315 [DOI] [PubMed] [Google Scholar]
- 20. Wacker J. L., Huang S. Y., Steele A. D., Aron R., Lotz G. P., Nguyen Q., Giorgini F., Roberson E. D., Lindquist S., Masliah E., Muchowski P. J. (2009) Loss of Hsp70 exacerbates pathogenesis but not levels of fibrillar aggregates in a mouse model of Huntington's disease. J. Neurosci. 29, 9104–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paganetti P., Weiss A., Trapp M., Hammerl I., Bleckmann D., Bodner R. A., Coven-Easter S., Housman D. E., Parker C. N. (2009) Development of a method for the high-throughput quantification of cellular proteins. Chembiochem 10, 1678–1688 [DOI] [PubMed] [Google Scholar]
- 22. Weiss A., Roscic A., Paganetti P. (2009) Inducible mutant huntingtin expression in HN10 cells reproduces Huntington's disease-like neuronal dysfunction. Mol. Neurodegener. 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin C. H., Tallaksen-Greene S., Chien W. M., Cearley J. A., Jackson W. S., Crouse A. B., Ren S., Li X. J., Albin R. L., Detloff P. J. (2001) Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 10, 137–144 [DOI] [PubMed] [Google Scholar]
- 24. Ying Q. L., Wray J., Nichols J., Batlle-Morera L., Doble B., Woodgett J., Cohen P., Smith A. (2008) The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bibel M., Richter J., Lacroix E., Barde Y. A. (2007) Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat. Protoc. 2, 1034–1043 [DOI] [PubMed] [Google Scholar]
- 26. Heldens L., Dirks R. P., Hensen S. M., Onnekink C., van Genesen S. T., Rustenburg F., Lubsen N. H. (2010) Co-chaperones are limiting in a depleted chaperone network. Cell. Mol. Life Sci. 67, 4035–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ko J., Ou S., Patterson P. H. (2001) New anti-huntingtin monoclonal antibodies: implications for huntingtin conformation and its binding proteins. Brain Res. Bull. 56, 319–329 [DOI] [PubMed] [Google Scholar]
- 28. Weiss A., Abramowski D., Bibel M., Bodner R., Chopra V., DiFiglia M., Fox J., Kegel K., Klein C., Grueninger S., Hersch S., Housman D., Régulier E., Rosas H. D., Stefani M., Zeitlin S., Bilbe G., Paganetti P. (2009) Single-step detection of mutant huntingtin in animal and human tissues: a bioassay for Huntington's disease. Anal. Biochem. 395, 8–15 [DOI] [PubMed] [Google Scholar]
- 29. Weiss A., Grueninger S., Abramowski D., Giorgio F. P., Lopatin M. M., Rosas H. D., Hersch S., Paganetti P. (2011) Microtiter plate quantification of mutant and wild-type huntingtin normalized to cell count. Anal. Biochem. 410, 304–306 [DOI] [PubMed] [Google Scholar]
- 30. Schilb A., Riou V., Schoepfer J., Ottl J., Müller K., Chene P., Mayr L. M., Filipuzzi I. (2004) Development and implementation of a highly miniaturized confocal 2D-FIDA-based high-throughput screening assay to search for active site modulators of the human heat shock protein 90beta. J. Biomol. Screen. 9, 569–577 [DOI] [PubMed] [Google Scholar]
- 31. Brough P. A., Aherne W., Barril X., Borgognoni J., Boxall K., Cansfield J. E., Cheung K. M., Collins I., Davies N. G., Drysdale M. J., Dymock B., Eccles S. A., Finch H., Fink A., Hayes A., Howes R., Hubbard R. E., James K., Jordan A. M., Lockie A., Martins V., Massey A., Matthews T. P., McDonald E., Northfield C. J., Pearl L. H., Prodromou C., Ray S., Raynaud F. I., Roughley S. D., Sharp S. Y., Surgenor A., Walmsley D. L., Webb P., Wood M., Workman P., Wright L. (2008) 4,5-Diarylisoxazole Hsp90 chaperone inhibitors: potential therapeutic agents for the treatment of cancer. J. Med. Chem. 51, 196–218 [DOI] [PubMed] [Google Scholar]
- 32. Eccles S. A., Massey A., Raynaud F. I., Sharp S. Y., Box G., Valenti M., Patterson L., de Haven B. A., Gowan S., Boxall F., Aherne W., Rowlands M., Hayes A., Martins V., Urban F., Boxall K., Prodromou C., Pearl L., James K., Matthews T. P., Cheung K. M., Kalusa A., Jones K., McDonald E., Barril X., Brough P. A., Cansfield J. E., Dymock B., Drysdale M. J., Finch H., Howes R., Hubbard R. E., Surgenor A., Webb P., Wood M., Wright L., Workman P. (2008) NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 68, 2850–2860 [DOI] [PubMed] [Google Scholar]
- 33. Johnson J. L., Toft D. O. (1995) Binding of p23 and hsp90 during assembly with the progesterone receptor. Mol. Endocrinol. 9, 670–678 [DOI] [PubMed] [Google Scholar]
- 34. Duyao M. P., Auerbach A. B., Ryan A., Persichetti F., Barnes G. T., McNeil S. M., Ge P., Vonsattel J. P., Gusella J. F., Joyner A. L. (1995) Inactivation of the mouse Huntington's disease gene homolog Hdh. Science 269, 407–410 [DOI] [PubMed] [Google Scholar]
- 35. Kaarniranta K., Oksala N., Karjalainen H. M., Suuronen T., Sistonen L., Helminen H. J., Salminen A., Lammi M. J. (2002) Neuronal cells show regulatory differences in the hsp70 gene response. Brain Res. Mol. Brain Res. 101, 136–140 [DOI] [PubMed] [Google Scholar]
- 36. Waza M., Adachi H., Katsuno M., Minamiyama M., Sang C., Tanaka F., Inukai A., Doyu M., Sobue G. (2005) 17-AAG, an Hsp90 inhibitor, ameliorates polyglutamine-mediated motor neuron degeneration. Nat. Med. 11, 1088–1095 [DOI] [PubMed] [Google Scholar]
- 37. Tokui K., Adachi H., Waza M., Katsuno M., Minamiyama M., Doi H., Tanaka K., Hamazaki J., Murata S., Tanaka F., Sobue G. (2009) 17-DMAG ameliorates polyglutamine-mediated motor neuron degeneration through well-preserved proteasome function in an SBMA model mouse. Hum. Mol. Genet. 18, 898–910 [DOI] [PubMed] [Google Scholar]
- 38. Thomas M., Harrell J. M., Morishima Y., Peng H. M., Pratt W. B., Lieberman A. P. (2006) Pharmacologic and genetic inhibition of hsp90-dependent trafficking reduces aggregation and promotes degradation of the expanded glutamine androgen receptor without stress protein induction. Hum. Mol. Genet. 15, 1876–1883 [DOI] [PubMed] [Google Scholar]
- 39. Labbadia J., Cunliffe H., Weiss A., Katsyuba E., Sathasivam K., Seredenina T., Woodman B., Moussaoui S., Frentzel S., Luthi-Carter R., Paganetti P., Bates G. P. (2011) Altered chromatin architecture underlies progressive impairment of the heat shock response in mouse models of Huntington disease. J. Clin. Investig. 121, 3306–3319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 41. Bowman A. B., Yoo S. Y., Dantuma N. P., Zoghbi H. Y. (2005) Neuronal dysfunction in a polyglutamine disease model occurs in the absence of ubiquitin-proteasome system impairment and inversely correlates with the degree of nuclear inclusion formation. Hum. Mol. Genet. 14, 679–691 [DOI] [PubMed] [Google Scholar]
- 42. Hunter J. M., Lesort M., Johnson G. V. (2007) Ubiquitin-proteasome system alterations in a striatal cell model of Huntington's disease. J. Neurosci. Res. 85, 1774–1788 [DOI] [PubMed] [Google Scholar]
- 43. Ortega Z., Díaz-Hernández M., Maynard C. J., Hernández F., Dantuma N. P., Lucas J. J. (2010) Acute polyglutamine expression in inducible mouse model unravels ubiquitin/proteasome system impairment and permanent recovery attributable to aggregate formation. J. Neurosci. 30, 3675–3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bett J. S., Goellner G. M., Woodman B., Pratt G., Rechsteiner M., Bates G. P. (2006) Proteasome impairment does not contribute to pathogenesis in R6/2 Huntington's disease mice: exclusion of proteasome activator REGgamma as a therapeutic target. Hum. Mol. Genet. 15, 33–44 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.