Background: Known indole prenyltransferases catalyzed regioselective prenylations at N-1, C-2, C-3, C-4, C-6, and C-7 of the indole ring.
Results: Recombinant 5-DMATS was assayed with tryptophan and derivatives in the presence of DMAPP.
Conclusion: 5-DMATS prenylated indole derivatives at C-5.
Significance: 5-DMATS fills the last prenylation gap of indole derivatives and could be used as a potential catalyst for chemoenzymatic synthesis.
Keywords: Aspergillus, Biotechnology, Enzyme Catalysis, Genomics, Natural Product Biosynthesis, DMATS, Dimethylallyltryptophan Synthase, Indole Prenyltransferase
Abstract
The putative prenyltransferase gene ACLA_031240 belonging to the dimethylallyltryptophan synthase superfamily was identified in the genome sequence of Aspergillus clavatus and overexpressed in Escherichia coli. The soluble His-tagged protein EAW08391 was purified to near homogeneity and used for biochemical investigation with diverse aromatic substrates in the presence of different prenyl diphosphates. It has shown that in the presence of dimethylallyl diphosphate (DMAPP), the recombinant enzyme accepted very well simple indole derivatives with l-tryptophan as the best substrate. Product formation was also observed for tryptophan-containing cyclic dipeptides but with much lower conversion yields. In contrast, no product formation was detected in the reaction mixtures of l-tryptophan with geranyl or farnesyl diphosphate. Structure elucidation of the enzyme products by NMR and MS analyses proved unequivocally the highly regiospecific regular prenylation at C-5 of the indole nucleus of the simple indole derivatives. EAW08391 was therefore termed 5-dimethylallyltryptophan synthase, and it filled the last gap in the toolbox of indole prenyltransferases regarding their prenylation positions. Km values of 5-dimethylallyltryptophan synthase were determined for l-tryptophan and DMAPP at 34 and 76 μm, respectively. Average turnover number (kcat) at 1.1 s−1 was calculated from kinetic data of l-tryptophan and DMAPP. Catalytic efficiencies of 5-dimethylallyltryptophan synthase for l-tryptophan at 25,588 s−1·m−1 and for other 11 simple indole derivatives up to 1538 s−1·m−1 provided evidence for its potential usage as a catalyst for chemoenzymatic synthesis.
Introduction
Prenylated indole alkaloids represent a group of natural products with diverse chemical structures and are widely distributed in bacteria, fungi, plants, and marine organisms (1, 2). Because of their impressive pharmacological and biological activities as drugs or as toxins (1, 3), prenylated indole alkaloids attract the attention of scientists from different scientific disciplines, including chemistry, ecology, biology, pharmacology, and biochemistry (1, 4–7). These compounds are hybrid molecules containing prenyl moieties derived from prenyl diphosphates and indole or indoline ring from tryptophan or its precursors (1, 5). Indole prenyltransferases catalyze the connection of these two characteristic structural features and contribute significantly to the structural diversity of the prenylated indole alkaloids. Significant progress has been achieved for molecular biological, biochemical, and structural biological investigations on different prenyltransferase groups, including indole prenyltransferases (1, 8, 9). By the end of October 2011, 20 indole prenyltransferases from bacteria and fungi have been characterized biochemically (1, 10–15). These enzymes catalyzed the transfer of prenyl moieties onto nitrogen or carbon atoms at the indole ring resulting in formation of “regularly” or “reversely” prenylated derivatives (15). More interestingly, indole prenyltransferases showed the usually broad substrate specificity but catalyzed regiospecific prenylation at different positions of indole or indoline rings (Fig. 1) (15). CymD from the marine actinobacterium Salinispora arenicola catalyzed the reverse prenylation at the indole nitrogen of l-tryptophan (12), whereas FtmPT2 from the fungus Aspergillus fumigatus catalyzed the regular prenylation of 12,13-dihydroxyfumitremorgin C at this position (16). Regular and reverse C2-prenylations were observed for FtmPT1 (17) and FgaPT1 (18), both from A. fumigatus, respectively. Prenylations at C-3 of cyclic dipeptides by AnaPT (19) and CdpC3PT (10), both from the fungus Neosartorya fischeri, lead to the formation of indoline derivatives with an α- and β-configured reverse prenyl moiety, respectively. FgaPT2 and its orthologues from different fungi represent the first pathway-specific enzyme in the biosynthesis of ergot alkaloids and catalyzed the regular prenylation of l-tryptophan at C-4 (3, 20). CpaD from Aspergillus oryzae catalyzed regular C4-prenylation as well but used cyclo-acetoacetyl-l-tryptophan as substrate (21). IptA from a soil bacterium Streptomyces sp. was reported to prenylate l-tryptophan at C-6 (13) and 7-DMATS4 from A. fumigatus at C-7 (22). An additional example of C7-prenyltransferase is CTrpPT from A. oryzae, which used cyclo-l-Trp-l-Trp as the best substrate (11).
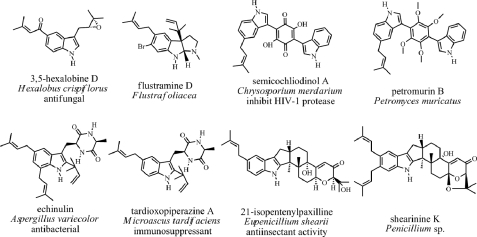
FIGURE 1.
Examples of indole prenyltransferases with different prenylation positions.
In summary, indole prenyltransferases with regioselectivity for N-1, C-2, C-3, C-4, C-6, and C-7 have already been identified and characterized in detail (Fig. 1). However, a prenyltransferase responsible for transferring a prenyl moiety to C-5 of the indole ring has not been reported prior to this study. However, database searching revealed the presence of a number of biologically active indole alkaloids carrying a prenyl moiety at C-5 in nature (Fig. 2). These compounds include simple prenylated indole derivatives like the antifungal compound 3,5-hexalobine D from the plant Hexalobus crispiflorus (23) or the brominated tryptamine derivative flustramine D from the bryozoan Flustra foliacea (24). Semicochliodinol A from the fungus Chrysosporium merdarium (25) and petromurin B from the fungus Petromyces muricatus (26) are C5-prenylated indolyl benzoquinones derived from two tryptophan molecules (27). Semicochliodinol A was reported to inhibit HIV-1 protease (25). C5-prenylated derivatives were also found for tryptophan-containing cyclic dipeptides like cyclo-l-Trp-l-Ala, e.g. echinulin from Aspergillus strains (28) or tardioxopiperazine A from the fungus Microascus tardifaciens (29). A relatively large indole alkaloid group are the indole diterpenes from Penicillium (30) and other Ascomycetes (31). The members of this group carry prenyl moieties at C-5 (21-isopentenylpaxilline) (32), C-6, or at both positions (shearinine K) (30), or modified structures thereof.
FIGURE 2.
Examples of naturally occurring C5-prenylated indole alkaloids.
The discrepancy between the natural occurrence of a large number of C5-prenylated indole derivatives on the one hand and undiscovered C5-prenyltransferases on the other hand prompted us to search for such enzymes. In this study, we reported the identification and characterization of the first C5-prenyltransferase of indoles, i.e. 5-dimethylallyltryptophan synthase (5-DMATS) from Aspergillus clavatus and its potential usage as a catalyst for chemoenzymatic synthesis.
EXPERIMENTAL PROCEDURES
Computer-assisted Sequence Analysis
Sequence identities were obtained by alignments of amino acid sequences using the program “BLAST 2 SEQUENCES” (www.ncbi.nlm.nih.gov). FGENESH from Softberry and the DNASIS software package (version 2.1, Hitachi Software Engineering, San Bruno, CA) were used for exon prediction and sequence analysis, respectively.
Chemicals
Dimethylallyl diphosphate (DMAPP), geranyl diphosphate (GPP), and farnesyl diphosphate (FPP) were prepared according to the method described for geranyl diphosphate by Woodside et al. (33). Indole derivatives of the highest available purity were purchased from TCI, Acros Organics, Aldrich, Sigma, Bachem, and Alfa Aesar.
Bacterial Strains, Plasmids, and Culture Conditions
pGEM-T Easy and pQE70 were obtained from Promega and Qiagen (Hilden, Germany), respectively. Escherichia coli XL1 Blue MRF′ (Stratagene, Amsterdam, the Netherlands) and M15 [pREP4] (Qiagen) were used for cloning and expression experiments, respectively. They were grown in liquid Terrific-Broth (TB) or Luria-Bertani (LB) medium and on solid LB medium with 1.5% (w/v) agar at 37 or 22 °C. 50 μg·ml−1 of carbenicillin were used for selection of recombinant E. coli XL1 Blue MRF′ strains. Addition of carbenicillin at 50 μg·ml−1 and kanamycin at 25 μg·ml−1 was used for selection of recombinant E. coli M15 [pREP4] strains.
Cultivation of A. clavatus, RNA Isolation, and cDNA Synthesis
A. clavatus NRRL1 was kindly provided by Agricultural Research Service Culture Collection, United States Department of Agriculture, and was cultivated on solid YME media consisting of 0.4% (w/v) yeast extract, 1% (w/v) malt extract, 0.4% (w/v) glucose, pH 7.3, and 2% (w/v) agar at 26 °C. For RNA isolation, mycelia of A. clavatus NRRL1 from plates were inoculated into a 300-ml Erlenmeyer flask containing 100 ml of liquid YME media and cultivated at 26 °C and 160 rpm for 7 days. After separation of mycelia from the medium, RNA was isolated by using the High Pure RNA isolation kit (Roche Diagnostics) according to the manufacturer's protocol. cDNA was synthesized with the Transcriptor High Fidelity cDNA synthesis kit (Roche Diagnostics).
PCR Amplification and Gene Cloning
PCR amplification was carried out on a MiniCycler from Bio-Rad. A PCR fragment of 1300 bp containing the entire coding sequence of 5-dmats was amplified from cDNA by using Expand high fidelity kit (Roche Diagnostics). The primers were 5-DMATS_for (5′- GCCCAGCATGCCTCACCAAAACAGC-3′) at the 5′-end, and 5-DMATS_rev (5′- GGTCGAAGATCTCAATTTCCAAGACTT-3′) at the 3′-end of the gene. Bold letters represent mutations inserted in comparison with the original genome sequence to give the underlined restriction site SphI at the start codon in 5-DMATS_for and BglII located at the predicted stop codon in 5-DMATS_rev. A program of 30 cycles with annealing at 58 °C for 50 s and elongation at 72 °C for 90 s was used for PCR amplification. The PCR fragment was cloned into pGEM-T easy vector resulting in plasmid pYL08, which was subsequently sequenced (Eurofins MWG Operon, Ebersberg, Germany) to confirm the sequence. Plasmid pYL08 was digested with BglII alone or together with SphI to obtain BglII-BglII fragment of 748 bp and SphI-BglII fragment of 531 bp, respectively. To get the expression construct pYL09, these two fragments were cloned into pQE70 subsequently.
Overproduction and Purification of His6-5-DMATS
For overproduction of 5-DMATS, E. coli M15 [pREP4] cells harboring the plasmid pYL09 were cultivated in 2000-ml Erlenmeyer flasks containing 1000 ml of liquid TB medium, supplemented with carbenicillin (50 μg·ml−1) and kanamycin (25 μg·ml−1), and then grown at 37 °C to an absorption at 600 nm of 0.6. For induction, isopropyl thiogalactoside was added to a final concentration of 0.4 mm, and the cells were cultivated for a further 16 h at 22 °C before harvest. The bacterial cultures were centrifuged, and the pellets were resuspended in lysis buffer (10 mm imidazole, 50 mm NaH2PO4, 300 mm NaCl, pH 8.0) at 2–5 ml/g wet weight. After addition of 1 mg·ml−1 lysozyme and incubation on ice for 30 min, the cells were sonicated six times for 10 s each at 200 watts. To separate the cellular debris from the soluble proteins, the lysate was centrifuged at 13,000 × g for 30 min at 4 °C. One-step purification of the recombinant His6-tagged fusion protein by affinity chromatography with nickel-nitrilotriacetic acid-agarose resin (Qiagen) was carried out according to the manufacturer's instructions. The protein was eluted with 250 mm imidazole in 50 mm NaH2PO4, 300 mm NaCl, pH 8.0. To change the buffer, the protein fraction was passed through a PD-10 column (GE Healthcare), which had been equilibrated with 50 mm Tris-HCl, 15% (v/v) of glycerol, pH 7.5, previously. 5-DMATS was eluted with the same buffer and frozen at −80 °C for enzyme assays.
Protein Analysis and Determination of Relative Molecular Mass of Active His6-5-DMATS
Proteins were analyzed by 12% (w/v) SDS-polyacrylamide gels according to the method of Laemmli (34) and stained with Coomassie Brilliant Blue G-250. The Mr value of the recombinant active His6-5-DMATS was determined by size exclusion chromatography on a HiLoad 16/60 Superdex 200 column (GE Healthcare), which had been equilibrated with 50 mm Tris-HCl buffer, pH 7.5, containing 150 mm NaCl. The column was calibrated with dextran blue 2000 (2000 kDa), ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), carbonic anhydrase (29 kDa), and ribonuclease A (13.7 kDa) (GE Healthcare). The proteins were eluted with the same buffer as for equilibration.
Enzyme Assays with 5-DMATS
The enzyme reaction mixtures for determination of the relative activities with different indole derivatives or l-tyrosine (100 μl) contained 50 mm Tris-HCl, pH 7.5, 5 mm CaCl2, 1 mm aromatic substrate, 2 mm DMAPP, GPP, or FPP, 0.15–5% (v/v) glycerol, 0–5% (v/v) DMSO, and 1 μm purified recombinant protein. The reaction mixtures were incubated at 37 °C for 7 h (with DMAPP) or 24 h (with GPP or FPP). The enzyme reactions were terminated by addition of 100 μl of methanol per 100 μl reaction mixture.
For determination of kinetic parameters of DMAPP, l-tryptophan at 1 mm and DMAPP at final concentrations of up to 3 mm were used as substrates. For determination of kinetic parameters of l-tryptophan and derivatives, the assays contained 2 mm DMAPP. Because of the difference of solubility in the aqueous system, various concentrations were used for aromatic substrates as follows: for 8a and 12a up to 5 mm, for 1a–3a, 7a, and 9a–11a up to 2 mm, and for 4a–6a up to 1 mm. The protein concentration was 20 nm (1a), 40 nm (DMAPP), or 200 nm (other substrates), and the incubation time was 30 min (1a), 40 min (DMAPP) or 60 min (other substrates).
Preparative Synthesis of Enzyme Products for Structure Elucidation
For isolation of the enzyme products, reactions were carried out in large scale (10 ml) containing each of the 12 substrates 1a–12a (1 mm), DMAPP (2 mm), CaCl2 (5 mm), Tris-HCl (50 mm, pH 7.5), glycerol 0.15–5% (v/v), and 5-DMATS (1.4 μm). After incubation for 16 h, the reactions were terminated by addition of 10 ml of methanol each. After removal of the precipitated protein by centrifugation at 6000 rpm for 30 min, the reaction mixtures were concentrated on a rotating vacuum evaporator at 30 °C to a final volume of 1 ml before injection onto HPLC.
HPLC Conditions for Analysis and Isolation of Enzyme Products
The enzyme products of the incubation mixtures were analyzed by HPLC on an Agilent series 1200 by using a Multiphor 120 RP-18 column (250 × 4 mm, 5 μm, C+S Chromatographie Service, Langerwehe, Germany) at a flow rate of 1 ml·min−1. Water (solvent A) and methanol (solvent B) each with 0.5% (v/v) trifluoroacetic acid were used as solvents. For analysis of enzyme products of tryptophan, simple indole derivatives and l-tyrosine, a linear gradient of 20–100% (v/v) solvent B, for 15 min were used. The column was then washed with 100% (v/v) solvent B for 5 min and equilibrated with 20% (v/v) solvent B for 5 min. For analysis of enzyme products of tryptophan-containing cyclic dipeptides, the gradient began with 40% (v/v) solvent B and increased from 40 to 100% (v/v) solvent B in 15 min. After washing with 100% (v/v) solvent B for 5 min, the column was equilibrated with 40% (v/v) solvent B after each run. For assays with GPP or FPP, a linear gradient of 20–84% (v/v) solvent B for 12 min and then 84–100% (v/v) solvent B for 18 min was used. The column was then washed with 100% (v/v) solvent B for 10 min and equilibrated with 20% (v/v) solvent B for 5 min. Detection was carried out with a Photo Diode Array Detector.
For isolation of the enzyme products, the same HPLC equipment with a Multospher 120 RP-18 column (250 × 10 mm, 5 μm, C+S Chromatographie Service) was used. The flow rate was 2.5 ml·min−1. Water (solvent C) and methanol (solvent D) without acid were used as solvents. Gradients of 20–100% (v/v) solvent D for different times were used for isolation. The column was then washed with 100% (v/v) solvent D for 8 min and equilibrated with 20% (v/v) solvent D for 8 min.
NMR Spectroscopic Analysis and High Resolution ESI-MS
1H NMR spectra were recorded on a Bruker Avance 500 MHz spectrometer or a JEOL ECX-400 spectrometer. The HSQC and HMBC spectra were recorded with standard methods (35) on the Bruker Avance 500 MHz spectrometer. Chemical shifts were referenced to the signal of CD3OD at 3.31 ppm or DMSO-d6 at 2.50 ppm. All spectra were processed with MestReNova 5.2.2.
The isolated products were also analyzed by high resolution ESI-MS with a Q-Trap Quantum (Applied Biosystems). Positive ESI-MS data are given in supplemental Table S3.
Nucleotide Sequence Accession Number
The coding sequence of 5-dmats (ACLA_031240) is available at GenBankTM under the accession number XM_001269816.
RESULTS
Sequence Analysis and Cloning of 5-Dimethylallyltryptophan Synthase Gene 5-dmats
In the course of our search for prenyltransferases responsible for prenylation of tryptophan or indole derivatives at C-5, one putative gene ACLA_031240 from the genome sequence of A. clavatus NRRL1 raised our interest. The deduced gene product EAW08391 consists of 427 amino acids and shares high sequence similarities with C4-prenyltransferases of tryptophan, e.g. 52% identity with FgaPT2 from A. fumigatus (20) at the amino acid level. Based on this homology, it can be expected that EAW08391 catalyzes a similar reaction as FgaPT2.
Inspection of down- and upstream genes of ACLA_031240 in the genome sequence of A. clavatus (www.ncbi.nlm.nih.gov) revealed that the enzymes encoded by these genes are very likely involved in the fungal development and biosynthesis of primary rather than secondary metabolites, e.g. prenylated indole alkaloids. This means that ACLA_031240 is very likely not clustered with genes for the biosynthesis of secondary metabolites. For example, in the downstream sequence of ACLA_031240, the putative gene ACLA_031250 is separated from ACLA_031240 by a segment of 13 kb and encodes a putative MYB family conidiophore development protein. ACLA_031260, downstream of ACLA_031250, was predicted to be a glycosyl hydrolase gene. In the upstream sequence of ACLA_031240, ACLA_031230 was predicted to encode a UDP-N-acetylglucosaminyltransferase involved in the cell wall biosynthesis. ACLA_031220 is likely a 60 S ribosomal protein gene.
Blast search with EAW08391 from A. clavatus in GenBankTM indicated the presence of three homologues as follows: XP_001818057 encoded by AOR_1_1880174 from A. oryzae RIB40; EED57628 encoded by AFLA_083250 from Aspergillus flavus NRRL3357, and EEQ32624 encoded by MCYG_05443 from Arthroderma otae CBS113480. EAW08391 shares on the amino acid level sequence identities of 76, 75, and 74% with XP_001818057, EED57628, and EEQ32624, respectively. It can be expected that these three enzymes catalyze the same reaction as EAW08391.
Inspection of down- and upstream genes of AOR_1_1880174 in the genome sequence of A. oryzae, AFLA_083250 of A. flavus, and MCYG_05443 of A. otae (www.ncbi.nlm.nih.gov) indicated also their nonclustering with other secondary metabolite biosynthesis genes. The direct neighboring genes of AOR_1_1880174 were predicted to encode l-arabitol/xylitol dehydrogenase (AOR_1_1878174) and a putative transporter gene for choline, γ-aminobutyric acid, or other amino acids (AOR_1_1882174). Orthologues of these two genes (AFLA_083240 and AFLA_083270) were also found directly at AFLA_083250. A gene AFLA_083260 coding for a small hypothetical protein with 84 amino acids was located between AFLA_083250 and AFLA_083270. In the genome of A. otae, MCYG_05443 was found between an l-allothreonine aldolase gene (MCYG_05442) and a hypercellular protein HypA gene (MCYG_05444).
To prove the function of EAW08391, the coding region of ACLA_031240, termed 5-dmats in this study, was amplified by PCR from cDNA synthesized from mRNA. The PCR product was cloned via pGEM-T easy vector into pQE70, resulting in the expression construct pYL09. For overproduction of His6-5-DMATS, E. coli M15 cells harboring pYL09 were cultivated in TB medium and induced with 0.4 mm isopropyl thiogalactoside at 22 °C for 16 h. A significant band with migration near the 45-kDa size marker was observed on SDS-PAGE of the purified protein (see supplemental Fig. S1), corresponding to the calculated Mr value of 50,411 for His6-5-DMATS. The yield was calculated to be 1.8 mg of purified protein/liter of culture. The Mr value of the native recombinant His6-5-DMATS was determined by size exclusion chromatography at about 79,000. This indicated that 5-DMATS acted likely as a homodimer.
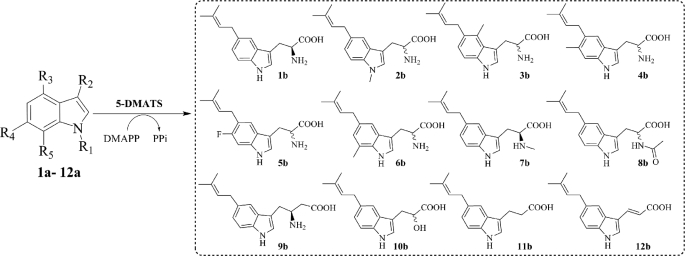
5-DMATS Accepted Well l-Tryptophan and Simple Indole Derivatives as Substrates
Because of the high sequence similarity with tryptophan C4-prenyltransferases mentioned above, l-tryptophan and 17 simple indole derivatives (Table 1 and supplemental Table S1) were incubated with the purified 5-DMATS in the presence of DMAPP (2 mm). These substances included eight tryptophan derivatives with modification at the indole ring and nine at the side chain. The reaction mixtures were incubated with 5-DMATS at a final concentration of 1 μm for 7 h. HPLC analysis was used for monitoring the enzyme product formation. Assays with heat-inactivated protein by boiling for 20 min were used as negative control.
TABLE 1.
Enzyme activity of 5-DMATS towards tryptophan and other simple indole derivatives (1a–12a)
The reaction mixtures containing indole derivatives and DMAPP were incubated with 1 μm protein for 7 h. Under this condition, l-tryptophan was completely consumed.
HPLC analysis of the incubation mixtures showed clear product formation for 17 of the 18 tested indole derivatives with l-tryptophan as the best substrate (Table 1 and supplemental Table S1). HPLCs of incubation mixtures of 12 substrates (1a–12a, Table 1) showed clearly the presence of one product peak for each substrate. Similar behavior was also observed for other five accepted substrates (data not shown). Under this condition, l-tryptophan was almost completely consumed, and the 11 other substrates (2a–12a) were accepted with total yields between 38 and 91%. Even in the incubation mixtures with 0.40 μm protein for 1 h, the conversion yield for l-tryptophan was calculated to be 95.4% (see supplemental Table S2).
Inspection of the activities of the tested substances (Table 1 and supplemental Table S1) revealed that, with the exceptions for C5-substituted derivatives, all of the substances with modifications by fluoro or methyl group at the indole ring were well accepted by 5-DMATS. These results could indicate the prenylation position at C-5 of the indole rings of the accepted substrates. 5-Bromo-dl-tryptophan was not accepted by 5-DMATS. The conversion yields of 5-fluoro-l-tryptophan and 5-methyl-dl-tryptophan at 7.4 and 1.7%, respectively, were significantly lower than those with substitution at other positions of the indole ring. The prenylation of these substances had very likely taken place at other positions rather than C-5 (see under “Discussion”). Modification at the side chain of tryptophan reduced enzyme activity, especially by shortening the chain length, as in the case of indole-3-acetic acid and tryptamine. In comparison, changes at the amino group such as N-methylation (l-abrine, 7a), replacement by a hydroxyl group (dl-indole-3-lactic acid, 10a), or deamination (indole-3-propionic acid, 11a) showed less influence on the enzyme activity.
A previous study (36) showed that the tryptophan C4-prenyltransferase FgaPT2 also accepted tryptophan-containing cyclic dipeptides as substrates. 5-DMATS was therefore also assayed with five such cyclic dipeptides and analyzed on HPLC. It has been shown that these compounds were also substrates for 5-DMATS but were accepted with significantly lower yields (<10%) than most of the simple indole derivatives (see supplemental Table S1). Incubations of 5-DMATS with l-tyrosine in the presence of DMAPP or with l-tryptophan in the presence of GPP or FPP did not result in the formation of any enzyme product, even after incubation with 1 μm protein for 24 h (see supplemental Table S2).
5-DMATS Catalyzed the Regular C5 Prenylation at the Indole Ring
To confirm the prenylation position, enzyme products of 12 substrates (1a–12a, Table 1) were isolated on HPLC and subjected to high resolution MS and NMR analyses. High resolution ESI-MS (see supplemental Table S3) confirmed the presence of one dimethylallyl moiety each in the products of 1a–12a by detection of masses, which are 68 daltons larger than those of the respective substrates.
In the 1H NMR spectra of all the enzyme products (taken in CD3OD or DMSO-d6), signals at δH 3.34–3.47 (d, 2H-1′), 5.17–5.40 (t or m, H-2′), 1.71–1.78 (s, 3H-4′), and 1.67–1.77 (s, 3H-5′) were observed (see supplemental Table S4 and supplemental Figs. S3–S14), proving unequivocally the attachment of a regular dimethylallyl moiety to a carbon atom (37, 38).
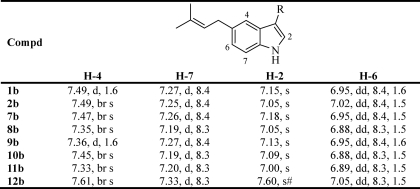
Substrates 2a and 7a–11a are derivatives of l-tryptophan (1a) with modifications at N-1 at the indole ring or at the side chain. Characteristic signals of the four coupling protons at the indole ring (H-4, -5, -6, and -7) appeared as two doublets and two triplets, all with coupling constants of 7–9 Hz in the 1H NMR spectra of 1a, 2a, and 7a–11a (data not shown). In comparison, the two triplets had disappeared in the 1H NMR spectra of their enzyme products 1b, 2b, and 7b–11b. One additional singlet or doublet with a coupling constant smaller than 2 Hz was observed instead. These changes indicated that prenylations had taken place at C-5 or C-6. As given in Table 2, the signals of the remaining three protons at H-4, H-5 or H-6 and H-7 in the 1H NMR spectra (all taken in CD3OD) were found to be in the same order, i.e. (from low to high magnetic field) doublet (1.6 Hz) or broad singlet, doublet (8.3–8.4 Hz), and double doublet (8.3–8.4 and 1.5–1.6 Hz). The singlets for H-2 were found between the doublet (8.3–8.4 Hz) and double doublet. It is plausible that the structures of these compounds have the same prenylation position.
TABLE 2.
Signals for aromatic protons in the 1H NMR spectra of the selected products (CD3OD)
For structures, see Fig. 3. The signals are arranged in an order from low to high magnetic field so that the relative signal positions caused by C5-prenylation can be better compared.
#,Due to the presence of a double bond between C-10 and C-11, the signal of H-2 was downshifted in the 1H NMR spectrum.
Enzyme products of 1a and 7a with prenylation at C-6 were reported by Takahashi et al. (13). The NMR data of 6-dimethylallyl-l-tryptophan and 6-dimethylallyl-l-abrine (also taken in CD3OD) differed clearly from those of 1b and 7b, especially in the region for aromatic protons. The aromatic protons in the 1H NMR spectra of 6-dimethylallyl-l-tryptophan and 6-dimethylallyl-l-abrine appeared (from low to high magnetic field) in a different order than in the spectra of 1b and 7b, i.e. doublet (7.8–8.2 Hz), two singlets, and double doublet. Therefore, the prenylation position in 1b and 7b must be C-5. This conclusion was also confirmed by interpretation of the two-dimensional NMR spectra of 1b (in DMSO-d6) and 7b (in CD3OD). In the HMBC spectrum of 1b (see supplemental Figs. S2 and S3), connectivity from δH 7.32 (1H, br. s, H-4) to C-1′ of the prenyl moiety at δC 34.1 proved unequivocally the attachment of the dimethylallyl moiety to C-5. The signal at δH 7.32 (1H, s) for proton H-4 was unambiguously confirmed by the detected connectivities between this signal and C-8 at 134.9 ppm as well as C-3 at 109.3 ppm. For 7b, similar phenomena were also observed in the HMBC spectrum (see supplemental Figs. S2 and S9). It can be concluded that the prenyl moieties in 1b, 2b, and 7b–11b are attached to C-5 of the indole rings.
Substrate 12a is also a derivative of l-tryptophan with alteration at the side chain. Because of the presence of a double bond between C-10 and C-11, the signal of H-2 was downshifted in 1H NMR spectrum to δH 7.60 (1H, s), in comparison with those of 1b, 2b, and 7b–11b between 7.00 and 7.18 ppm. However, the signals for H-4, H-6, and H-7 of 12b appeared in the same order as in the spectra of 1b, 2b, and 7b–11b (Table 2), proving the C5-prenylation in the structure of 12b.
In the 1H NMR spectrum of 3b (see supplemental Fig. S5), the two doublets at δH 6.85 (1H, d, 8.2 Hz, H-6) and δH 7.08 (1H, d, 8.2 Hz, H-7) represent signals for two protons at the ortho-position and indicated the prenylation at C-5 or C-7. The connectivity from H-6 to δC 31.4 of C-1′ and from δH 2.55 (3H, s, H-13) of the methyl group at C-4 to δC 128.9 of C-5 in the HMBC spectrum confirmed the prenylation at C-5 in 3b (see supplemental Fig. S2).
The structures of 4b, 5b, and 6b were elucidated by interpretation of their 1H NMR spectra (see supplemental Figs. S6–S8). Three singlets observed at δH 7.14 (1H, s, H-2), δH 7.44 (1H, s, H-4), and δH 7.08 (1H, s, H-7) in the 1H NMR spectrum of 4b proved the prenylation at C-5. In the 1H NMR spectrum of 5b, the two doublets at δH 7.02 (1H, d, 10.8 Hz, H-7) and δH 7.49 (1H, d, 7.3 Hz, H-4) with clearly different coupling constants are caused by the different distances of the protons to fluoro atom at C-6. In the 1H NMR spectrum of the enzyme product of 6a, signals for two products 6b and 6c with a ratio of 1.5:1 were detected. Unfortunately, these two compounds could not be separated from each other. Based on their different contents in the mixture, we were able to identify the major product 6b as C5-prenylated derivative, which is characteristic of the presence of three singlets at δH 7.14 (1H, br. s), 7.14 (1H, br. s), and 6.69 (1H, br. s) for H-2, H-4, and H-6, respectively.
In conclusion, 5-DMATS catalyzed the C5-prenylation of l-tryptophan and simple indole derivatives (Fig. 3). To the best of our knowledge, the structures 1b–12b were not described previously. The structure of 6c could not be unequivocally elucidated in this study. The presence of two doublets with a coupling constant of 8.1 Hz in the 1H NMR spectrum indicated a prenylation at C-4 or C-6.
FIGURE 3.
Prenyl transfer reactions catalyzed by 5-DMATS, exemplified with simple indole derivatives 1a–12a.
Biochemical Characterization and Kinetic Parameters of 5-DMATS
For determination of the metal ion dependence of 5-DMATS, incubations of l-tryptophan (1a) with DMAPP were carried out in the presence of different metal ions at a final concentration of 5 mm. Incubations with the chelating agent EDTA or without additives were used as controls. In the incubation mixture with EDTA, no decrease of the enzyme activity was observed, in comparison with that of incubation without additives. As observed for other members of the DMATS superfamily (15, 39), several divalent metal ions enhanced slightly the enzyme activity of 5-DMATS. For example, the enzyme activities with Ca2+ and Mg2+ were found to be 250 and 204% of that without additives, respectively.
To study the behavior of 5-DMATS toward 12 indole derivatives (1a–12a) and DMAPP in detail, kinetic parameters, including Michaelis-Menten constants (Km) and turnover numbers (kcat), were determined by Hanes-Woolf and Eadie-Hofstee plots and are given in Table 3. The reactions catalyzed by 5-DMATS apparently followed Michaelis-Menten kinetics. The Km values for DMAPP and l-tryptophan (1a) were found to be 76 and 34 μm, respectively, whereas Km values of 0.10–1.0 mm were determined for 2a–12a, much higher than that observed for 1a. The turnover numbers were calculated for DMAPP and 1a at 1.3 and 0.87 s−1, respectively. Lower turnover numbers of 0.059–0.76 s−1 were determined for 2a–12a. The catalytic efficiencies (kcat/Km) of 5-DMATS toward DMAPP and 1a were calculated to be 17,105 and 25,588 s−1·m−1, respectively. In comparison, lower catalytic efficiencies at 155–1538 s−1·mm−1 were found for 2a–12a, only 0.6–6.0% that of 1a.
TABLE 3.
Kinetic parameters of 5-DMATS for selected substrates
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| mm | s−1 | s−1·m−1 | |
| l-Tryptophan (1a) | 0.034 | 0.87 | 25,588 |
| 1-Methyl-dl-tryptophan (2a) | 0.10 | 0.059 | 590 |
| 4-Methyl-dl-tryptophan (3a) | 0.25 | 0.29 | 1160 |
| 6-Methyl-dl-tryptophan (4a) | 1.0 | 0.76 | 760 |
| 6-Fluoro-dl-tryptophan (5a) | 0.27 | 0.29 | 1074 |
| 7-Methyl-dl-tryptophan (6a) | 0.47 | 0.40 | 851 |
| l-Abrine (7a) | 0.26 | 0.40 | 1538 |
| N-Acetyl-dl-tryptophan (8a) | 0.35 | 0.10 | 286 |
| l-β-Homotryptophan (9a) | 0.97 | 0.15 | 155 |
| dl-Indole-3-lactic acid (10a) | 0.67 | 0.43 | 642 |
| Indole-3-propionic acid (11a) | 0.39 | 0.39 | 1000 |
| trans-Indole-3-acrylic acid (12a) | 0.40 | 0.14 | 350 |
| DMAPP | 0.076 | 1.3 | 17,105 |
DISCUSSION
Prenyltransferases catalyze transfer reactions of prenyl moieties from prenyl diphosphates to diverse aliphatic or aromatic receptors, including proteins, terpenes, benzoic acids, naphthalenes, flavonoids, and indole alkaloids (1, 8, 9, 40, 41). These enzymes are involved in both primary and secondary metabolism, play an important role in living organisms (42), and contribute significantly to the structural diversity of natural products (43). Prenylated indole alkaloids represent a large group of natural products, predominantly identified as mycotoxins in Ascomycetes (1, 44). Indole prenyltransferases catalyze transfer reactions of prenyl moieties onto the indole nucleus and are involved in the biosynthesis of diverse natural products, especially mycotoxins (1, 15). A large number of indole prenyltransferases, mainly belonging to the DMATS superfamily from fungi of Ascomycetes, have been identified in the last years and characterized biochemically (1, 10, 11). The members of the DMATS superfamily are soluble proteins, showed usually broad substrate specificity and accepted tryptophan, simple indole derivatives, tryptophan-containing cyclic dipeptides, or other indole-containing structures as aromatic substrates and DMAPP as prenyl donor (1, 10, 11). A few examples of soluble indole prenyltransferases have also been identified in bacteria (13, 45). One of the important features of indole prenyltransferases is the regioselectivity of their prenylation reactions. With the exception for C-5, diverse indole prenyltransferases for the other six positions (N-1, C-2, C-3, C-4, C-6, and C-7) have been identified and characterized (Fig. 1). No enzyme for prenylation at C-5 of the indole ring was reported prior to this study, although a number of C5-prenylated derivatives have been isolated from different organisms (Fig. 2). Therefore, there is a need to find enzymes for C5-prenylation at the indole nucleus, so that these enzymes could be better used as tools for chemoenzymatic synthesis of prenylated indole derivatives or even for synthesis of prenylated hydroxynaphthalenes and flavonoids, which have been very recently described for several members of the DMATS superfamily (46, 47).
In this study, we identified and characterized the first tryptophan C5-prenyltransferase 5-DMATS from A. clavatus, which catalyzes the regiospecific C5-prenylation of indole derivatives and fills herewith the last gap in the search for indole prenyltransferases regarding their prenylation positions. Blast searching in the database with 5-DMATS from A. clavatus NRRL1 revealed the presence of three orthologues in the genome sequences of A. oryzae RIB40 (48), A. flavus NRRL3357 (49), and A. otae CBS 113480 (GenBankTM). Analysis of genes in the genomic region of these prenyltransferase genes in all four strains did not provide any indication for their clustering with genes for secondary metabolite biosynthesis. No C5-prenylated derivative was reported for these fungi. Therefore, their roles in these strains could not be predicted in this study. It seems that these genes are the results of redundant copies in the evolution. Nevertheless, considering the low Km values of 34 and 76 μm for l-tryptophan and DMAPP, respectively, as well as the high turnover number of 1.1 s−1 (Table 3), it can be speculated that l-tryptophan and DMAPP are very likely the natural substrates of 5-DMATS in the four fungal strains. Inactivation of these genes in the fungal strains could provide detailed information about their roles in nature, if the genes are expressed and the gene products involved in the biosynthesis of certain substance. Therefore, we tried to detect the production of C5-prenylated indole derivatives by A. clavatus NRRL1. For this purpose, the fungus was cultivated in different liquid media like YME. Culture filtrates and mycelia were extracted with ethyl acetate and methanol, respectively. In the 1H NMR spectra of both extracts, no signal for aromatic protons was observed, which could be assigned to C5-substituted indole rings. Signals for prenyl moieties were also absent in their 1H NMR spectra (data not shown). This means that C5-substituted indole derivatives were not or only in a very low yield produced by this fungus under the tested conditions. These results could indicate that 5-DMATS is not involved in the biosynthesis of secondary metabolites in A. clavatus. It cannot be excluded, however, that 5-DMATS catalyzed a C5-prenylation in the biosynthesis of a fungal product. It is plausible that the expression level of 5-dmats and other related genes would be too low to produce a substantial amount of prenylated derivative. In both cases, cultivation of A. clavatus NRRL1 and 5-dmats-defective mutants would very likely not result in significant changes of secondary metabolite accumulation, which prohibited the potential usage of knock-out experiments to prove gene function. Therefore, optimization of culture conditions should be carried out to improve the level of gene expression in A. clavatus. Cultivation of A. oryzae, A. flavus, or A. otae under different conditions and proof of the accumulated C5-prenylated secondary metabolites, e.g. by NMR analysis, after purification or as extracts, would also be a prerequisite for gene knock-out experiments in these strains. This work is now in progress.
In addition to 5-DMATS described in this study, several DMATSs using l-tryptophan as natural or best aromatic substrate with different regioselectivity have been studied biochemically in detail, e.g. the 4-DMATS FgaPT2 from A. fumigatus (20) and its orthologues MaPT from Malbranchea aurantiaca (50) and DmaW from a clavicipitalean fungus (51, 52), the 6-DMATS IptA from Streptomyces sp. SN-593 (13), and the 7-DMATS from A. fumigatus (22). All of these enzymes catalyzed regular C-prenylation (13, 20, 22, 50, 51). As mentioned above, 5-DMATS shares high sequence similarities on the amino acid level with 4-DMATSs, e.g. 52% with FgaPT2 from A. fumigatus, 50% with MaPT from M. aurantiaca, and 47% with DwaW from the clavicipitalean fungus. As observed for the low sequence similarity between FgaPT2 and 7-DMATS (22), 5-DMATS also showed low sequence similarity (27%) to 7-DMATS. No meaningful similarity was found for 5-DMATS and IptA. These data proved again that substrates of indole prenyltransferases and their prenylation positions could not be predicted by sequence analysis and comparison (15).
As observed for other members of the DMATS superfamily (15, 39), the enzyme activity of 5-DMATS was enhanced by some metal ions such as Ca2+ or Mg2+. 5-DMATS accepted only DMAPP but not GPP or FPP as prenyl donor. It showed, however, similar to many members of the DMATS superfamily, broad promiscuity toward its aromatic substrates. With the exception for C5-substituted derivatives, the most tested simple indole derivatives with modifications at the indole ring or side chain have been well accepted by 5-DMATS (Table 1 and supplemental Table S1). It is not surprising that C5-substituted derivatives were poor substrates for a 5-DMATS with a high regioselectivity at C-5. No product formation was observed for 5-bromo-dl-tryptophan, although low conversion yields of 7.4 and 1.7% were detected for 5-fluoro-l-tryptophan and 5-methyl-dl-tryptophan, respectively. These results indicated a limited feasibility of 5-DMATS to prenylate C5-substituted tryptophan derivatives. Similar phenomena were also observed for the tryptophan C6-prenyltransferase IptA, which also accepted 6-methyl-dl-tryptophan as substrate and catalyzed a C7-prenylation (13). Because of the low amounts, the enzyme products of 5-fluoro-l-tryptophan and 5-methyl-dl-tryptophan were not identified in this study. It could be speculated, however, that prenylation had taken place at another position rather than C-5. We have shown in this study that 5-DMATS prenylated predominantly tryptophan and derivatives at C-5 but occasionally also catalyzed prenylations at other positions, as observed in the incubation mixture of 7-methyl-dl-tryptophan. Both C5- and C4- or C6-prenylated derivatives were identified in this case (supplemental Table S4). The acceptance of C5-substituted tryptophan derivatives by 5-DMATS could also be explained by the substrate promiscuity of indole prenyltransferases of the DMATS superfamily, which accepted even hydroxynaphthalenes and flavonoids as substrates and catalyzed C-, O-, or both prenylations (46, 47).
As given in Table 1, high conversion yields of more than 38% were detected for 12 simple indole derivatives after incubation with 1 μm 5-DMATS for 7 h. NMR and MS analyses proved unequivocally that the reaction catalyzed by 5-DMATS was highly regioselective regular prenylation at C-5 of the indole nucleus in most cases. It can therefore be expected that 5-DMATS could serve as an effective catalyst for chemoenzymatic synthesis of prenylated derivatives in the program of drug discovery and development. Acceptance of tryptophan-containing cyclic dipeptides by 5-DMATS could also be used to increase the structural diversity of prenylated derivatives, although the conversion yields detected in this study were still very low, which perhaps could be improved in the future by mutagenesis experiments.
Supplementary Material
Acknowledgments
We thank Dr. Laufenberg for taking mass spectra and Marco Matuschek and Edyta Stec for synthesis of DMAPP, GPP, and FPP.
This work was supported in part by Deutsche Forschungsgemeinschaft Grant Li844/4-1 (to S.-M. L.) and by the Deutscher Akademischer Austauschdienst within the Programme des Projektbezogenen Personenaustauschs.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) XM_001269816.

This article contains supplemental Tables S1–S4 and Figs. S1–S14.
- 7-DMATS
- 7-dimethylallyltryptophan synthase
- DMAPP
- dimethylallyl diphosphate
- 5-DMATS
- 5-dimethylallyltryptophan synthase
- HMBC
- heteronuclear multiple-bond correlation spectroscopy
- FPP
- farnesyl diphosphate
- GPP
- geranyl diphosphate.
REFERENCES
- 1. Li S. M. (2010) Prenylated indole derivatives from fungi. Structure diversity, biological activities, biosynthesis, and chemoenzymatic synthesis. Nat. Prod. Rep. 27, 57–78 [DOI] [PubMed] [Google Scholar]
- 2. Ruiz-Sanchis P., Savina S. A., Albericio F., Álvarez M. (2011) Structure, bioactivity, and synthesis of natural products with hexahydropyrrolo[2,3-b]indole. Chemistry 17, 1388–1408 [DOI] [PubMed] [Google Scholar]
- 3. Wallwey C., Li S. M. (2011) Ergot alkaloids. Structure diversity, biosynthetic gene clusters, and functional proof of biosynthetic genes. Nat. Prod. Rep. 28, 496–510 [DOI] [PubMed] [Google Scholar]
- 4. Lindel T., Marsch N., Adla S. K. (2011) Indole prenylation in alkaloid synthesis. Top. Curr. Chem. DOI: 10.1007/128_2011_204 [DOI] [PubMed] [Google Scholar]
- 5. Williams R. M., Stocking E. M., Sanz-Cervera J. F. (2000) Biosynthesis of prenylated alkaloids derived from tryptophan. Top. Curr. Chem. 209, 97–173 [Google Scholar]
- 6. Schardl C. L., Panaccione D. G., Tudzynski P. (2006) Ergot alkaloids, biology and molecular biology. Alkaloids Chem. Biol. 63, 45–86 [DOI] [PubMed] [Google Scholar]
- 7. Uhlig S., Botha C. J., Vrålstad T., Rolén E., Miles C. O. (2009) Indole-diterpenes and ergot alkaloids in Cynodon dactylon (Bermuda grass) infected with Claviceps cynodontis from an outbreak of tremors in cattle. J. Agric. Food Chem. 57, 11112–11119 [DOI] [PubMed] [Google Scholar]
- 8. Heide L. (2009) Prenyl transfer to aromatic substrates. Genetics and enzymology. Curr. Opin. Chem. Biol. 13, 171–179 [DOI] [PubMed] [Google Scholar]
- 9. Yazaki K., Sasaki K., Tsurumaru Y. (2009) Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 70, 1739–1745 [DOI] [PubMed] [Google Scholar]
- 10. Yin W. B., Yu X., Xie X. L., Li S. M. (2010) Preparation of pyrrolo[2,3-b]indoles carrying a β-configured reverse C3-dimethylallyl moiety by using a recombinant prenyltransferase CdpC3PT. Org. Biomol. Chem. 8, 2430–2438 [DOI] [PubMed] [Google Scholar]
- 11. Zou H. X., Xie X. L., Linne U., Zheng X. D., Li S. M. (2010) Simultaneous C7- and N1-prenylation of cyclo-l-Trp-l-Trp catalyzed by a prenyltransferase from Aspergillus oryzae. Org. Biomol. Chem. 8, 3037–3044 [DOI] [PubMed] [Google Scholar]
- 12. Schultz A. W., Lewis C. A., Luzung M. R., Baran P. S., Moore B. S. (2010) Functional characterization of the cyclomarin/cyclomarazine prenyltransferase CymD directs the biosynthesis of unnatural cyclic peptides. J. Nat. Prod. 73, 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Takahashi S., Takagi H., Toyoda A., Uramoto M., Nogawa T., Ueki M., Sakaki Y., Osada H. (2010) Biochemical characterization of a novel indole prenyltransferase from Streptomyces sp. SN-593. J. Bacteriol. 192, 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ding Y., de Wet J. R., Cavalcoli J., Li S., Greshock T. J., Miller K. A., Finefield J. M., Sunderhaus J. D., McAfoos T. J., Tsukamoto S., Williams R. M., Sherman D. H. (2010) Genome-based characterization of two prenylation steps in the assembly of the stephacidin and notoamide anticancer agents in a marine-derived Aspergillus sp. J. Am. Chem. Soc. 132, 12733–12740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li S. M. (2009) Evolution of aromatic prenyltransferases in the biosynthesis of indole derivatives. Phytochemistry 70, 1746–1757 [DOI] [PubMed] [Google Scholar]
- 16. Grundmann A., Kuznetsova T., Afiyatullov S. Sh., Li S. M. (2008) FtmPT2, an N-prenyltransferase from Aspergillus fumigatus, catalyzes the last step in the biosynthesis of fumitremorgin B. ChemBioChem 9, 2059–2063 [DOI] [PubMed] [Google Scholar]
- 17. Grundmann A., Li S. M. (2005) Overproduction, purification, and characterization of FtmPT1, a brevianamide F prenyltransferase from Aspergillus fumigatus. Microbiology 151, 2199–2207 [DOI] [PubMed] [Google Scholar]
- 18. Unsöld I. A., Li S. M. (2006) Reverse prenyltransferase in the biosynthesis of fumigaclavine C in Aspergillus fumigatus. Gene expression, purification, and characterization of fumigaclavine C synthase FGAPT1. ChemBioChem 7, 158–164 [DOI] [PubMed] [Google Scholar]
- 19. Yin W. B., Grundmann A., Cheng J., Li S. M. (2009) Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J. Biol. Chem. 284, 100–109 [DOI] [PubMed] [Google Scholar]
- 20. Unsöld I. A., Li S. M. (2005) Overproduction, purification, and characterization of FgaPT2, a dimethylallyltryptophan synthase from Aspergillus fumigatus. Microbiology 151, 1499–1505 [DOI] [PubMed] [Google Scholar]
- 21. Liu X., Walsh C. T. (2009) Characterization of cyclo-acetoacetyl-l-tryptophan dimethylallyltransferase in cyclopiazonic acid biosynthesis. Substrate promiscuity and site -directed mutagenesis studies. Biochemistry 48, 11032–11044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kremer A., Westrich L., Li S. M. (2007) A 7-dimethylallyltryptophan synthase from Aspergillus fumigatus. Overproduction, purification, and biochemical characterization. Microbiology 153, 3409–3416 [DOI] [PubMed] [Google Scholar]
- 23. Achenbach H., Renner C., Waibel R. (1995) Constituents of tropical medicinal plants, LXIX. The hexalobines, diprenylated indoles from Hexalobus crispiflorus and Hexalobus monopetalus. Liebigs Ann. Recl. 1995, 1327–1337 [Google Scholar]
- 24. Peters L., König G. M., Terlau H., Wright A. D. (2002) Four new bromotryptamine derivatives from the marine bryozoan Flustra foliacea. J. Nat. Prod. 65, 1633–1637 [DOI] [PubMed] [Google Scholar]
- 25. Fredenhagen A., Petersen F., Tintelnot-Blomley M., Rösel J., Mett H., Hug P. (1997) Semicochliodinol A and B. Inhibitors of HIV-1 protease and EGF-R protein-tyrosine kinase related to asterriquinones produced by the fungus Chrysosporium merdarium. J. Antibiot. 50, 395–401 [DOI] [PubMed] [Google Scholar]
- 26. Ooike M., Nozawa K., Udagawa S I., Kawai K. I. (1997) Bisindolylbenzenoids from ascostromata of Petromyces muricatus. Can. J. Chem. 75, 625–628 [Google Scholar]
- 27. Balibar C. J., Howard-Jones A. R., Walsh C. T. (2007) Terrequinone A biosynthesis through l-tryptophan oxidation, dimerization and bisprenylation. Nat. Chem. Biol. 3, 584–592 [DOI] [PubMed] [Google Scholar]
- 28. Wang W. L., Lu Z. Y., Tao H. W., Zhu T. J., Fang Y. C., Gu Q. Q., Zhu W. M. (2007) Isoechinulin-type alkaloids, variecolorins A–L, from halotolerant Aspergillus variecolor. J. Nat. Prod. 70, 1558–1564 [DOI] [PubMed] [Google Scholar]
- 29. Fujimoto H., Fujimaki T., Okuyama E., Yamazaki M. (1999) Immunomodulatory constituents from an ascomycete, Microascus tardifaciens. Chem. Pharm. Bull. 47, 1426–1432 [DOI] [PubMed] [Google Scholar]
- 30. Xu M., Gessner G., Groth I., Lange C., Christner A., Bruhn T., Deng Z., Li X., Heinemann S. H., Grabley S., Bringmann G., Sattler I., Lin W. (2007) Shearinines D-K, new indole triterpenoids from an endophytic Penicillium sp. (strain HKI0459) with blocking activity on large-conductance calcium-activated potassium channels. Tetrahedron 63, 435–444 [Google Scholar]
- 31. Sings H., Singh S. (2003) Tremorgenic and nontremorgenic 2,3-fused indole diterpenoids. Alkaloids Chem. Biol. 60, 51–163 [DOI] [PubMed] [Google Scholar]
- 32. Belofsky G. N., Gloer J. B., Wicklow D. T., Dowd P. F. (1995) Antiinsectan alkaloids: Shearinines A-C and a new paxilline derivative from the ascostromata of Eupenicillium shearii. Tetrahedron 51, 3959–3968 [Google Scholar]
- 33. Woodside A. B., Huang Z., Poulter C. D. (1988) Trisammonium geranyl diphosphate. Org. Synth. 66, 211–215 [Google Scholar]
- 34. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 35. Berger S., Braun S. (2004) 200 and More NMR Experiments. A Practical Course, pp. 1–854, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 36. Steffan N., Li S. M. (2009) Increasing structure diversity of prenylated diketopiperazine derivatives by using a 4-dimethylallyltryptophan synthase. Arch. Microbiol. 191, 461–466 [DOI] [PubMed] [Google Scholar]
- 37. Steffan N., Unsöld I. A., Li S. M. (2007) Chemoenzymatic synthesis of prenylated indole derivatives by using a 4-dimethylallyltryptophan synthase from Aspergillus fumigatus. ChemBioChem 8, 1298–1307 [DOI] [PubMed] [Google Scholar]
- 38. Kremer A., Li S. M. (2008) Potential of a 7-dimethylallyltryptophan synthase as a tool for production of prenylated indole derivatives. Appl. Microbiol. Biotechnol. 79, 951–961 [DOI] [PubMed] [Google Scholar]
- 39. Steffan N., Grundmann A., Yin W. B., Kremer A., Li S. M. (2009) Indole prenyltransferases from fungi. A new enzyme group with high potential for the production of prenylated indole derivatives. Curr. Med. Chem. 16, 218–231 [DOI] [PubMed] [Google Scholar]
- 40. Li S. M. (2009) Applications of dimethylallyltryptophan synthases and other indole prenyltransferases for structural modification of natural products. Appl. Microbiol. Biotechnol. 84, 631–639 [DOI] [PubMed] [Google Scholar]
- 41. Liang P. H. (2009) Reaction kinetics, catalytic mechanisms, conformational changes, and inhibitor design for prenyltransferases. Biochemistry 48, 6562–6570 [DOI] [PubMed] [Google Scholar]
- 42. Turunen M., Olsson J., Dallner G. (2004) Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 1660, 171–199 [DOI] [PubMed] [Google Scholar]
- 43. Sacchettini J. C., Poulter C. D. (1997) Creating isoprenoid diversity. Science 277, 1788–1789 [DOI] [PubMed] [Google Scholar]
- 44. Nielsen K. F., Smedsgaard J. (2003) Fungal metabolite screening. Database of 474 mycotoxins and fungal metabolites for dereplication by standardized liquid chromatography-UV-mass spectrometry methodology. J. Chromatogr. A 1002, 111–136 [DOI] [PubMed] [Google Scholar]
- 45. Edwards D. J., Gerwick W. H. (2004) Lyngbyatoxin biosynthesis. Sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J. Am. Chem. Soc. 126, 11432–11433 [DOI] [PubMed] [Google Scholar]
- 46. Yu X., Xie X., Li S. M. (2011) Substrate promiscuity of secondary metabolite enzymes: prenylation of hydroxynaphthalenes by fungal indole prenyltransferases. Appl. Microbiol. Biotechnol. 92, 737–748 [DOI] [PubMed] [Google Scholar]
- 47. Yu X., Li S. M. (2011) Prenylation of flavonoids by using a dimethylallyltryptophan synthase 7-DMATS from Aspergillus fumigatus. ChemBioChem 12, 2280–2283 [DOI] [PubMed] [Google Scholar]
- 48. Machida M., Asai K., Sano M., Tanaka T., Kumagai T., Terai G., Kusumoto K., Arima T., Akita O., Kashiwagi Y., Abe K., Gomi K., Horiuchi H., Kitamoto K., Kobayashi T., Takeuchi M., Denning D. W., Galagan J. E., Nierman W. C., Yu J., Archer D. B., Bennett J. W., Bhatnagar D., Cleveland T. E., Fedorova N. D., Gotoh O., Horikawa H., Hosoyama A., Ichinomiya M., Igarashi R., Iwashita K., Juvvadi P. R., Kato M., Kato Y., Kin T., Kokubun A., Maeda H., Maeyama N., Maruyama J., Nagasaki H., Nakajima T., Oda K., Okada K., Paulsen I., Sakamoto K., Sawano T., Takahashi M., Takase K., Terabayashi Y., Wortman J. R., Yamada O., Yamagata Y., Anazawa H., Hata Y., Koide Y., Komori T., Koyama Y., Minetoki T., Suharnan S., Tanaka A., Isono K., Kuhara S., Ogasawara N., Kikuchi H. (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438, 1157–1161 [DOI] [PubMed] [Google Scholar]
- 49. Cleveland T. E., Yu J., Fedorova N., Bhatnagar D., Payne G. A., Nierman W. C., Bennett J. W. (2009) Potential of Aspergillus flavus genomics for applications in biotechnology. Trends Biotechnol. 27, 151–157 [DOI] [PubMed] [Google Scholar]
- 50. Ding Y., Williams R. M., Sherman D. H. (2008) Molecular analysis of a 4-dimethylallyltryptophan synthase from Malbranchea aurantiaca. J. Biol. Chem. 283, 16068–16076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Markert A., Steffan N., Ploss K., Hellwig S., Steiner U., Drewke C., Li S. M., Boland W., Leistner E. (2008) Biosynthesis and accumulation of ergoline alkaloids in a mutualistic association between Ipomoea asarifolia (Convolvulaceae) and a clavicipitalean fungus. Plant Physiol. 147, 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steiner U., Leibner S., Schardl C. L., Leuchtmann A., Leistner E. (2011) Periglandula, a new fungal genus within the Clavicipitaceae and its association with Convolvulaceae. Mycologia 103, 1133–1145 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.