Background: In the malaria parasite, Plasmodium falciparum, a phosphoethanolamine methyltransferase (PfPMT) is critical for membrane biogenesis.
Results: Structures and mutagenesis of PfPMT suggest Tyr-19 and His-132 as a catalytic dyad.
Conclusion: The reaction sequence of PfPMT likely involves structural changes with the Tyr-19-His-132 forming an active site latch.
Significance: This is the first structure of an enzyme essential for the survival of the malaria parasite.
Keywords: Crystal Structure, Enzyme Catalysis, Enzyme Structure, Parasite, S-Adenosylmethionine (SAM)
Abstract
In the malarial parasite Plasmodium falciparum, a multifunctional phosphoethanolamine methyltransferase (PfPMT) catalyzes the methylation of phosphoethanolamine (pEA) to phosphocholine for membrane biogenesis. This pathway is also found in plant and nematodes, but PMT from these organisms use multiple methyltransferase domains for the S-adenosylmethionine (AdoMet) reactions. Because PfPMT is essential for normal growth and survival of Plasmodium and is not found in humans, it is an antiparasitic target. Here we describe the 1.55 Å resolution crystal structure of PfPMT in complex with AdoMet by single-wavelength anomalous dispersion phasing. In addition, 1.19–1.52 Å resolution structures of PfPMT with pEA (substrate), phosphocholine (product), sinefungin (inhibitor), and both pEA and S-adenosylhomocysteine bound were determined. These structures suggest that domain rearrangements occur upon ligand binding and provide insight on active site architecture defining the AdoMet and phosphobase binding sites. Functional characterization of 27 site-directed mutants identifies critical active site residues and suggests that Tyr-19 and His-132 form a catalytic dyad. Kinetic analysis, isothermal titration calorimetry, and protein crystallography of the Y19F and H132A mutants suggest a reaction mechanism for the PMT. Not only are Tyr-19 and His-132 required for phosphobase methylation, but they also form a “catalytic” latch that locks ligands in the active site and orders the site for catalysis. This study provides the first insight on this antiparasitic target enzyme essential for survival of the malaria parasite; however, further studies of the multidomain PMT from plants and nematodes are needed to understand the evolutionary division of metabolic function in the phosphobase pathway of these organisms.
Introduction
Malaria is a major worldwide health threat as this disease, caused by different species of Plasmodium parasites, results in over 1 million deaths and 300 million clinical cases each year (1). In particular, infections by Plasmodium falciparum are the most severe with the highest rates of mortality and morbidity. The global prevalence of malaria, the fact that ∼40% of mankind lives in endemic areas, and the emergence of drug-resistant strains drive the need to identify new biochemical targets for the development of therapeutics targeting this protozoan parasite (2).
The rapid growth and replication of Plasmodium in human erythrocytes requires a significant source of phospholipids to support membrane biogenesis, and phospholipid biosynthesis has been explored as a target for antimalarial compounds (3, 4). In mammals, phosphatidylcholine is the major phospholipid in cellular membranes and is synthesized either by the de novo choline (or Kennedy) pathway, which converts dietary choline to phosphocholine (pCho)2 and then to the phospholipid via CDP intermediates, or by methylation of phosphatidylethanolamine to phosphatidylcholine through the Bremer-Greenberg pathway (5). In contrast, plants methylate phosphoethanolamine (pEA) into pCho (Fig. 1A), which then enters the Kennedy pathway, through the action of S-adenosylmethionine (AdoMet)-dependent phosphoethanolamine methyltransferases (PMT) (6). Plasmodium use a plant-like phosphobase pathway for the biosynthesis of pCho as a metabolic precursor for phospholipid synthesis (7–10). Similarly, the plant-like phosphobase methylation pathway is also essential for the normal growth and development of the free-living nematode Caenorhabditis elegans (11–14). Importantly, this phosphobase methylation pathway, which is required by Plasmodium and nematodes, is not found in mammals.
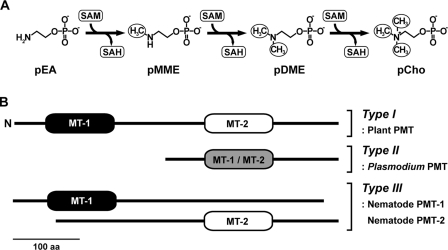
FIGURE 1.
Phosphobase methylation and domain arrangement in the PMT family. A, the PMT pathway showing the sequential methylation of pEA to pCho. SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine. B, organization of the methyltransferase (MT) domains in the plant, Plasmodium, and nematode PMT is shown. MT-1 domains catalyze the conversion of pEA to pMME, and MT-2 domains catalyze the reactions from pMME to pCho. 100 aa, 100 amino acids.
The PMT from plants, nematodes, and Plasmodium utilize differing protein architectures to catalyze the sequential methylation of pEA to phospho-monomethylethanolamine (pMME), pMME to phospho-dimethylethanolamine (pDME), and pDME to pCho (Fig. 1B). In plants, a bifunctional PMT catalyzes these reactions with an N-terminal methyltransferase domain converting pEA to pMME and a C-terminal methyltransferase domain performing the last two methylation reactions of the pathway (6). The function of these domains is divided into two monofunctional PMT in C. elegans (11, 12), whereas P. falciparum uses a single-domain PMT (PfPMT) for all three methylation reactions (7).
PfPMT was shown to use pEA as a substrate to synthesize pCho as a precursor for phosphatidylcholine synthesis (7, 8). A genetic knock-out of the PfPMT gene completely abolishes phosphatidylcholine synthesis via the phosphobase pathway and shows that the Bremer-Greenberg pathway does not compensate for the loss of the primary metabolic route to the phospholipid (9). Moreover, disruption of the PfPMT gene leads to significant defects in growth, reproduction, and viability, suggesting a critical role for phosphobase methylation in the pathogenesis of the parasite (9). Although in vivo studies demonstrate the metabolic role of the enzyme and suggest it as a potential therapeutic target, the molecular understanding of how PfPMT functions is limited. Site-directed mutagenesis of conserved residues in the canonical AdoMet binding motif revealed the importance of this motif (15) but did not suggest functional roles of these residues. Extensive kinetic analysis of the C. elegans and wheat PMT showed that these enzymes use a random bi bi mechanism (11, 12, 16), but do not provide information about the active sites of these proteins. There have also been initial efforts to identify inhibitors of PfPMT, which include the AdoMet analog sinefungin, the choline analog miltefosine (hexadecyltrimethylammonium), and the 4-aminoquinoline amodiaquine (7, 17).
Here we describe the 1.19–1.55 Å resolution structures of PfPMT in complex with AdoMet (substrate), pEA (substrate), pCho (product), and sinefungin (inhibitor) and as a dead-end complex with pEA and S-adenosylhomocysteine (AdoCys). Functional characterization of site-directed mutants targeting active site residues and crystallographic analysis of the Y19F and H132A variants investigate the roles of key residues in catalysis and substrate recognition. Overall, this study provides insight on the reaction mechanism and the active site structure of an enzyme that is essential for survival of the malaria parasite P. falciparum and is a potential target for the development of antiparasitic compounds.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Mutagenesis
For bacterial expression, the PfPMT cDNA (7) was synthesized (GenScript) with Escherichia coli codon optimization and introduction of NdeI and BamHI sites at the 5′- and 3′-ends of the gene, respectively. The NdeI/BamHI fragment from the synthetic gene was excised and subcloned into pET-28a (Novagen). Expression in E. coli BL21(DE3) and purification by nickel-affinity and size-exclusion chromatographies were as described for the C. elegans PMT (11, 12). Selenomethionine (Se-Met)-substituted protein was produced by inhibition of the E. coli methionine biosynthesis pathway with the AdoMet vector and bacterial strain used for native protein expression (18). Incorporation of Se-Met was confirmed by mass spectrometry to compare intact molecular masses of native and derivatized protein. Purification of Se-Met-PfPMT was as for native protein. Site-directed mutants were generated using the QuikChange PCR method (Stratagene) with expression and purification as above.
Enzyme Kinetics and Isothermal Titration Calorimetry
Activity assays were performed using the standard PMT radiochemical assay at fixed concentrations of AdoMet (0.5 mm) and pEA (2 mm) (11, 12). For determination of kinetic parameters, reactions were performed either with fixed AdoMet (0.5 mm) and varied phosphobase (0.010–2 mm) or with fixed phosphobase (2 mm) and varied AdoMet (5–500 μm). All data were fit to the Michaelis-Menten equation in SigmaPlot. Calorimetric analysis of AdoCys and pCho binding to PfPMT was performed, and data were analyzed as described previously for the nematode PMT (19).
Protein Crystallography
Crystals of Se-Met-PfPMT in complex with AdoMet were grown by the vapor diffusion method in hanging drops of a 1:1 mixture of protein (13.5 mg ml−1) and crystallization buffer (20% PEG-8000, 0.1 m sodium cacodylate, pH 6.5, 0.2 m sodium acetate, 20 mm tris(2-carboxyethyl)phosphine, and 5 mm AdoMet). Crystals of native and mutant PfPMT in complex with various ligands (5 mm for each ligand except 0.5 mm AdoCys) were obtained in similar conditions (20–30% PEG8000, 0.1 m sodium cacodylate, pH 6.5, 0.2 m sodium acetate, 5 mm β-mercaptoethanol).
The PfPMT structure was solved by single-wavelength anomalous dispersion phasing. Diffraction data collected at beamline 19ID of the Argonne National Laboratory Advanced Photon Source were indexed, integrated, and scaled using HKL3000 (20). SHELX (21) was used to determine initial Se-Met positions and to estimate initial phases from the peak wavelength data set. Se-Met positions and parameters were refined in MLPHARE (22). Solvent flattening was performed with DM (23), and ARP/wARP (24) was used to build an initial model. Iterative rounds of manual model building and refinement were performed with COOT (25) and PHENIX (26), respectively. The resulting model was used for molecular replacement into the higher resolution native data set using PHASER (27). Structures of the PfPMT·sinefungin, PfPMT·pEA, PfPMT·pCho, PfPMT·pEA·AdoCys, H132A·pCho, and Y19F·pCho complexes were solved by the difference Fourier method. Data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Summary of PfPMT crystallographic statistics
| Crystal | PfPMT (Se-Met) + AdoMet, PO42− | PfPMT + AdoMet, PO42− | PfPMT + sinefungin, PO42− | PfPMT + pCho | PfPMT + pEA | PfPMT + AdoCys, pEA | PfPMT-H132A + pCho | PfPMT-Y19F + pCho |
| Space group | C2 | P2 | C2 | C2 | C2 | P2 | C2 | C2 |
| Cell dimensions | a = 88.39 Å, b = 41.15 Å, c = 77.06 Å; β = 108.6° | a = 77.04 Å, b = 44.13 Å, c = 88.41 Å; β = 108.5° | a = 89.31 Å, b = 43.89 Å, c = 77.13 Å; β = 108.6° | a = 90.07 Å, b = 43.98 Å, c = 77.03 Å; β = 109.1° | a = 89.66 Å, b = 44.07 Å, c = 77.05 Å; β = 108.6° | a = 76.80 Å, b = 44.16 Å, c = 88.99 Å; β = 108.3° | a = 89.30 Å, b = 44.19 Å, c = 76.95 Å; β = 108.8° | a = 89.23 Å, b = 44.06 Å, c = 76.97 Å; β = 108.4° |
| Data collection | ||||||||
| Wavelength (Å) | 0.979 | 0.979 | 0.979 | 0.979 | 0.979 | 0.979 | 0.979 | 0.979 |
| Resolution (Å) (highest shell) | 36.5-1.97 (2.00-1.97) | 37.8-1.55 (1.60-1.55) | 36.7-1.35 (1.37-1.35) | 24.9-1.24 (1.26-1.24) | 32.5-1.47 (1.50-1.47) | 38.1-1.52 (1.55-1.52) | 28.9-1.19 (1.21-1.19) | 43.0-1.50 (1.53–1.50) |
| Reflections (total/unique) | 61,355/19,867 | 230,099/74,087 | 217,888/59,825 | 274,651/76,023 | 166,772/48,655 | 314,964/87,067 | 318,728/90,748 | 146,444/45,420 |
| Completeness (highest shell) | 97.3% (67.6%) | 99.1% (99.1%) | 96.1% (78.8%) | 94.3% (88.5%) | 99.4% (94.2%) | 99.7% (99.2%) | 99.6% (95.2%) | 99.5% (98.6%) |
| <I/σ> (highest shell) | 25.2 (11.1) | 18.1 (1.8) | 25.2 (1.9) | 29.3 (3.0) | 31.3 (2.2) | 19.9 (2.0) | 27.4 (2.0) | 26.2 (2.0) |
| Rsym (highest shell) | 6.3 (10.7)% | 6.7 (38.2)% | 4.7 (35.5)% | 7.8 (35.9)% | 6.3 (40.6)% | 8.2 (63.8)% | 4.7 (40.9)% | 6.0 (50.4)% |
| Refinement | ||||||||
| Rcryst/Rfree | 14.9/18.5 | 16.7/18.5 | 16.3/17.8 | 14.7/17.3 | 16.5/17.9 | 17.1/19.7 | 14.5/16.7 | 16.2/18.3 |
| No. of protein atoms | 2,140 | 4,371 | 2,163 | 2,228 | 2,172 | 4,287 | 2,228 | 2,172 |
| No. of water molecules | 325 | 875 | 388 | 516 | 404 | 808 | 521 | 378 |
| No. of ligand atoms | 32 | 64 | 32 | 11 | 8 | 68 | 11 | 11 |
| r.m.s. deviation, bond lengths (Å) | 0.006 | 0.006 | 0.006 | 0.005 | 0.006 | 0.006 | 0.005 | 0.006 |
| r.m.s. deviation, bond angles (°) | 1.015 | 1.030 | 1.014 | 0.947 | 0.966 | 1.016 | 0.980 | 0.975 |
| Average B-factor (Å2), protein, water, ligand | 16.9, 30.6, 20.4 | 16.1, 33.2, 20.3 | 16.2, 29.7, 13.4 | 14.3, 32.1, 9.5 | 21.0, 36.6, 16.9 | 15.5, 31.7, 12.8 | 13.3, 29.9, 8.1 | 18.4, 35.0, 13.9 |
| Stereochemistry: favored and allowed | 98.8, 1.2% | 98.5, 1.5% | 98.1, 1.9% | 98.9, 1.1% | 98.5, 1.5% | 98.5, 1.5% | 98.6, 1.4% | 98.9, 1.1% |
RESULTS
Steady-state Kinetic Analysis
Previously, only pEA was used as a substrate for PfPMT (7). To examine phosphobase specificity, PfPMT was overexpressed in E. coli as an N-terminal hexahistidine-tagged protein and purified by Ni2+-affinity and size-exclusion chromatographies. As observed for the C. elegans PMT (11, 12), PfPMT was monomeric upon gel filtration. Kinetic analysis of PfPMT revealed less than 2-fold differences in kcat/Km for pEA, pMME, and pDME (Table 2), which contrasts with the 6–20-fold higher efficiency of CePMT1 versus CePMT2 (11, 12). These results confirm that PfPMT efficiently catalyzes all reactions in the phosphobase pathway.
TABLE 2.
Kinetic parameters of PfPMT
All values are expressed as a mean ± S.E. (n = 3).
| Substrate | kcat | Km | kcat/Km |
|---|---|---|---|
| min−1 | μm | m−1s−1 | |
| pEA | 109 ± 2 | 54.2 ± 3.8 | 33,518 |
| pMME | 218 ± 24 | 181 ± 45 | 20,074 |
| pDME | 185 ± 14 | 66.8 ± 12 | 46,158 |
| AdoMet (pEA) | 23.5 ± 0.9 | 29.9 ± 3.8 | 13,099 |
| AdoMet (pDME) | 29.2 ± 1.2 | 33.1 ± 4.2 | 14,703 |
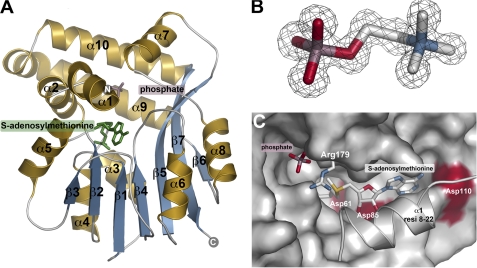
Overall Structure of PfPMT
The x-ray crystal structure of PfPMT in complex with AdoMet and PO42− was determined by single-wavelength anomalous dispersion phasing using the anomalous signal from three Se-Met. The resulting model used to solve, build, and refine a native 1.55 Å resolution structure (Table 1 and Fig. 2A). The PfPMT monomer consists of an α-helical “lid” domain (α1, α2, α7, α9-α10) and a canonical AdoMet binding domain defined by a central β-sheet (β1-β7) flanked by two α-helical regions (α3/α4/α5 and α6/α8). The overall fold of PfPMT was most similar to those of cyclopropane-fatty-acyl-phospholipid synthase (2.4 Å r.m.s.d.; 17% identity) and methoxymycolic acid synthase (2.5 Å r.m.s.d.; 16% identity), both N-methyltransferases from Mycobacterium (28, 29). In addition, other methyltransferases, including glycine N-methyltransferase (2.9 Å r.m.s.d.; 13% identity), histamine N-methyltransferase (3.3 Å r.m.s.d.; 14% identity), phenylethanolamine N-methyltransferase (3.0 Å r.m.s.d.; 13% identity), and guanidinoacetate N-methyltransferase (3.3 Å r.m.s.d.; 12% identity), were also identified by a DALI search as structurally related (30–33). All of these proteins share a conserved AdoMet binding domain with major variations in the topology and residues of the lid domain (supplemental Fig. S1).
FIGURE 2.
PfPMT structure. A, ribbon diagram of the PfPMT·AdoMet·PO42− complex with α-helices and β-strands colored gold and blue, respectively. Positions of AdoMet (green) and PO42− (rose) are indicated by the stick molecules. B, representative electron density. The 2Fo − Fc omit map (1.5 σ) for pCho in the 1.19 Å resolution PfPMT·pCho complex is shown. C, view of the AdoMet binding site and the position of the α1 lid helix. PfPMT is shown as a surface rendering with the exception of Arg-179 and residues 8–22 (resi 8–22) of α1. Surfaces for Asp-61, Asp-85, and Asp-110 are highlighted in red.
In addition to the PfPMT·AdoMet·PO42− complex, structures of the pEA, pCho, sinefungin·PO42−, and pEA·AdoCys complexes, and structures of the Y19F and H132A mutants, each in complex with pCho, were also determined (Table 1). Electron density for each ligand in these structures was unambiguous, as illustrated by a representative omit map for pCho (Fig. 2B).
PfPMT Active Site
In the PfPMT·AdoMet·PO42− complex, the position of AdoMet at the interface of the two domains clearly delineates the active site (Fig. 2, A and C). Asp-61, Asp-85, and Asp-110 define the interior side of the AdoMet binding site along the top of the central β-sheet with α1 of the lid domain locking the ligand into the site and occluding solvent access. Binding of PO42− identifies a likely phosphobase binding pocket with Arg-179 capping the ligand. This structure suggests that conformational changes occur upon ligand binding as both sites are inaccessible to substrates without structural rearrangements. Subsequent structures of PfPMT in complex with pEA, pCho, sinefungin·PO42−, and pEA·AdoCys further define the active site architecture.
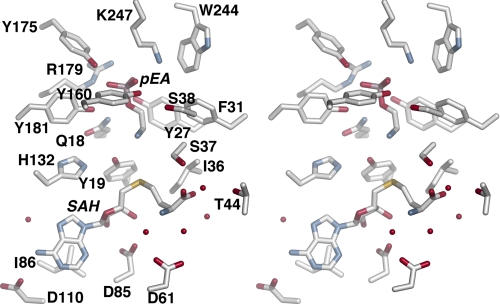
The structure of AdoCys and pEA as a dead-end complex provides an overall view of the active site, which can be divided into the AdoMet/AdoCys binding site, the phosphobase binding site, and potential catalytic residues (Fig. 3). Within the AdoMet/AdoCys binding site (Fig. 3 and supplemental Fig. S2A), the adenine ring stacks with Ile-86 and forms hydrogen bonds with the backbone nitrogen of Ile-111, the side-chain carboxylate of Asp-110, and a water, which in turn contacts Asp-10. Asp-85 forms a central bidentate contact with the two hydroxyl groups of the ribose moiety. The backbone nitrogen of Ile-36, the hydroxyl group of Ser-37, and a water-mediated interaction with Thr-44 form interactions with the carboxylate of the homocysteine moiety. The amino group of the homocysteine moiety interacts with the backbone carbonyl of Gly-63 and two water molecules that are anchored to Asp-61. These interactions orient the homocysteine Sδ toward the nitrogen of pEA.
FIGURE 3.
Stereo view of active site residues in PfPMT·AdoCys·pEA complex. Waters forming hydrogen bonds with the ligands are shown as spheres. SAH, S-adenosylhomocysteine.
Extensive interactions with the phosphate group of pEA positions the substrate in the active site with the primary amine ∼4 Å from the Sδ of AdoCys (Fig. 3 and supplemental Fig. S2B). Arg-179 and Lys-247 form charge-charge interactions, and the hydroxyl groups of Tyr-27, Tyr-160, Tyr-175, and Tyr-181 provide hydrogen bonds to the phosphate group. The nitrogen of the Gln-18 side chain contacts the bridging oxygen of pEA. Between the AdoCys and pEA binding sites, the Nϵ of His-132 interacts with the hydroxyl group of Tyr-19, which is 2.5 Å away from the substrate amine. Based on the position of Tyr-19 and His-132, these residues may function as a catalytic dyad in the phosphobase methylation reaction.
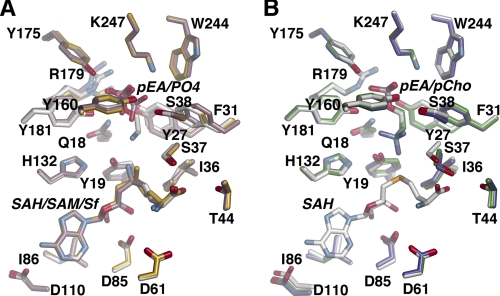
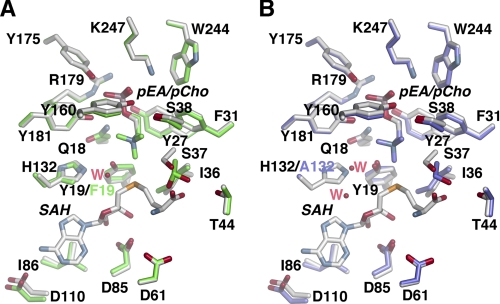
Structural comparison of the AdoMet, AdoCys, and sinefungin complexes shows that residues interacting with these ligands do not vary in position (Fig. 4A); however, the Nδ of sinefungin is oriented toward Tyr-19 (2.84 Å), and not the phosphobase site, as observed for the methyl group of AdoMet. Likewise, the conformations of residues and ligands in the phosphobase site are essentially identical in the pEA, pCho, and AdoCys·pEA complexes (Fig. 4B), although Tyr-27, Phe-31, and Arg-179 differ in position depending on whether PO42− or a phosphobase is bound in the site (Fig. 4). The largest conformational change occurs in Arg-179, which is oriented into the active site when either pEA or pCho are bound, but shifts away from the active site when PO42− is bound.
FIGURE 4.
Active site comparisons between different complexes. A, overlay of the active sites in the AdoCys·pEA (white), AdoMet·PO4 (gold), and sinefungin (Sf)·PO4 (rose) complexes. SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine. B, overlay of the active sites of PfPMT in the AdoCys·pEA (white), pEA (green), and pCho (purple) complexes.
Site-directed Mutagenesis of AdoMet/AdoCys and Phosphobase Binding Site Residues
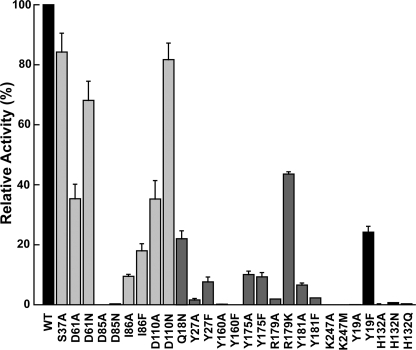
A series of site-directed mutants targeting residues in the AdoMet binding site (S37A, D61N, D61A, D85N, D85A, I86A, I86F, D110N, D110A), the phosphobase binding site (Q18N, Y27F, Y27A, Y160F, Y160A, Y175F, Y175A, R179K, R179A, Y181F, Y181A, K247M, K247A), and the putative catalytic dyad (Y19F, Y19A, H132A, H132Q, H132N) were generated by PCR mutagenesis. All 27 mutants were expressed and purified as soluble monomeric proteins with yields comparable with wild-type PfPMT for activity assays. Using standard assay conditions, the methylation reaction was not detected for the Y19A, D85A, D85N, H132A, H132Q, H132N, Y160A, Y160F, R179A, K247A, and K247M mutants, but varying levels of activity were observed for other mutants (Fig. 5). Deviations in relative activity can result from either changes in turnover rates and/or elevated Km values.
FIGURE 5.
Comparison of wild-type and mutant PfPMT activities. Mutants proteins are grouped and colored by active site location, as follows: light gray = AdoMet binding site; dark gray = phosphobase site; and black = catalytic dyad. Standard assay conditions were used. Average values (n = 3) with S.E. are plotted relative to wild-type activity.
Within the AdoMet site, Asp-85 and Ile-86 provide critical interactions with the substrate as mutations of either residue (D85N/D85A or I86F/I86A) abrogated activity, likely through a loss of AdoMet binding. Mutation of Ser-37, Asp-61, and Asp-110 resulted in mutants with ∼35–85% of wild-type activity (Fig. 5). The higher activity in the D61N (68%) and D110N (82%) mutants suggests that substitution of the side-chain carboxylate for an amide retains hydrogen-bonding patterns with AdoMet, whereas the D61A (35%) and D110A (35%) mutants disrupt these interactions.
Mutant analysis of residues in the phosphobase site indicate that the electrostatic interactions provided by Arg-179 and Lys-247 are required for activity as the K247A and K247M mutants were inactive and the R179A and R179K mutants displayed 1 and 43% activity, respectively (Fig. 5). In addition, each of the tyrosine residues in the site (Tyr-27, Tyr-160, Tyr-175, and Tyr-181) were also important for PfPMT activity as removal of either the hydroxyl or the phenol moieties severely disrupted methylation of pEA, yielding mutants that were inactive (Y160F/Y160A) or with 1–10% of wild-type activity (Fig. 5). Similarly, the Q18N mutant also reduced activity to 22% (Fig. 5). These data indicate that the phosphobase site is highly organized with multiple specific interactions for efficient catalysis, most likely through binding interactions with substrate.
Structural and Functional Analysis of the Catalytic Dyad
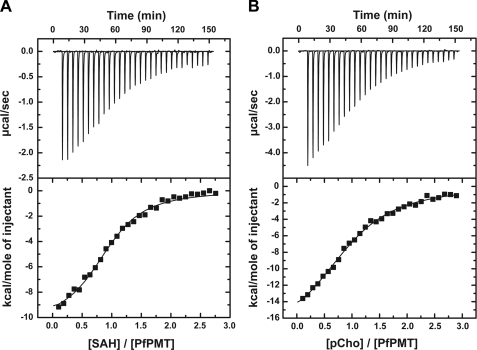
Mutagenesis of either Tyr-19 or His-132 in the putative catalytic dyad severely affects PfPMT activity. The H132N, H132Q, H132A, and Y19A mutants displayed no detectable activity, and the Y19F mutant retained 24% activity (Fig. 5). Because of the potential role of these residues in the reaction mechanism of PfPMT, a more detailed analysis of the Y19F and H132A mutants was performed, which included kinetic analysis of the Y19F mutant, isothermal titration calorimetry (ITC) of AdoCys and pCho binding to wild-type, Y19F, and H132A PfPMT, and crystallographic analysis of Y19F and H132A mutants, both in complex with pCho.
Determination of the steady-state kinetic parameters for the Y19F mutant (kcat = 1.85 ± 0.3 min−1; KmpEA = 644 ± 40 μm) showed that removal of the side-chain hydroxyl group reduced the turnover rate 60-fold and increased the Km for pEA 12-fold when compared with wild-type enzyme. Evaluation of AdoCys and pCho binding to the wild-type, Y19F, and H132A PfPMT was performed using ITC (Fig. 6 and Table 3) (24). This analysis shows that these proteins bind each ligand with comparable Kd values; however, the enthalpic and entropic contributions to ligand binding change in the mutants (Table 3). Crystallographic analysis of the Y19F and H132A mutants (Fig. 7) reveal that the overall active site structures are similar to that of the PfPMT·AdoCys·pEA complex with differences localized where the amino acid substitutions are. In the Y19F active site (Fig. 7A), mutation of Tyr-19 to a phenylalanine provides space for a water molecule to enter the site and interact with the Nϵ of His-132 (2.7 Å), which shifts slightly in position when compared with wild type. This water approximates the position of the hydroxyl group of Tyr-19 and maintains the overall structure of the catalytic dyad, albeit with a reduction in reaction rate. In the H132A active site (Fig. 7B), two water molecules enter the space formed by removal of the imidazole group, one of which is 2.7 Å from the hydroxyl group of Tyr-19. In the H132A mutant structure, the position of Tyr-19 remains in the same position as observed in the wild-type active site. Structural and functional analysis of the Tyr-19 and His-132 PfPMT mutants suggests a likely reaction mechanism for the PMT family of enzymes (Fig. 8).
FIGURE 6.
ITC analysis of ligand binding. A, titration of PfPMT with AdoCys. The top panel shows data plotted as heat signal (μcal sec−1) versus time (min). The experiment consisted of 30 injections of 10 μl AdoCys (650 μm) into a solution containing PMT protein (51.5 μm) at 20 °C. The bottom panel shows the integrated heat response per injection from the upper panel plotted as normalized heat per mol of injectant. SAH, S-adenosylhomocysteine. The solid line represents the fit to the data. B, titration of PfPMT (65.1 μm) with pCho (850 μm). Top and bottom panels are as in A.
TABLE 3.
ITC analysis of AdoCys and pCho binding
Titrations were performed at 20 °C and data fit to a single-site binding model as described under “Experimental Procedures.”
| Protein-Ligand | Kd | ΔG | ΔH | −TΔS |
|---|---|---|---|---|
| μm | kcal mol−1 | kcal mol−1 | kcal mol−1 | |
| PfPMT-AdoCys | 7.9 ± 0.8 | −6.8 ± 0.7 | −11.0 ± 0.3 | 4.2 |
| Y19F-AdoCys | 12.0 ± 1.8 | −6.6 ± 1.0 | −5.3 ± 0.2 | −1.3 |
| H132A-AdoCys | 12.2 ± 2.4 | −6.6 ± 1.3 | −1.6 ± 0.1 | −5.0 |
| PfPMT-pCho | 23.4 ± 1.6 | −6.2 ± 0.4 | −22.3 ± 0.7 | 16.1 |
| Y19F-pCho | 20.2 ± 6.8 | −6.3 ± 2.1 | −1.9 ± 0.2 | −4.4 |
| H132A-pCho | 26.0 ± 11.3 | −6.2 ± 2.7 | −1.6 ± 0.2 | −4.5 |
FIGURE 7.
Active site comparisons between wild-type and mutant PfPMT. A, overlay of the active sites in the AdoCys·pEA (white) and Y19F·pCho (green) complexes. SAH, S-adenosylhomocysteine. B, overlay of the active sites in the AdoCys·pEA (white) and H132A·pCho (purple) complexes.
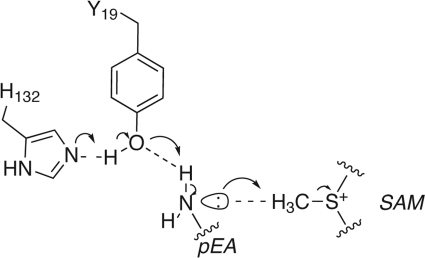
FIGURE 8.
Proposed reaction mechanism of PfPMT. SAM, S-adenosylmethionine.
DISCUSSION
The phosphobase methylation pathway from pEA to pCho (Fig. 1A) provides Plasmodium with a novel and essential metabolic route for membrane biogenesis that is not found in mammals (7–10). In addition, this pathway is also required for growth and development in C. elegans (11–14, 19). Given the biological function of the PMT in Plasmodium and nematodes and that these enzymes are not found in mammals, the PMT are potential targets for the development of antiparasitic therapeutics (10, 13, 14, 17). The PfPMT structure (Fig. 2) provides the first three-dimensional view of this enzyme from any species, yields new insight on catalysis and ligand binding in this group of methyltransferases, and may be useful for the development of inhibitors targeting this enzyme from P. falciparum, the major causative agent of malaria.
Comparisons of PfPMT with other N-methyltransferases show structural conservation of the canonical AdoMet binding fold in PfPMT but also highlight the diversification of the lid domain, most likely for recognition of chemically distinct substrates (supplemental Fig. S1). Although the organization of methyltransferase domains in the Plasmodium, nematode, and plant PMT differs (Fig. 1B), sequence alignment of PfPMT with the C-terminal methyltransferase domains of C. elegans PMT2 (11), Haemonchus contortus (sheep barber pole worm) PMT2 (19), and Arabidopsis thaliana PMT (34) shows that residues forming the sequence motifs (I, post-I, II, and III) defining the AdoMet binding region in the C terminus are conserved and located in the β-strands at the core of this scaffold (supplemental Fig. S3). Low sequence identity between PfPMT and the N-terminal methyltransferase domains from C. elegans PMT1, H. contortus PMT1, and A. thaliana PMT preclude accurate sequence alignments. Previously, mutagenesis of PfPMT targeting residues in the conserved motifs yielded inactive proteins (15), which likely resulted from disruption of protein folding and not binding and/or catalysis. Within the AdoMet binding site, Ser-37, Gly-63, Asp-61, Asp-85, and Asp-110 are conserved in PfPMT, CePMT2, HcPMT2, and AtPMT, but other residues (Asp-10, Ile-36, Thr-44, Ile-86, and Ile-111) that are generally at the periphery of the AdoMet binding site vary. Functional analysis of PfPMT mutants in the AdoMet binding site showed that Asp-85 and Ile-86 are important for activity with other positions of varied importance (Fig. 5).
All of the residues forming the phosphobase site are invariant between PfPMT, CePMT2, HcPMT2, and the second domain of AtPMT (Figs. 3 and 4 and supplemental Fig. S3). Site-directed mutagenesis of amino acids in the phosphobase site also highlights the functional importance of interactions with the substrate phosphate group for activity (Fig. 5). Interestingly, conservation of residues in the phosphobase binding site of the PMT raises the question of what determines substrate specificity in these enzymes as PfPMT accepts pEA, pMME, and pDME as substrates (Table 2), but the nematode PMT2 and the C-terminal domain of the plant PMT do not use pEA as a substrate (6, 11, 19). Structural comparison of PfPMT with pEA-, pCho-, and AdoCys·pEA-bound forms shows that the active site can readily accommodate pEA and its methylated products (Fig. 4B). The molecular basis for substrate specificity in the different types of PMT is currently unclear; however, it is possible that the multidomain organization of the plant and nematode PMT alters the active site structure to favor pMME and pDME versus pEA as substrates. Further structural studies of the PMT from nematodes and plants are needed to understand the evolutionary division of metabolic function in the phosphobase pathway of these organisms.
Structural and functional analysis of PfPMT, together with previous work on the nematode and plant PMT, suggests a plausible reaction sequence for phosphobase methylation. The nematode and plant PMT use a random bi bi kinetic mechanism in which binding of either AdoMet or phosphobase occurs first followed by binding of the other ligand (11, 12, 16). Crystal structures of PfPMT in complex with pEA, pCho, AdoMet, and AdoCys·pEA are consistent with this kinetic mechanism (Figs. 2–4). However, these structures suggest that conformational changes between apoenzyme- and ligand-bound forms likely occur as both phosphobase and AdoMet binding sites are largely solvent-inaccessible (Fig. 2C). In particular, movement of the N-terminal region of PfPMT, including α1, upon ligand binding appears important for locking of AdoMet/AdoCys in the active site. Similar movement of the N-terminal regions in other N-methyltransferases upon ligand binding has been observed (30–33). Previously, thermodynamic analysis of AdoCys binding to the two PMT from H. contortus suggested that ligand binding is accompanied by changes in protein structure (19). Moreover, localized conformational changes in the PfPMT active site are also likely as the conformations of Tyr-27, Phe-31, and Arg-179 differ between PO42−-, pEA-, and pCho-bound forms (Fig. 4) Thus, structural changes likely order the active site for catalysis.
Within the PfPMT active site, Tyr-19 and His-132 are positioned between the AdoMet and phosphobase sites to function as a catalytic dyad in the SN2 transfer of a methyl group from AdoMet to the amine on pEA, pMME, or pDME (Figs. 3 and 8). These two amino acids are also invariant in the nematode and plant PMT (supplemental Fig. S3). In the PfPMT·AdoCys·pEA complex, the hydroxyl group of Tyr-19 is 3.5 Å away from the Sδ of AdoCys, 2.5 Å from the amine of pEA, and 2.8 Å from the Nϵ of His-132. In the proposed reaction mechanism (Fig. 8), His-132 functions as a general base to abstract a proton from the hydroxyl group of Tyr-19 to activate the residue. The negatively charged oxygen interacts with a hydrogen on the substrate amine to distort the electron configuration from sp2 to sp3, which orients the lone pair electrons toward the methyl group of AdoMet to facilitate methylation of the phosphobase.
In this mechanism, His-132 is the critical driver for catalysis. The loss of activity in the H132A, H132Q, and H132N mutants is consistent with the proposed mechanism. Binding analysis of the H132A mutant by ITC demonstrates that this substitution does not significantly change the Kd values for AdoCys and pCho when compared with wild type (Table 3), which indicates that the loss of activity does not result from an inability to bind ligands. In addition, the 1.19 Å resolution structure of H132A shows that the active site structure is essentially identical to that of wild-type with the exception of two water molecules that fit in the space left by removal of the side-chain imidazole in the mutant (Fig. 7B). This structure also indicates that the presence and positioning of Tyr-19 alone are insufficient to drive the methylation reaction.
Although the lack of activity in the Y19A mutant agrees with the reaction mechanism, the reduced activity in the Y19F mutant appears inconsistent. Removal of the hydroxyl group from Tyr-19 reduces activity; however, the Y19F mutant binds AdoCys and pCho with affinities similar to wild type (Table 3). Structural analysis of the Y19F mutant reveals the presence of a water molecule that approximates the position of the Tyr-19 hydroxyl group and forms a hydrogen bond with His-132. Activation of this water molecule by His-132 in the Y19F mutant likely substitutes for the wild-type hydroxyl group, albeit with compromised efficiency.
ITC analysis of AdoCys and pCho binding to the wild-type, Y19F, and H132A PfPMT also suggests a possible role for the Tyr-19-His-132 interaction as a “catalytic latch.” As noted above, the structures of PfPMT show that the active site is occluded by α1, which includes Tyr-19. We suggest that movement of the N-terminal helix upon AdoMet/AdoCys binding not only locks the substrate in the active site, but also positions the tyrosine for interaction with His-132 to form the catalytic dyad. ITC analysis of AdoCys and pCho binding to the Y19F and H132A mutants indicates that these mutations do not change binding affinity; however, the enthalpic and entropic contributions decrease and increase, respectively, when compared with wild-type enzyme (Table 3). The greater entropy contribution in the mutants suggests that alteration of either residue in the dyad increases flexibility that becomes more ordered upon ligand binding.
The overall reaction catalyzed by the PMT resembles that of other N-methyltransferases, including the well studied glycine and guanidinoacetate methyltransferases; however, how different methyltransferases chemically perform the transfer reaction differs (30–33, 35–39). The structure of the glycine methyltransferase in complex with AdoMet and acetate led to the proposal that an active site glutamate facilitated the reaction as a general acid (35). Later structures of this enzyme suggested that an arginine drives catalysis primarily by proximity and orientation (30), but computational modeling of the reaction energetics suggests that this is unlikely (36, 37). In guanidinoacetate methyltransferase, a critical aspartate residue is positioned to abstract a proton from the substrate amine (33, 38, 39). Although all these enzymes bind AdoMet using an evolutionarily conserved scaffold, the residues responsible for recognizing substrates, positioning the amine for methylation, and facilitating catalysis differ.
The structural and mechanistic differences between the PMT and other methyltransferases suggest that the development of specific inhibitors is possible. Although the identification of compounds targeting the Plasmodium and nematode PMT is in the early stages, a handful of known compounds, such as sinefungin (an AdoMet analog), miltefosine (a choline analog), and amodiaquine (a 4-aminoquinoline), are inhibitors (7, 17, 19). Previous work on the inhibition of PfPMT by amodiaquine suggested a possible binding site near Gly-32 and Gly-68 for this compound based on NMR titration studies (17). In the PfPMT crystal structure, Gly-32 is in a loop after α2 and Gly-68 in α4, which is also positioned near α2. Given the possible structural changes in PfPMT upon ligand binding, the shifts observed in the NMR spectra in the presence and absence of amodiaquine may result not from direct interaction with the molecule but from conformation changes in the N-terminal region upon binding. Further structural studies with inhibitors are required to elucidate how these molecules interact with PfPMT.
Supplementary Material
Acknowledgments
Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center of the Advanced Photon Source, a national user facility operated by the University of Chicago for the United States Department of Energy Office of Biological and Environmental Research (Grant DE-AC02-06CH11357).
This work was supported by funds from Washington University and the Washington University Summer Undergraduate Research Fellowship Program (to T. D. A.) and from the Knox College Howard Hughes Medical Institute (HHMI)-SURF Program (to A. N.).
This article was selected as a Paper of the Week.

This article contains supplemental Figs. S1–S3.
- pCho
- phosphocholine
- pDME
- phospho-dimethylethanolamine
- pEA
- phosphoethanolamine
- pMME
- phospho-monomethylethanolamine
- AdoCys
- S-adenosylhomocysteine
- AdoMet
- S-adenosylmethionine
- Se-Met
- selenomethionine
- ITC
- isothermal titration calorimetry
- PMT
- phosphoethanolamine methyltransferase(s)
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. World Health Organization Expert Committee on Malaria (2000) WHO Technical Report Series - 892, World Health Organization, Geneva, Switzerland: [PubMed] [Google Scholar]
- 2. Hunter W. N. (2009) Structure-based ligand design and the promise held for antiprotozoan drug discovery. J. Biol. Chem. 284, 11749–11753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wengelnik K., Vidal V., Ancelin M. L., Cathiard A. M., Morgat J. L., Kocken C. H., Calas M., Herrera S., Thomas A. W., Vial H. J. (2002) A class of potent antimalarials and their specific accumulation in infected erythrocytes. Science 295, 1311–1314 [DOI] [PubMed] [Google Scholar]
- 4. Calas M., Ancelin M. L., Cordina G., Portefaix P., Piquet G., Vidal-Sailhan V., Vial H. (2000) Antimalarial activity of compounds interfering with Plasmodium falciparum phospholipid metabolism: comparison between mono- and bisquaternary ammonium salts. J. Med. Chem. 43, 505–516 [DOI] [PubMed] [Google Scholar]
- 5. Kent C. (1995) Eukaryotic phospholipid biosynthesis. Annu. Rev. Biochem. 64, 315–343 [DOI] [PubMed] [Google Scholar]
- 6. Nuccio M. L., Ziemak M. J., Henry S. A., Weretilnyk E. A., Hanson A. D. (2000) cDNA cloning of phosphoethanolamine N-methyltransferase from spinach by complementation in Schizosaccharomyces pombe and characterization of the recombinant enzyme. J. Biol. Chem. 275, 14095–14101 [DOI] [PubMed] [Google Scholar]
- 7. Pessi G., Kociubinski G., Mamoun C. B. (2004) A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc. Natl. Acad. Sci. U.S.A. 101, 6206–6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pessi G., Choi J. Y., Reynolds J. M., Voelker D. R., Mamoun C. B. (2005) In vivo evidence for the specificity of Plasmodium falciparum phosphoethanolamine methyltransferase and its coupling to the Kennedy pathway. J. Biol. Chem. 280, 12461–12466 [DOI] [PubMed] [Google Scholar]
- 9. Witola W. H., El Bissati K., Pessi G., Xie C., Roepe P. D., Mamoun C. B. (2008) Disruption of the Plasmodium falciparum PfPMT gene results in a complete loss of phosphatidylcholine biosynthesis via the serine-decarboxylase-phosphoethanolamine-methyltransferase pathway and severe growth and survival defects. J. Biol. Chem. 283, 27636–27643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bobenchik A. M., Augagneur Y., Hao B., Hoch J. C., Ben Mamoun C. (2011) Phosphoethanolamine methyltransferases in phosphocholine biosynthesis: functions and potential for antiparasite therapy. FEMS Microbiol. Rev. 35, 609–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palavalli L. H., Brendza K. M., Haakenson W., Cahoon R. E., McLaird M., Hicks L. M., McCarter J. P., Williams D. J., Hresko M. C., Jez J. M. (2006) Defining the role of phosphomethylethanolamine N-methyltransferase from Caenorhabditis elegans in phosphocholine biosynthesis by biochemical and kinetic analysis. Biochemistry 45, 6056–6065 [DOI] [PubMed] [Google Scholar]
- 12. Brendza K. M., Haakenson W., Cahoon R. E., Hicks L. M., Palavalli L. H., Chiapelli B. J., McLaird M., McCarter J. P., Williams D. J., Hresko M. C., Jez J. M. (2007) Phosphoethanolamine N-methyltransferase (PMT-1) catalyzes the first reaction of a new pathway for phosphocholine biosynthesis in Caenorhabditis elegans. Biochem. J. 404, 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jez J. M. (2007) Phosphatidylcholine biosynthesis as a potential target for inhibition of metabolism in parasitic nematodes. Curr. Enzym. Inhib. 3, 133–142 [Google Scholar]
- 14. Lee S. G., Jez J. M. (2011) The phosphobase methylation pathway in Caenorhabditis elegans: a new route to phospholipids in animals. Curr. Chem. Biol. 5, 183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reynolds J. M., Takebe S., Choi J. Y., El Bissati K., Witola W. H., Bobenchik A. M., Hoch J. C., Voelker D. R., Mamoun C. B. (2008) Biochemical and genetic analysis of the phosphoethanolamine methyltransferase of the human malaria parasite Plasmodium falciparum. J. Biol. Chem. 283, 7894–7900 [DOI] [PubMed] [Google Scholar]
- 16. Jost R., Berkowitz O., Shaw J., Masle J. (2009) Biochemical characterization of two wheat phosphoethanolamine N-methyltransferase isoforms with different sensitivities to inhibition by phosphatidic acid. J. Biol. Chem. 284, 31962–31971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bobenchik A. M., Choi J. Y., Mishra A., Rujan I. N., Hao B., Voelker D. R., Hoch J. C., Mamoun C. B. (2010) Identification of inhibitors of Plasmodium falciparum phosphoethanolamine methyltransferase using an enzyme-coupled transmethylation assay. BMC Biochem. 11, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van Duyne G. D., Standaert R. F., Karplus P. A., Schreiber S. L., Clardy J. (1993) Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 [DOI] [PubMed] [Google Scholar]
- 19. Lee S. G., Haakenson W., McCarter J. P., Williams D. J., Hresko M. C., Jez J. M. (2011) Thermodynamic evaluation of ligand binding in the plant-like phosphoethanolamine methyltransferases of the parasitic nematode Haemonchus contortus. J. Biol. Chem. 286, 38060–38068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 21. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 22. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 23. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Morris R. J., Perrakis A., Lamzin V. S. (2003) ARP/wARP and automatic interpretation of protein electron density maps. Methods Enzymol. 374, 229–244 [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huang C. C., Smith C. V., Glickman M. S., Jacobs W. R., Jr., Sacchettini J. C. (2002) Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis. J. Biol. Chem. 277, 11559–11569 [DOI] [PubMed] [Google Scholar]
- 29. Boissier F., Bardou F., Guillet V., Uttenweiler-Joseph S., Daffé M., Quémard A., Mourey L. (2006) Further insight into S-adenosylmethionine-dependent methyltransferases: structural characterization of Hma, an enzyme essential for the biosynthesis of oxygenated mycolic acids in Mycobacterium tuberculosis. J. Biol. Chem. 281, 4434–4445 [DOI] [PubMed] [Google Scholar]
- 30. Takata Y., Huang Y., Komoto J., Yamada T., Konishi K., Ogawa H., Gomi T., Fujioka M., Takusagawa F. (2003) Catalytic mechanism of glycine N-methyltransferase. Biochemistry 42, 8394–8402 [DOI] [PubMed] [Google Scholar]
- 31. Horton J. R., Sawada K., Nishibori M., Zhang X., Cheng X. (2001) Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons. Structure 9, 837–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu Q., Gee C. L., Lin F., Tyndall J. D., Martin J. L., Grunewald G. L., McLeish M. J. (2005) Structural, mutagenic, and kinetic analysis of the binding of substrates and inhibitors of human phenylethanolamine N-methyltransferase. J. Med. Chem. 48, 7243–7252 [DOI] [PubMed] [Google Scholar]
- 33. Komoto J., Yamada T., Takata Y., Konishi K., Ogawa H., Gomi T., Fujioka M., Takusagawa F. (2004) Catalytic mechanism of guanidinoacetate methyltransferase: crystal structures of guanidinoacetate methyltransferase ternary complexes. Biochemistry 43, 14385–14394 [DOI] [PubMed] [Google Scholar]
- 34. Bolognese C. P., McGraw P. (2000) The isolation and characterization in yeast of a gene for Arabidopsis S-adenosylmethionine:phospho-ethanolamine N-methyltransferase. Plant Physiol. 124, 1800–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fu Z., Hu Y., Konishi K., Takata Y., Ogawa H., Gomi T., Fujioka M., Takusagawa F. (1996) Crystal structure of glycine N-methyltransferase from rat liver. Biochemistry 35, 11985–11993 [DOI] [PubMed] [Google Scholar]
- 36. Soriano A., Castillo R., Christov C., Andrés J., Moliner V., Tuñón I. (2006) Catalysis in glycine N-methyltransferase: testing the electrostatic stabilization and compression hypothesis. Biochemistry 45, 14917–14925 [DOI] [PubMed] [Google Scholar]
- 37. Velichkova P., Himo F. (2005) Methyl transfer in glycine N-methyltransferase. A theoretical study. J. Phys. Chem. B 109, 8216–8219 [DOI] [PubMed] [Google Scholar]
- 38. Komoto J., Huang Y., Takata Y., Yamada T., Konishi K., Ogawa H., Gomi T., Fujioka M., Takusagawa F. (2002) Crystal structure of guanidinoacetate methyltransferase from rat liver: a model structure of protein arginine methyltransferase. J. Mol. Biol. 320, 223–235 [DOI] [PubMed] [Google Scholar]
- 39. Velichkova P., Himo F. (2006) Theoretical study of the methyl transfer in guanidinoacetate methyltransferase. J. Phys. Chem. B 110, 16–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.