Background: Homeostatic cell competitive system between cancerous cells and non-cancerous cells is considered as the reason for tumor initiation.
Results: Exosomal tumor-suppressive microRNAs secreted by non-cancerous cells inhibit the proliferation of cancerous cells.
Conclusion: Exosomal tumor-suppressive microRNAs act as an inhibitory signal for cancer cells in a cell-competitive process.
Significance: This provides a novel insight into a tumor initiation mechanism.
Keywords: Cancer Biology, Cell-Cell Interaction, Exosomes, MicroRNA, Sphingomyelinase, Tumor Microenvironment, Cell Competition, Secretory MicroRNAs, Tumor-suppressive MicroRNAs, Tumor Initiation
Abstract
Normal epithelial cells regulate the secretion of autocrine and paracrine factors that prevent aberrant growth of neighboring cells, leading to healthy development and normal metabolism. One reason for tumor initiation is considered to be a failure of this homeostatic cell competitive system. Here we identify tumor-suppressive microRNAs (miRNAs) secreted by normal cells as anti-proliferative signal entities. Culture supernatant of normal epithelial prostate PNT-2 cells attenuated proliferation of PC-3M-luc cells, prostate cancer cells. Global analysis of miRNA expression signature revealed that a variety of tumor-suppressive miRNAs are released from PNT-2 cells. Of these miRNAs, secretory miR-143 could induce growth inhibition exclusively in cancer cells in vitro and in vivo. These results suggest that secretory tumor-suppressive miRNAs can act as a death signal in a cell competitive process. This study provides a novel insight into a tumor initiation mechanism.
Introduction
Competitive interactions among cells are the basis of many homeostatic processes in biology. In Drosophila, normal epithelial cells compete with transformed ones for individual survival, which is a process called cell competition (1, 2). If a given group of cells was exposed to some stress, it would be separated into subpopulations of cells with different levels of damage. In noncompetitive conditions, cells with severe damage die in a short time, whereas moderately damaged cells survive to the next generation, indicative of the transduction of a negative phenotype. On the other hand, in competitive conditions even slightly damaged cells are eliminated from the cell group because healthy cells, the “winners,” convey death signals to damaged cells, the “losers,” and the losers reciprocally confer growth signals to the winners. This feed-forward regulation enables the cell population to eradicate abnormal cells and maintain the same number of normal cells in a limited niche.
Oncogenesis is characterized by genetic and metabolic changes reprogramming living cells to undergo uncontrolled proliferation (3). This suggests that the abnormal cells that are originally destined for elimination can survive and expand against the cell competitive regulation, leading to the formation of a tumor mass. Consistently with this concept, Bondar and Medzhitov (4) showed that the cell competition process involves p53, a tumor-suppressive gene, between the hematopoietic stem cells and progenitor cells, suggesting that gene modifications of p53 could disturb the homeostatic mechanism and give rise to tumor initiation. It is conceivable that p53 target genes could be associated with intercellular communication between winners and losers; however, this literature has not answered the question of whether this regulatory system is mediated by contact-dependent or contact-independent manner. More than 10 years ago a pioneer study suggested that non-cancerous cells co-cultured with cancer cells inhibit the growth of cancer cells in vitro (5). This result indicated that humoral factors could be involved in cell competition as intercellular communicators (6).
As recently as a few years ago it was believed that RNAs could not behave as extracellular signal molecules because of their vulnerability to the attack of ribonucleases largely existing in body fluid. Evidence is presently increasing to show that miRNAs4 contained in exosomes are released from mammalian cells and act as a signal transducer (7). It is important that many different tumor-suppressive miRNAs, such as miR-16 and miR-143, are down-regulated in cancer cells, resulting in tumorigenesis, tumor progression, and metastasis (8–11). Taken together, these findings suggest that secretory miRNAs may have favorable aspects for anti-proliferative signals mediating cell competition.
In this report we show that miR-143 expression in normal prostate cells, PNT-2 cells, is higher than that in prostate cancer cells, PC-3M-luc cells, and that miR-143 released from non-cancerous cells transfers growth-inhibitory signals to cancerous cells in vitro and in vivo. These results suggest that secretory tumor-suppressive miRNAs might be a death signal from winners to losers in the context of cell competition. Secretory miRNAs can be conducive to the maintenance of normal growth and development.
EXPERIMENTAL PROCEDURES
Reagents
Mouse monoclonal anti-KRAS (F234) (sc-30) was purchased from Santa Cruz. Rabbit polyclonal anti-ERK5 (#3372) was purchased from Cell Signaling. Mouse monoclonal anti-actin, clone C4 (MAB1501), was obtained from Millipore. Mouse monoclonal ant-human-CD63 antibody (556019) was purchased from BD Pharmingen. Peroxidase-labeled anti-mouse and anti-rabbit antibodies were included in the Amersham Biosciences ECL PLUS Western blotting Reagents Pack (RPN2124) (GE Healthcare). Synthetic Caenorhabditis elegans miRNA cel-miR-39 was synthesized by Qiagen (Valencia, CA). Synthetic hsa-miR-143 (pre-miR-143), the negative control 1 (NC1), has-miR-143 inhibitor molecule (anti-miR-143), and the negative control inhibitor molecule (anti-NC) were purchased from Ambion (Austin, TX). GW4869 was purchased from Calbiochem. Geneticin was purchased from Invitrogen.
Cell Culture
PNT-2 cells, immortalized normal adult prostatic epithelial cell line, were purchased from the DS Pharma Biomedical Co., Ltd. (Osaka, Japan). HEK293 cells, a human embryonic kidney cell line (CRL-1573), were obtained from American Type Culture Collection (Manassas, VA). HEK293 cells were cultured in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum (FBS) and an antibiotic-antimycotic (Invitrogen) at 37 °C in 5% CO2. PNT-2 and the prostate cancer cell line, PC-3M-luc cells, continuously expressing firefly luciferase (Xenogen, Alameda, CA), were cultured in RPMI containing 10% heat-inactivated FBS and an antibiotic-antimycotic at 37 °C in 5% CO2.
Preparation of Conditioned Medium and Exosomes
Before the collection of culture medium, cells were washed 3 times with Advanced RPMI containing an antibiotic-antimycotic and 2 mm l-glutamine (medium A), and the medium was switched to fresh medium A. After incubation for 3 days, medium A was collected and centrifuged at 2000 × g for 10 min at room temperature. To thoroughly remove cellular debris, the supernatant was centrifuged again at 12,000 × g for 30 min at room temperature or filtered through a 0.22-μm filter (Millipore). The conditioned medium (CM) was then used for miRNA extraction and functional assays as well as exosome isolation.
For exosome preparation the CM was ultracentrifuged at 110,000 × g for 70 min at 4 °C. The pellets were washed with 11 ml of PBS, and after ultracentrifugation they were resuspended in PBS. The exosome fraction was measured for its protein content using the Micro BCA Protein Assay kit (Thermo Scientific, Wilmington, DE).
Isolation of MicroRNAs
Isolation of extracellular and cellular miRNAs was performed using the miRNeasy Mini Kit (Qiagen). Two hundred microliters of conditioned medium or cell lysate was diluted with 1 ml of Qiazol Solution. After 5 min of incubation, 10 μl of 0.1 nm cel-miR-39 was added to each aliquot followed by vortexing for 30 s. Subsequent extraction and filter cartridge work were carried out according to the manufacturer's protocol.
Quantitative Real Time PCR (QRT-PCR)
The method for QRT-PCR has been previously described (7). PCR was carried out in 96-well plates using the 7300 Real Time PCR System (Applied Biosystems). All reactions were done in triplicate. All TaqMan MicroRNA Assays were purchased from Applied Biosystems. Cel-miR-39 and RNU6 were used as an invariant control for the CM and cells, respectively.
Immunoblot Analysis
SDS-PAGE gels, SuperSep Ace 5–20% (194–15021) (Wako), were calibrated with Precision Plus Protein Standards (161–0375) (Bio-Rad), and anti-KRAS (1:100), anti-ERK5 (1:1000), anti-CD63 (1:200), and anti-actin (1:1000) were used as primary antibodies. The dilution ratio of each antibody is indicated in parentheses. Two secondary antibodies (peroxidase-labeled anti-mouse and anti-rabbit antibodies) were used at a dilution of 1:10,000. Bound antibodies were visualized by chemiluminescence using the ECL PLUS Western blotting detection System (RPN2132) (GE Healthcare), and luminescent images were analyzed by a LuminoImager (LAS-3000; Fuji Film, Inc.). Only gels for CD63 (BD Biosciences) detection were run under non-reducing conditions.
Plasmids
The primary-miR-143 expression vector was purchased from TaKaRa BIO. For luciferase-based reporter gene assays, pLucNeo was constructed by inserting a firefly luciferase gene derived from the pGL3-control (Promega) into the pEYFP-1 vector (Clontech) at BglII and AflII sites. The sensor vector for miR-143 was constructed by introducing tandem binding sites with perfect complementarity to miR-143 separated by a four-nucleotide spacer into the NotI site of psiCHECK2 (Promega). The sequences of the binding site are as follows: 5′-AAACCTAGAGCGGCCGCGAGCTACAGTGCTTCATCTCAAAGAATTCTTGAGCTACAGTGCTTCATCTCAGCGGCCGCTGGCCGCAA-3′ (sense) and 5′-TTGCGGCCAGCGGCCGCTGAGATGAAGCACTGTAGCTCAAGAATTCTTTGAGATGAAGCACTGTAGCTCGCGGCCGCTCTAGGTTT-3′ (antisense). The “seed” sequence of miR-143 is indicated by bold italics. In a mutated miR-143 sensor vector, the seed sequence, TCATCTC, was displaced with GACGAGA. All the plasmids were verified by DNA sequencing.
Transient Transfection Assays
Transfections of 10 nm miR-143 mimic and 3 nm anti-miR-143 were accomplished with the DharmaFECT Transfection Reagent (Thermo Scientific) according to the manufacturer's protocol. The total amounts of miRNAs for each transfection were equally adjusted by the addition of NC1 and anti-NC, respectively.
Establishment of Stable Cell Lines
Stable HEK293 cell lines that express miR-143 were generated by selection with 300 μg/ml Geneticin. HEK293 cells were transfected with 0.5 μg of the pri-miR-143 expression vector at 90% confluency in 24-well dishes using a Lipofectamine LTX reagent in accordance with the manufacturer's instructions. Twelve hours after the transfection, the cells were re-plated in a 10-cm dish followed by a 3-week selection with the antibiotic. Ten surviving single colonies were picked up from each transfectant and then cultured for another 2 weeks. The cells expressing the largest amount of miR-143 among transfectants were used as miR-143 stably expressing cells.
Luciferase Reporter Assay
HEK293 cells were cultured at a density of 1 × 104 cells/well in 96-well tissue culture plates overnight, and miRNA transfections or the addition of CM was performed. The cells were harvested, and renilla luciferase activity was measured and normalized by firefly luciferase activity (10). All assays were performed in triplicate and repeated at least three times, and the most representative results are shown.
Cell Growth Assay
PC-3M-luc cells were seeded at a density of 2 × 103 cells/well in a 96-well plate. The following day the cells were transfected with mature miRNAs or incubated with a CM. Twenty-four hours later the culture medium of the transfected cells was switched to medium A, whereas the conditioned medium was not changed. After a 3-day culture, cells were harvested for the measurement of firefly luciferase activity. To know the cellular proliferation by the tetrazolium-based colorimetric MTT assay, 20 μl CM of TetraColor ONE (SEIKAGAKU Corp., Tokyo, Japan) was added to each well after 72 h of culture. After 2–4 h of incubation at 37 °C, the optical density was measured at a wavelength of 450 nm using a microplate reader.
PKH67-labeled Exosome Transfer
Purified exosomes derived from PNT-2 CM were labeled with a PKH67 green fluorescent labeling kit (Sigma). Exosomes were incubated with 2 μm PKH67 for 5 min, washed 4 times using a 100-kDa filter (Microcon YM-100, Millipore) to remove excess dye, and incubated with PC-3M-luc cells at 37 °C.
Co-culture Experiment
In co-culture experiments, 2 × 105 cells/well of PNT-2 cells were plated in 6-well plates. To stain the PNT-2 cells with BODIPY-TR-ceramide (Invitrogen), 5 μm BODIPY-TR-ceramide in a non-serum culture medium was added and incubated with the cells at 37 °C. After 30 min the cells were rinsed several times with a non-serum culture medium and incubated in a fresh medium at 37 °C for an additional 30 min. After the staining of PNT-2 cells by BODIPY-TR-ceramide, labeling of PC-3M-luc cells with PKH67 was performed in accordance with the manufacturer's instructions. After that, labeled PC-3M-luc cells were added and co-cultured with PNT-2 cells for 12 h at 37 °C.
Microarray Analysis
To detect the miRNAs in exosomes and cells derived from PNT-2 and PC-3M-luc cells, 100 ng of total RNA was labeled and hybridized using a human microRNA microarray kit (Agilent Technologies) according to the manufacturer's protocol (Protocol for Use with Agilent MicroRNA Microarrays Version 1.5). Hybridization signals were detected using a DNA microarray scanner (Agilent Technologies), and the scanned images were analyzed using Agilent Feature Extraction software.
Evaluation of Tumor-suppressive miRNA Delivery to Subcutaneously Implanted Prostate Cancer Cell Line in Mice
Animal experiments in this study were performed in compliance with the guidelines of the Institute for Laboratory Animal Research, National Cancer Center Research Institute. Seven-week-old male Balb/c athymic nude mice (CLEA Japan, Shizuoka, Japan) were anesthetized by exposure to 3% isoflurane for injections and in vivo imaging. Four days ahead of the first CM injection, the anesthetized animals were subcutaneously injected with 5 × 105 PC-3M-luc cells suspended in 100 μl of sterile Dulbecco's phosphate-buffered saline into each dorsal region. Five hundred μl of CM derived from miR-143-overexpressing HEK293 cells and control cells were daily injected into each tumor from day 0 to 6. For in vivo imaging, the mice were administered d-luciferin (150 mg/kg, Promega) by intraperitoneal injection. Ten minutes later, photons from animal whole bodies were counted using the IVIS imaging system (Xenogen) according to the manufacturer's instructions. Data were analyzed using LIVINGIMAGE 2.50 software (Xenogen).
RESULTS
Suppression of Prostate Cancer Cell Proliferation by Conditioned Medium Isolated from Non-cancerous Prostatic Cell
Cell competition is a homeostatic mechanism for the accommodation of an appropriate number of cells in a limited niche or stroma (1). Based on this idea it is possible that the cell competition between normal and abnormal cells frequently occurs in a precancerous state. Of note is that non-cancerous cells suppress cancer cell development by contact-independent interaction (12). For instance, endothelial cells provide the major extracellular heparan sulfate proteoglycan as anti-proliferative signals (12); however, the molecular mechanism by which the other types of cells in a tumor environment associate with cancer cells is not fully understood.
To analyze the mechanism, we treated a hormone-insensitive prostatic carcinoma cell line, PC-3M-luc cells, with a CM from the non-cancerous prostate cell line PNT-2 cells. After a 3-day incubation, the PNT-2 CM inhibited the growth of the PC-3M-luc cells up to ∼10% compared with the cell growth treated by fresh culture medium (Fig. 1A; compare lanes 1 and 3). In contrast, the growth of PC-3M-luc cells incubated in the CM of PC-3M-luc cells themselves showed no inhibitory effect (Fig. 1A; compare lanes 1 and 2). To determine that the performed treatments did not affect the luciferase activity, we also used the colorimetric MTT assay to measure the cell growth of PC-3M-luc cells. As shown in supplemental Fig. 1A, not only luciferase assay but also MTT assay show the inhibition of PC-3M-luc cell proliferation by the addition of PNT-2 cells derived CM, indicating that our treatment did not affect the luciferase activity. These results indicate that the non-cancerous cells may secrete some molecules that can suppress cancer cell proliferation.
FIGURE 1.
Suppression of cancerous cell proliferation by exosome isolated from non-cancerous cells. A, cell growth inhibition by a conditioned medium derived from PNT-2 cells is shown. PC-3M-luc cells were incubated for 3 days in a conditioned medium isolated from PC-3M-luc cells, PNT-2 cells, or a culture medium followed by a cell growth assay as described under “Experimental Procedures.” The values on the y axis are depicted relative to the normalized luciferase activity of culture medium-treated cells, which is defined as 1. Each bar is presented as the mean S.E. (n = 3). *, p < 0.05 as compared with culture medium-treated PC-3M-luc cells; Student's t test. B, treatment with GW4869 to donor cells restored the reduced cell growth by the PNT-2-derived CM is shown. Donor PNT-2 cells were incubated in the presence or absence of 10 μm GW4869 for 2 days. The conditioned medium from PC-3M-luc cells was used as a control. The values on the y axis are depicted relative to the normalized luciferase activity of PC-3M-luc-conditioned medium-treated cells, which is defined as 1. Each bar is presented as the mean S.E. (n = 3). *, p < 0.05; Student's t test. C, cell growth inhibition by exosomes derived from PNT-2 cells is shown. PC-3M-luc cells were incubated in exosomes isolated from PNT-2 cells or PC-3M-luc cells followed by a cell growth assay, as described under “Experimental Procedures.” The values on the y axis are depicted relative to the normalized luciferase activity of cells treated with exosomes derived from PC-3M-luc cells is defined as 1. Each bar is presented as the mean S.E. (n = 3). **, p < 0.005, as compared with exosomes isolated from PC-3M-luc cells; Student's t test. D, shown are fluorescent photos of BODIPY-ceramide-labeled PNT-2 and PC-3M-luc cells marked by PKH67. PNT-2 cells and PC-3M-luc cells were labeled with red fluorescent BODIPY-ceramide and green fluorescent PKH67, respectively, as described under “Experimental Procedures.” After treatment of PNT-2 by BODIPY-ceramide, PKH67-labeled PC-3M-luc cells were added. After co-culturing for 3 or 12 h, images were obtained. Fluorescent photos were detected with the Eclipse TE 2000 Inverted Research Microscope, and images were produced using NIS-Elements BR software. Arrowheads show yellow colored cancer cells. The size bar indicates 100 μm.
In a recent report we showed that miRNAs contained in exosomes are secreted and that their secretion is tightly regulated by neutral sphingomyelinase 2, which is known to hydrolyze sphingomyelins to generate ceramides and trigger the budding of exosomes. We collected two separate aliquots of CM from PNT-2 cells incubated with or without GW4869, a specific inhibitor for neutral sphingomyelinase 2. The isolated exosomes were verified by the detection of CD63 protein, a well established exosome marker, with immunoblotting (supplemental Fig. 1B), and the activity of GW4869 was confirmed by the decreased amount of exosomal protein (supplemental Fig. 1C). The CM prepared in the presence of the GW4869 compound cancelled most tumor-suppressive activity of the non-treated PNT-2 CM (Fig. 1B; compare lanes 1–3). Furthermore, proliferation of PC-3M-luc cells was inhibited by the addition of the exosome fraction isolated from the PNT-2 CM by ultracentrifugation (Fig. 1C). These observations suggest that exosomal miRNAs derived from non-cancerous cells were transferred to cancerous cells, resulting in the inhibition of their proliferation.
To visualize the transfer of ceramide-containing exosome from PNT-2 to PC-3M-luc in vitro, a co-culture experiment was performed. Before the co-culture, 2 × 105 PNT-2 cells were incubated for 30 min with red fluorescent BODIPY-ceramide dye, which can label the exosomes inside the cells (13, 14). After washing five times with PBS, equal numbers of PC-3M-luc cells labeled by green fluorescent PKH67, a cellular membrane indicator, were added into the culture dishes. Three hours later we did not observe any PC-3M-luc cells with a yellow color (Merged photo in upper panel of Fig. 1D), indicating that carried-over red dyes were thoroughly removed as 3 h is enough time for the dye to be incorporated directly into the cells. By contrast, after 12 h of co-culture, yellow fluorescence was observed in green-labeled PC-3M-luc cells (indicated by arrowheads in Merged photo in the lower panel of Fig. 1D), suggesting that ceramide-containing exosomes from PNT-2 cells were transferred to the PC-3M-luc cells. This result is corroborated by the uptake experiment using the PKH67-labeled exosomes purified from PNT-2 culture medium (supplemental Fig. 1D). Green fluorescence was detected in PC-3M-luc cells after 16 h of incubation, providing a direct evidence for exosome uptake by cancerous cells.
Tumor-suppressive miRNAs Down-regulated in Cancerous Cells Were Secreted from Non-cancerous Cells
We propose a hypothetical model of tumor initiation involving cell competition and anti-proliferative secretory miRNAs (Fig. 2A). In a cell competition cycle, as illustrated in the bottom part of Fig. 2A, growth inhibitory miRNAs are actively released from non-cancerous cells to kill abnormal cells with a partial oncogenic ability, thereby restoring them to a healthy state. Indeed, inhibitory capacity of these miRNAs appears to be limited in the setting of single treatment with the PNT-2 CM (Fig. 1A); however, they can potentially prevent emergence of tumor cells in a physiological condition. Because abundantly existing healthy cells continuously provide nascent overproliferative cells with tumor-suppressive miRNAs for a long period, a local concentration of secretory miRNAs can become high enough to restrain a tumor initiation. A dashed arrow in Fig. 2A indicates the way whereby the disruption of the homeostatic system leads to tumor expansion. If precancerous cells acquire resistance to anti-proliferative secretory miRNAs or normal cells cannot supply an adequate amount of miRNAs, then this defensive system will fail to maintain the healthy condition.
FIGURE 2.
Down-regulation of cellular and extracellular tumor-suppressive miRNAs in PC-3M-luc cells. A, shown is a schematic representation of hypothetical tumor initiation process. Neighboring healthy cells (blue) secrete tumor-suppressive miRNAs (light yellow) to inhibit the proliferation of abnormal cells (gray), and this cell population returns to the initial healthy condition (a homeostatic cycle). Once the cell competitive cycle is compromised, this niche become susceptible to tumor initiation (indicated by a dashed arrow). B, comparison of cellular and extracellular miRNAs expression in PNT-2 and PC-3M-luc cells is shown. miRNA expression levels were determined by a Taq-Man QRT-PCR. The values on the y axis are depicted relative to the normalized expression level of PNT-2 cells, which is defined as 1. C, secretion of miR-143 was suppressed by the treatment with GW4869. PNT-2 cells were seeded and cultured in a 24-well plate for 48 h in the indicated concentrations of GW4869. After the incubation, the medium was subjected to QRT-PCR for miR-143. The values on the y axis are depicted relative to the amount of miR-143 at 0 μm GW4869, which is defined as 1. D, shown is cell growth inhibition by miR-143 in PC-3M-luc cells but not in PNT-2 cells. PNT-2 and PC-3M-luc cells were transfected with 10 nm miR-143 molecules (miR-143) or 10 nm negative control molecules (control) or without RNA molecules (Mock). The values on the y axis are depicted relative to the normalized luciferase activity of untreated cells (Mock), which is defined as 1. Each bar is presented as the mean S.E. (n = 3). *, p < 0.05; **, p < 0.005, as compared with untreated PC-3M-luc cells; Student's t test.
To test this hypothesis we checked the secretion amount of representative tumor-suppressive miRNAs by comparing PNT-2 and PC-3M-luc cells with Taq-Man QRT-PCR analysis. As shown in Fig. 2B, miR-16, miR-205, and miR-143, which are already reported to be dysregulated in prostate cancer (10, 15, 16), were down-regulated in PC-3M-luc cells at a cellular and extracellular level. The GW4869 inhibitor suppressed the secretion of miR-143 from PNT-2 cells in a dose-dependent manner (Fig. 2C), whereas its cellular level was not altered (supplemental Fig. 2A). Additionally, the application of small interfering RNAs specific for human neutral sphingomyelinase 2 gene knocked down its mRNAs, resulting in profound decrease in miR-143 secretion (supplemental Fig. 2, B and C). On the contrary, the expression of miR-143 in the cells was not changed after the transfection of neutral sphingomyelinase 2 siRNA (supplemental Fig. 2D). Taken with the result of Fig. 1B, these results suggest that the secreted tumor-suppressive miRNAs are implicated in the process of growth inhibition by PNT-2 CM.
For a global understanding of the expression change of non-cancerous and cancerous cells, we performed an miRNA microarray analysis against cellular and exosomal RNAs purified from PNT-2 and PC-3M-luc cells. In the sub-dataset of secretory exosomal miRNAs from PNT-2 cells, we found 40 miRNAs whose cellular amounts were lowered by one-half in PC-3M-luc cells (Table 1). The selected miRNAs expectedly include several types of tumor-suppressive miRNAs, such as miR-15a, miR-200 family, miR-148a, miR-193b, miR-126, and miR-205 (10, 15, 17–20). This observation supports the idea that secretory tumor-suppressive miRNAs are transferred from non-cancerous cells to cancerous cells, in accordance with the concentration gradient of the miRNA.
TABLE 1.
A list of PNT-2-derived secretory miRNAs that were down-regulated less than 0.5-fold in PC-3M cells compared with PNT-2 cells
| miRNAs | Fold changea |
|---|---|
| hsa-miR-141 | 0.0 |
| hsa-miR-200c | 0.0 |
| hsa-miR-886–3p | 0.0 |
| hsa-miR-30a* | 0.0 |
| hsa-miR-155 | 0.0 |
| hsa-miR-205 | 0.0 |
| hsa-miR-224 | 0.0 |
| hsa-miR-148a | 0.0 |
| hsa-miR-130a | 0.0 |
| hsa-miR-30a | 0.1 |
| hsa-miR-663 | 0.1 |
| hsa-miR-181a-2* | 0.1 |
| hsa-miR-484 | 0.1 |
| hsa-miR-10a | 0.1 |
| hsa-miR-192 | 0.1 |
| hsa-miR-193b | 0.1 |
| hsa-miR-200a | 0.1 |
| hsa-miR-429 | 0.1 |
| hsa-miR-769–5p | 0.1 |
| hsa-miR-200b | 0.2 |
| hsa-miR-195 | 0.2 |
| hsa-miR-203 | 0.2 |
| hsa-miR-7 | 0.2 |
| hsa-miR-200a* | 0.2 |
| hsa-miR-200b* | 0.2 |
| hsa-miR-30c | 0.2 |
| hsa-miR-126 | 0.3 |
| hsa-miR-149 | 0.3 |
| hsa-miR-30d | 0.3 |
| hsa-miR-181a | 0.3 |
| hsa-miR-30e* | 0.3 |
| hsa-miR-365 | 0.4 |
| hsa-miR-135b | 0.4 |
| hsa-miR-454* | 0.4 |
| hsa-miR-129* | 0.4 |
| hsa-miR-30b | 0.4 |
| hsa-miR-181b | 0.4 |
| hsa-miR-210 | 0.4 |
| hsa-miR-455–3p | 0.5 |
| hsa-miR-15a | 0.5 |
a Fold change of the expression of miRNAs in PC-3M cells compared with PNT-2 cells is indicated.
We have so far demonstrated that normal cells have a higher secretion of tumor-suppressive miRNAs than cancerous cells; however, it remains unclear whether or not these secreted miRNAs affect the proliferation of cells of their origin. To answer this question, we introduced synthesized miR-143 to both PNT-2 and PC-3M-luc cells and assessed their proliferation rates. After 3 days of transfections, the miR-143 analog induced growth inhibition of PC-3M-luc cells compared with mock and control small RNA transfection (Fig. 2D, left panel). In contrast, the exogenously transduced miR-143 did not show its anti-proliferative effect in PNT-2 cells (Fig. 2D, right panel), indicating that excessive miR-143 did not confer an additional growth inhibitory effect on normal cells in which expression of miR-143 is maintained to a physiological level. This finding suggests that animal cells may have their own threshold amount for miRNA activity. The different sensitivity found in different cell types can help secretory miRNAs fulfill their purpose to combat exclusively precancerous cells. It is possible that secretory miRNAs, at least, derived from non-cancerous cells such as PNT-2 cells could supplement growth-suppressive signals that are decreased in cancerous cells. Thus, secreted miR-143 might be involved in the cell competitive regulatory system.
Secretory miR-143 Inhibited Prostate Cancer Cell Proliferation in Vitro
To examine whether miR-143 released from normal cells exert an anti-proliferative activity, we generated HEK293 cells overexpressing miR-143 by nearly 200-fold compared with control (supplemental Fig. 3A). After a 3-day incubation with the CM derived from the miR-143-overproducing HEK293 cells and control HEK293 cells, PC-3M-luc cells showed an ∼50% decrease in proliferation (Fig. 3A, lanes 1 and 3). Importantly, the decrease was recovered by the transfection of anti-miR-143 in PC-3M-luc cells (Fig. 3A, lane 3 and 4). These data indicate that the growth inhibition is attributable to secretory miR-143 contained in the supernatant of miR-143-overexpressing HEK293 cells. In agreement with the exosome-dependent machinery of miRNA secretion, we observed a similar result by using exosome fractions purified from miR-143-transduced HEK293 cells (Fig. 3B).
FIGURE 3.
Transfer of secretory miR-143 to PC-3M-luc cells in vitro. A, the transfection of anti-miR-143 to PC-3M-luc cells restored the reduced cell growth by the CM derived from miR-143 overproducing cells. After the transfection with 3 nm miR-143 inhibitor molecule (anti-miR-143) (lanes 2 and 4) or its control molecule (anti-NC) (lanes 1 and 3), PC-3M-luc cells were incubated for 3 days in a control conditioned medium (lanes 1 and 2) and CM containing extracellular miR-143 (lane 3 and 4) followed by a cell growth assay as described under “Experimental Procedures.” The values on the y axis are depicted relative to the normalized luciferase activity of cells treated in a culture medium, which is defined as 1. Each bar is presented as the mean S.E. (n = 3). (*, p < 0.05; Student's t test; n.s., not significant). B, cell growth inhibition by exosomes derived from miR-143-transduced HEK293 cells is shown. PC-3M-luc cells were incubated in the exosomes followed by cell growth assay as described under “Experimental Procedures.” The values on the y axis are depicted relative to the normalized luciferase activity of cells treated with exosomes derived from original HEK293 cells, defined as 1. Each bar is presented as the mean S.E. (n = 3). (**, p < 0.005; Student's t test). C, secretory miR-143-mediated KRAS suppression in PC-3M-luc cells is shown. Ten micrograms of protein of whole cell lysates prepared from PC-3M-luc cells treated with or without secretory miR-143 were applied to electrophoresis. Immunoblotting was performed with KRAS and actin antibodies and visualized by LAS-3000 system. D, extracellular miR-143 derived from HEK293 cells suppressed the luciferase activity of the sensor vector. HEK293 cells transfected with an miR-143 sensor vector were used as recipient cells. The recipient cells were incubated in a CM containing extracellular miRNAs. After a 2-day incubation, a luciferase reporter assay was performed as described under “Experimental Procedures.” The values on the y axis are depicted relative to the normalized luciferase activity of original HEK293-conditioned medium-treated cells, which is defined as 1. Each bar is presented as the mean S.E. (n = 3). *, p < 0.05; Student's t test). E, extracellular miR-143 did not reduce the luciferase activity of the mutated sensor vector. HEK293 cells transfected with the mutated miR-143 sensor vector were used as recipient cells. The recipient cells were incubated in a conditioned medium containing extracellular miRNAs. The luciferase assay was carried out as described above. The values on the y axis are depicted relative to the normalized renilla luciferase activity of control cells, which is defined as 1. Each bar is presented as the mean S.E. (n = 3). n.s. represents not significant.
To further study miRNA transfer on a molecular level, we performed a target gene expression analysis and an miRNA-responsive reporter assay. The immunoblotting analysis shows that the addition of the CM isolated from miR-143-overexpressing HEK293 cells significantly knocked down expression of KRAS, a target gene for miR-143 (21), in PC-3M-luc cells (Fig. 3C). In addition, we implemented luciferase analyses using a sensor vector harboring renilla luciferase fused in tandem with miR-143 seed sequence in the 3′-UTR. As shown in Fig. 3D, the normalized renilla luciferase activities were reduced by the treatment of miR-143-enriched CM derived from HEK293 cells stably expressing miR-143. In contrast, we did not detect any changes of luminescence by using a mutated vector instead of the intact sensor vector (Fig. 3E). Furthermore, we quantified cellular amounts of miR-143 in PC-3M-luc cells incubated with CM derived from HEK293 cells or miR-143 overproducing HEK293 cells by QRT-PCR. As shown in supplemental Fig. 3B, miR-143 was clearly increased at a cellular level by the treatment of the miR-143 enriched CM. These results indicate that secretory miR-143 exhibits its on-target growth-inhibitory effect in neighboring precancerous cells, thereby suppressing their disordered growth.
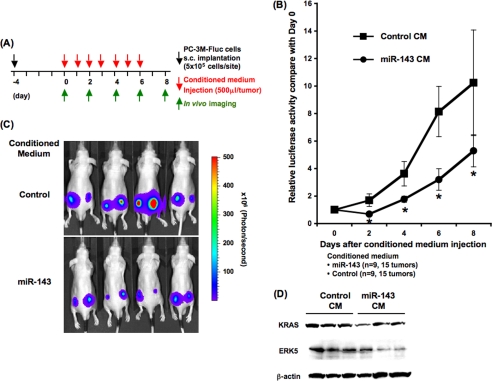
Secretory miR-143 Functions as Tumor Suppressor in Vivo
To our knowledge it has never been demonstrated that extracellular tumor-suppressive miRNAs can be transferred into living cells and induce phenotypic change in vivo. To address this possibility, we injected CM derived from miR-143 overproducing HEK293 cells or parental HEK293 cells into nude mice implanted with PC-3M-luc cells. Four days after the subcutaneous implantation, we carried out in vivo imaging and CM injections according to the timetable shown in Fig. 4A. Tumor expansions have been restrained for 8 days with intratumor administrations of miR-143 enriched CM, and consequently the tumor masses shrank by ∼0.5-fold on day 8 (Fig. 4B). The representative luminescent images of inoculated PC-3M-luc cells on day 8 were shown in Fig. 4C. Consistent with the finding that miR-143 did not impair growth activity of non-cancer cells in vitro (Fig. 2D), no toxicity was observed in these mice (data not shown). In addition, the expressions of miR-143 target genes, such as KRAS and ERK5 (16, 21), were decreased after miR-143-transduced CM injections, indicative of intercellular miRNA transfer in vivo (Fig. 4D). Thus, our prostate cancer xenograft model suggests that the tumor-suppressive miRNAs secreted from normal cells could be efficiently delivered into their neighboring tumors in vivo.
FIGURE 4.
Transfer of secretory miR-143 to PC-3M-luc cells in vivo. A, shown is the timetable for conditioned medium injections and in vivo imaging. B, shown are tumor growth ratios of the inoculated PC-3M-luc cells during the secretory miR-143 treatment. Closed circles and closed squares indicate the tumor mass administrated with CM from miR-143-overproducing HEK293 cells or parental HEK293 cells, respectively. The values on the y axis are depicted relative to the luciferase activity of each tumor on day 0, which is defined as 1. Each bar is presented as the mean S.E. (n = 9). *, p < 0.05; Student's t test. C, representative images are shown of tumor cells in the skin of mice. Bioluminescence of firefly luciferase from miR-143-enriched CM treated mice and control mice were detected on day 8 with IVIS imaging system. D, shown is secretory miR-143-mediated KRAS and ERK5 suppression in inoculated tumor cells. On day 8 the inoculated tumor masses were isolated and applied to immunoblotting analysis for the quantification of KRAS and ERK5 on a protein level.
DISCUSSION
In this study we documented that miR-143 derived from non-cancerous cells had the ability to suppress the growth of cancer cell proliferation not only in vitro but also in vivo. These observations suggest that tumor-suppressive miRNAs can be implicated in cell competition between cancer cells and non-cancer cells. In this context, normal cells attempt to prevent the outgrowth of precancerous cells by secreting anti-proliferative miRNAs and maintain a healthy condition; however, the abnormal cells can circumvent this inhibitory machinery, finally resulting in a tumor expansion (Fig. 2A). Cell competition could be a homeostatic mechanism that tumor cells need to overcome (1).
Here, we discuss two possible mechanisms by which cancer cells can gain resistance to secretory tumor-suppressive miRNAs. One is a blockade for the uptake of miRNAs, and the other is a cancellation of silencing activity of the incorporated miRNAs. As previously reported, miRNAs are loaded into exosomes and then secreted from living cells (7, 22, 23). If exosomes enriched in miRNAs are actively incorporated by recipient cells, cancer cells can impair the uptake mechanism to escape from the attack of secretory tumor-suppressive miRNAs. This scenario is supported by a recent publication regarding a Tim4 expected for an exosome receptor (24).
In the latter case cancer cells need to specifically compromise the incorporated tumor-suppressive miRNAs because there are some types of miRNAs that are indispensable for the expansion of cancer cells. A RISC assembly is composed of many protein families, such as the mammalian AGO family, GW182, and heat shock proteins (25). Moreover, each gene family also consists of many members, thereby generating diversity of RISC assemblies. The heterogeneity of RISC assemblies allows tumor-suppressive miRNAs to selectively bind with a RISC and silence their target genes on the complex. If cancer cells can exclusively destroy the tumor-suppressive RISC assembly, they can safely grow in a limited niche full of anti-proliferative miRNAs. The detailed mechanism of the resistance to cell competition remains unknown.
In addition to the acquired resistance, there is another possibility that normal cells will lose secretory capacity of exosomal miRNAs. p53 was shown to enhance exosome production in cells undergoing a p53 response to stress (26). In other words, dysfunction of p53 will result in decreased miRNA secretion. The tumor-suppressive ability of p53 can partly depend on the control of miRNA release from normal cells.
Numerous studies show a broad variety of reasons for tumor initiation, including gene amplification, cellular stress, metabolic alteration, and epigenetic changes. This work suggests that the disruption of the cell competitive process mediated by secretory miRNAs will result in the occurrence of neoplasm. Understanding the mechanism by which homeostasis is impaired leads to a novel therapeutic approach for cancer progression.
Supplementary Material
Acknowledgments
We thank Katsuyuki Hayashi and Ikuei Hiraka at DNA Chip Research Inc. for supporting the processing of microarray data. We thank Ayako Inoue for excellent technical assistance.
This work was supported in part by a grant-in-aid for the Third-Term Comprehensive 10-Year Strategy for Cancer Control, a grant-in-aid for Scientific Research on Priority Areas Cancer from the Ministry of Education, Culture, Sports, Science, and Technology, the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation, and the Japan Society for the Promotion of Science through the “Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program)” initiated by the Council for Science and Technology Policy.

This article contains supplemental Figs. 1–3.
- miRNA
- microRNA
- CM
- conditioned medium
- luc
- luciferase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- QRT-PCR
- quantitative real time PCR.
REFERENCES
- 1. Johnston L. A. (2009) Competitive interactions between cells: death, growth, and geography. Science 324, 1679–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Díaz B., Moreno E. (2005) The competitive nature of cells. Exp. Cell Res. 306, 317–322 [DOI] [PubMed] [Google Scholar]
- 3. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer. The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 4. Bondar T., Medzhitov R. (2010) p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell 6, 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong-Le Bourhis X., Berthois Y., Millot G., Degeorges A., Sylvi M., Martin P. M., Calvo F. (1997) Effect of stromal and epithelial cells derived from normal and timorous breast tissue on the proliferation of human breast cancer cell lines in co-culture. Int. J. Cancer 71, 42–48 [DOI] [PubMed] [Google Scholar]
- 6. Senoo-Matsuda N., Johnston L. A. (2007) Soluble factors mediate competitive and cooperative interactions between cells expressing different levels of Drosophila Myc. Proc. Natl. Acad. Sci. U.S.A. 104, 18543–18548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. (2010) Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 285, 17442–17452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Croce C. M. (2009) Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Suzuki H. I., Yamagata K., Sugimoto K., Iwamoto T., Kato S., Miyazono K. (2009) Modulation of microRNA processing by p53. Nature 460, 529–533 [DOI] [PubMed] [Google Scholar]
- 10. Takeshita F., Patrawala L., Osaki M., Takahashi R. U., Yamamoto Y., Kosaka N., Kawamata M., Kelnar K., Bader A. G., Brown D., Ochiya T. (2010) Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via down-regulation of multiple cell-cycle genes. Mol. Ther. 18, 181–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peng X., Guo W., Liu T., Wang X., Tu X., Xiong D., Chen S., Lai Y., Du H., Chen G., Liu G., Tang Y., Huang S., Zou X. (2011) Identification of miRs-143 and -145 that is associated with bone metastasis of prostate cancer and involved in the regulation of EMT. PLoS One 6, e20341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franses J. W., Baker A. B., Chitalia V. C., Edelman E. R. (2011) Sci. Transl. Med. 3, 66ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Savina A., Vidal M., Colombo M. I. (2002) The exosome pathway in K562 cells by Rab11. J. Cell Sci. 115, 2505–2515 [DOI] [PubMed] [Google Scholar]
- 14. Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. (2008) Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 [DOI] [PubMed] [Google Scholar]
- 15. Gandellini P., Folini M., Longoni N., Pennati M., Binda M., Colecchia M., Salvioni R., Supino R., Moretti R., Limonta P., Valdagni R., Daidone M. G., Zaffaroni N. (2009) miR-205 exerts tumor-suppressive functions in human prostate through down-regulation of protein kinase Cepsilon. Cancer Res. 69, 2287–2295 [DOI] [PubMed] [Google Scholar]
- 16. Clapé C., Fritz V., Henriquet C., Apparailly F., Fernandez P. L., Iborra F., Avancès C., Villalba M., Culine S., Fajas L. (2009) miR-143 interferes with ERK5 signaling and abrogates prostate cancer progression in mice. PLoS One 4, e7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong D., Li Y., Wang Z., Banerjee S., Ahmad A., Kim H. R., Sarkar F. H. (2009) miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells 27, 1712–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujita Y., Kojima K., Ohhashi R., Hamada N., Nozawa Y., Kitamoto A., Sato A., Kondo S., Kojima T., Deguchi T., Ito M. (2010) MiR-148a attenuates paclitaxel resistance of hormone-refractory, drug-resistant prostate cancer PC3 cells by regulating MSK1 expression. J. Biol. Chem. 285, 19076–19084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Saito Y., Friedman J. M., Chihara Y., Egger G., Chuang J. C., Liang G. (2009) Epigenetic therapy up-regulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem. Biophys. Res. Commun. 379, 726–731 [DOI] [PubMed] [Google Scholar]
- 20. Rauhala H. E., Jalava S. E., Isotalo J., Bracken H., Lehmusvaara S., Tammela T. L., Oja H., Visakorpi T. (2010) miR-193b is an epigenetically regulated putative tumor suppressor in prostate cancer. Int. J. Cancer 127, 1363–1372 [DOI] [PubMed] [Google Scholar]
- 21. Xu B., Niu X., Zhang X., Tao J., Wu D., Wang Z., Li P., Zhang W., Wu H., Feng N., Wang Z., Hua L., Wang X. (2011) miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol. Cell Biochem. 350, 207–213 [DOI] [PubMed] [Google Scholar]
- 22. Gibbings D. J., Ciaudo C., Erhardt M., Voinnet O. (2009) Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 11, 1143–1149 [DOI] [PubMed] [Google Scholar]
- 23. Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., van Eijndhoven M. A., Hopmans E. S., Lindenberg J. L., de Gruijl T. D., Würdinger T., Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miyanishi M., Tada K., Koike M., Uchiyama Y., Kitamura T., Nagata S. (2007) Identification of Tim4 as a phosphatidylserine receptor. Nature 450, 435–439 [DOI] [PubMed] [Google Scholar]
- 25. Kwak P. B., Iwasaki S., Tomari Y. (2010) The microRNA pathway and cancer. Cancer Sci. 101, 2309–2315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yu X., Harris S. L., Levine A. J. (2006) The regulation of exosome secretion. A novel function of the p53 protein. Cancer Res. 66, 4795–4801 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.