Abstract
SUMMARY
We have previously shown that expression of chiZ (Rv2719c), encoding a cell wall hydrolase, is upregulated in response to DNA damaging agents and exposure to cephalexin. Furthermore, increased levels of ChiZ lead to decreased viability, loss of membrane integrity and defects in FtsZ-GFP localization and cell division. We now show that ChiZ N′-terminal 110 amino acid region, containing the cell wall hydrolase activity, is sufficient to modulate FtsZ-GFP localization. Further, we found that FtsZ-GFP rings are stabilized in a chiZ deletion strain indicating that ChiZ activity regulates FtsZ assembly. Over-expression of ftsZ did not reverse the reduction in viability caused by overproduction of ChiZ indicating that ChiZ neither interacts with nor directly influences FtsZ assembly. Bacterial two-hybrid assays revealed that ChiZ interacts with FtsI and FtsQ, two other septasomal proteins, but not with FtsZ. Finally, we show that ChiZ is not required for virulence of Mycobacterium tuberculosis in murine macrophages and mice. Our data suggest that optimal levels and activity of the cell wall hydrolase ChiZ are required for regulated cell division in mycobacteria.
Keywords: Mycobacteria, Cell division, FtsZ, Cell-wall hydrolases, ChiZ
1. Introduction
Infections due to Mycobacterium tuberculosis are a leading cause of morbidity and mortality worldwide. M. tuberculosis is responsible for causing nearly 10 million new infections in the last year alone.1 Furthermore, the already prevalent multidrug resistant and the emerging extremely drug resistant strains of M. tuberculosis add urgency to the efforts to accelerate the discovery of novel drugs and efficient vaccines against this deadly global pathogen. The ability of M. tuberculosis to shift between active and persistent phases of infection is key to its success as an enduring pathogen. Pathways necessary for multiplication and survival of the pathogen during early, active and persistent phases of infection have been the focus of a number of recent studies2–5 and reviews.6,7 In addition, bacterial pathways necessary for proliferation such as cell division and cell wall remodeling and their regulation are considered important for understanding the fundamentals of entry into and exit from the persistent state.8–11
While great strides have been made in the field of cell division in Escherichia coli, Bacillus subtilis and Caulobacter crescentus, mycobacterial cell division studies are still in their infancy. M. tuberculosis FtsZ, a key cell division player, shows slow polymerization and GTP hydrolysis activities in vitro and FtsZ activities appear to be regulated during intracellular growth.12–14 Recent studies identified several proteins, some of which are novel, that interfere with FtsZ activities.15–21 Other studies have identified proteins involved in cell wall synthesis and cell division pathways in M. tuberculosis.22–25 Our previous studies characterized ChiZ, a small membrane protein, as a cell wall hydrolase that is induced upon DNA damage and intra-macrophage growth and interferes with cell division.15 These studies also showed that ChiZ overproduction led to destabilization of FtsZ-rings in vivo, but that purified ChiZ did not interfere with FtsZ assembly in vitro.15 It is undetermined if ChiZ interacts with FtsZ and/or other cell division proteins. It is also unknown whether the loss of ChiZ affects FtsZ assembly and finally, whether ChiZ is needed for virulence of M. tuberculosis. The present study was designed to address the above questions. Our results suggest that ChiZ interacts with other septasomal complex proteins and that optimal levels of ChiZ modulate normal progression of cell division in M. tuberculosis.
2. Materials and methods
2.1. Bacterial strains and culture conditions
E. coli Top 10 strain was used for cloning purposes and propagated in Luria Bertani (LB) broth or LB agar plates. Recombinant E. coli strains were grown in LB broth or LB agar supplemented with appropriate antibiotics as needed with kanamycin (Km) at 50 μg/ml, ampicillin (Amp) at 100 μg/ml and hygromycin (Hyg) at 10 μg/ml. Mycobacterium smegmatis and M. tuberculosis were propagated in Middlebrook 7H10 agar or broth supplemented with oleic acid, albumin, dextrose and sodium chloride (OADC). Transformants were selected on the same media supplemented with Km (15 μg/ml) or Hyg (10 μg/ml). Growth was monitored by measuring A600 of broth-grown cultures. For viability determinations, dilutions of exponential cultures were plated on 7H10 agar plates containing appropriate antibiotics and plates incubated for 3–5 days for M. smegmatis and ~ 3 weeks for M. tuberculosis. Colonies were counted and data plotted using Excel.
2.2. Molecular cloning and construction of recombinant strains
Polymerase chain reaction using deep vent or Phusion polymerases was performed to amplify various genomic DNA fragments. These full length or truncated coding regions were subsequently cloned into appropriate vectors to create the recombinant plasmids shown in Table 1. Restriction cloning sites were always included in the oligonucleotide primers used for PCR amplification. PCR products were digested with the appropriate restriction enzymes and cloned into corresponding sites of the desired vectors (see Table 1). All cloned fragments were confirmed by sequencing. Full length ChiZsmeg (MSMEG_2742) or its various truncations were cloned following PCR amplification of the appropriate regions (Table 1). Recombinant plasmids were used to transform M. smegmatis and M. tuberculosis as described before26 and transformants were selected on 7H10 agar plates containing appropriate antibiotics. Recombinant strains were confirmed by recovery of plasmids by bead-beating followed by restriction analysis or PCR as described.26
Table 1.
Strains and plasmids used in the study.
| Name | Description | Reference |
|---|---|---|
| Strains | ||
| Top10F′ | Escherichia coli strains | Invitrogen |
| H37Rv | Virulent wild type Mycobacterium tuberculosis | Laboratory stock |
| mC2155 | Mycobacterium smegmatis | Laboratory stock |
| Δ chiZsmeg | M. smegmatis chiZ deletion strain | 15 |
| Δ chiZTB | M. tuberculosis chiZ deletion strain | 15 |
| Cloning vectors | ||
| pJAM2 | E. coli-Mycobacterium shuttle vector with amidase promoter, replicating, Kmr | 37 |
| pMG103 | E. coli-Mycobacterium shuttle vector with amidase promoter, integrating, Kmr | 18 |
| pLR56 | E. coli – Mycobacterium shuttle vector, integrating, with tet promoter and tet repressor; Kmr | 18 |
| pMV306 | L5 integration vector, Hygr | MedImmune Inc. |
| pKT25 | E .coli expression vector allowing fusions to C-terminal of the T25 fragment of cyaA, Kmr | 27 |
| pUT18C | E. coli expression vector allowing fusions to C-terminal of the T18 fragment of cyaA, Ampr | 27 |
| pKNT25 | E. coli expression vector allowing fusions to N-terminal of the T25 fragment of cyaA, Kmr | 27 |
| Plasmids* used in this study | ||
| pPP79 | gfp-FtsITB cloned in pMG103, Kmr | 19 |
| pRR13 | Pami::ftsZsmeg-gfp in pJFR19 | 15 |
| pRD3 | Ptet::ftsZTB-gfp in pLR56 | 18 |
| pKNT25ftsZ | ftsZTB cloned as C-terminal fusion into pKNT25 low copy replicating, Kmr | 18 |
| pUT18ftsZ | ftsZTB cloned as C-terminal fusion into pUT18 high copy replicating vector, Ampr | 18 |
| pRD60 | wag31 cloned as N-terminal fusion into pUT18C vector, Ampr | This study |
| pRD64 | wag31 cloned as N-terminal fusion into pKT25 vector, Kmr | This study |
| pRD76 | ftsW cloned in pUT18, Ampr | This study |
| pRD77 | ftsW cloned in pKT25, Kmr | This study |
| pMK18 | ftsITB cloned in pUT18C, Ampr | 18 |
| pMK19 | ftsITB cloned in pKT25, Kmr | 18 |
| pMK20 | ftsQTB cloned in pUT18C, Ampr | 19 |
| pMK21 | ftsQTB cloned in pKT25, Kmr | 19 |
| pLR54 | PdnaA::chiZsmeg Δ LysM | 15 |
| pLR55 | chiZΔN-term encoding ChiZsmeg aa 91-end, cloned under Pami in pJAM2 | This study |
| pJFR78 | Pami::ftsZsmeg cloned in pMV306 | 18 |
| pJFR84 | gfp-ftsQsmeg cloned in pJAM2, Kmr | 18 |
| pACR9 | chiZLψσM encoding ChiZsmeg aa 105 to end, cloned under Pami in pJAM2 | This study |
| pACR24 | chiZN-term encoding ChiZsmeg aa 1–90, cloned under Pami in pJAM2 | This study |
| pSAR58 | PdnaA::chiZsmeg | 15 |
Sequences of primers used for PCR amplification of cloned inserts are available upon request.
2.3. Microscopy
For microscopy, wild type and recombinant M. smegmatis and M. tuberculosis strains were grown to exponential phase, cells pelleted by centrifugation at 3000× g and resuspended in PBS. GFP-FtsI could not be visualized in M. tuberculosis due to bleaching of fluorescent signal from paraformaldehyde fixation.19 Therefore, some fusion protein localization experiments were carried out in the surrogate host, M. smegmatis.19 The cells were visualized by brightfield and fluorescence microscopy using a Nikon Eclipse E600 microscope equipped with a CoolSnap ES CCDcamera (Photometrics) and a high-pressure mercury lamp (Nikon). A Nikon B2A filter set (Ex450–490/Em515) was used for GFP strains. The YFP and mCherry fusion were imaged using a Nikon YFP filter (Ex426–446/Em460–500) and a mCherry filter (Ex560/Em630) set from Chroma, respectively. Images were analyzed using MetaMorph 6.2 software (Universal Imaging Corporation). At least 100 cells from each set were scored for localization patterns and cell length measurements. Images were optimized using Adobe Photoshop CS4.
2.4. Bacterial two hybrid assays
Bacterial Adenylate Cyclase-based Two-Hybrid (BACTH) assay kit from Euromedex was used to examine interactions of ChiZ with various cell division proteins. Genes encoding the tested proteins were fused in-frame to the C-terminus or N-terminus of T25 and T18 fragments of Bordetella pertussis adenylate cyclase. For detection of interactions, recombinant T25 and T18 plasmid pairs were used to transform E. coli cya strain. Transformation mix was plated on LB agar containing Amp, Km, 100 μM IPTG and 40 μg/ml X-gal. Plates were incubated at 28 ° C for 24–48 h. Blue colonies indicated positive interactions and were subsequently grown in broth and assayed for β-galactosidase activity as described.18,27 The enzymatic activity was defined as units per milliliter: 200 × [(OD420 of the culture - OD420 in the control tube)/minutes of incubation] × dilution factor. The specific activity of β-galactosidase is defined as units/mg dry weight bacteria and 1 unit corresponds to 1 nmol of ONPG hydrolyzed per min at 28 ° C. At least 5-fold higher β–galactosidase activity than that measured for BTH101 strain carrying a single gene and an empty vector was considered indicative of an interaction. E. coli BTH101 transformants obtained with pKT25 - GCN4 and pUT18C-GCN4 served as positive controls for complementation.
2.5. Macrophage infection and viability determination
Murine macrophage cell line J774A.1 (ATCC) was propagated in RPMI-1640 media supplemented with 2 mM l-glutamic acid, sodium bicarbonate, 1 mM sodium pyruvate and 10% fetal bovine serum (HyClone) at 37 ° C and 5% carbon dioxide and adherent monolayers were established in 24-well plates. Macrophage monolayers were infected with M. tuberculosis at a moi of 1:2 for 3 h to allow phagocytosis.28 Media was then removed and macrophages washed and incubated with fresh media. Three days post-infection the macrophages were collected, lysed, resuspended in Middlebrook 7H9 media and lysates plated on Middlebrook 7H10 media to determine cfu counts. Viability of macrophages was assessed by staining with Trypan-blue.
2.6. Mice infection experiments
C57BL/6 female mice (Jackson laboratories, Bar Harbor, ME) were aerosol infected in an aerosol exposure chamber (Glascol, Terre Haute, IN). Briefly, a sonically dispersed suspension of M. tuberculosis at 106 cfu ml−1 in saline was nebulized for 30 min and implanted into the lungs at ~ 100 cfu mouse−1.28 Mice were sacrificed at select time points, and lungs and spleens were removed aseptically. Organs were homogenized in PBS with 0.05% Tween 80, and 10-fold dilutions plated on 7H11 agar for cfu counts.
3. Results and discussion
3.1. N-terminal 110 aa of ChiZ are sufficient for interfering with FtsZ assembly
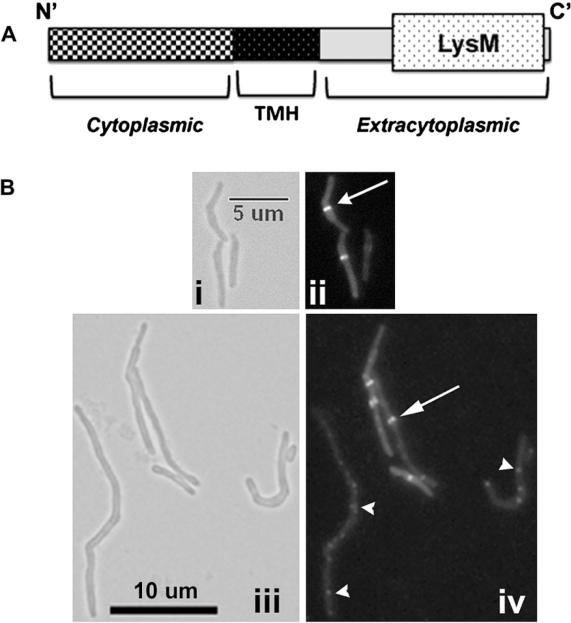
M. tuberculosis ChiZ, encoded by Rv2719c, is a 17.3 kDa membrane protein that contains a unique N′-terminal (N′-term) hydrophilic region, a short transmembrane segment and a C-terminal (C′-term) end containing the peptidoglycan (PG) binding domain, LysM (Figure 1A). Previously we showed that over-expression of ChiZ inhibits FtsZ ring assembly and cell division in M. tuberculosis and M. smegmatis.15 Furthermore, the cell wall hydrolytic activity is confined to the N′-term region and overproduction of ChiZTBΔLysM causes filamentation indicating interference with cell division. To ascertain that the filamentation is due to disruption of FtsZ-ring assembly, we expressed PdnaA‷chiZsmegΔLysM (pLR54, Table 1) in M. smegmatis Pami‷ftsZ-yfp strain and examined the FtsZ assembly in exponential cultures. These cells were filamentous and showed defects in FtsZ ring assembly (Figure 1B, compare panels ii and iv; Table 1), as with the full-length protein.15 Notably, the filamentous cells showed an increase in the percentage of cells with aberrant FtsZ localization. Overexpression of PdnaA‷chiZsmegΔLysM led to a decrease in the number of midcell FtsZ-GFP rings (6% vs WT = 33%) and an increase in the number of aberrantly localized FtsZ-GFP structures (22% vs WT 1.5%). Twenty seven percent of the cells with FtsZ structures contained 2 or more closely spaced FtsZ-rings (Figure 1B). Merodiploids over-producing either C′-term LysM domain (pACR9 in Table 1) or TMH + C′-term regions (pLR55 in Table 1) did not affect cell division and FtsZ assembly (data not shown). Interestingly, overproduction of the N′-term domain (pACR24) alone lacking the TMH region had no effect on cell length (not shown), suggesting that combined activity of N′-term domain + TMH regions is necessary for inhibiting the FtsZ-ring assembly and cell division.
Figure 1.
ChiZ regions and FtsZ-GFP localization. (A) Cartoon showing domains of ChiZTB protein. Amino acids 1–69 comprise the N0-term soluble end, aa 70–87 encompass the transmembrane domain and aa 88–165 comprise the C′-term extracytoplasmic part that contains the PG binding LysM domain (aa 115–163). ChiZsmeg is 185 aa long with N′-term cytoplasmic end = 1–90 aa; TMH = 91–114 aa, and C′ –term extracytoplasmic end = 105 to end (B) N′-term 110 aa of ChiZsmeg are sufficient for interference with FtsZ-ring assembly. M. smegmatis Pami∷ftsZsmeg-yfp cells were transformed with plasmid pLR54 overexpressing a chiZsmegΔLysM (Table 1) from the amidase promoter. The C′-term truncated protein, ChiZΔLysM, was overproduced following induction with 0.2% acetamide for 2 h and induced cells examined by brightfield (i, iii) and fluorescent (ii, iv) microscopy. Panels: i, ii – M. smegmatis Pami∷ftsZsmeg-yfp; iii, iv - M. smegmatis Pami∷ftsZsmeg-yfp with ChiZN110 overproduction. Arrow – midcell FtsZ-YFP rings. Arrowhead – aberrant FtsZ-YFP localization. Size bar as indicated.
3.2. M. tuberculosis ΔchiZ cells have stable FtsZ rings
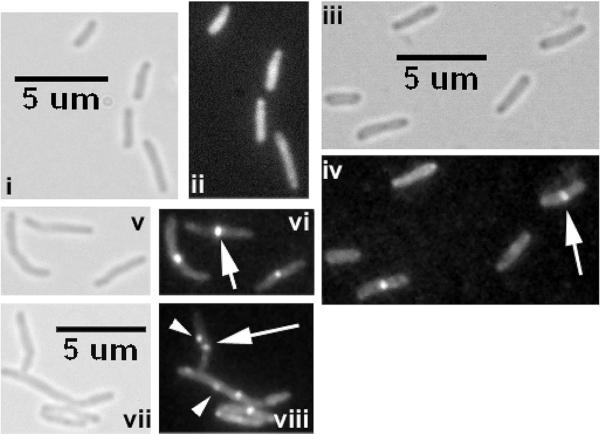
We showed that ChiZ-GFP localizes to sites of nascent PG synthesis, viz. midcell and cell poles.15 Since increased levels of ChiZ led to inhibition of FtsZ assembly, we asked if FtsZ-ring stability is altered in ΔchiZ strain. Therefore, we electroporated ftsZ-yfp (pRR13 in Table 1) and ftsZ-gfp (pRD3, Table 1) expressing plasmids, respectively into M. smegmatis and M. tuberculosis ΔchiZ strains and examined the recombinant strains by fluorescence and brightfield microscopy. Expression of Ptet-ftsZ-gfp in M. tuberculosis ΔchiZ led to an ~ 2-fold increase in number of FtsZ-GFP rings (WT = 11%; ΔchiZ = 21.1%). The FtsZ rings in this strain were more vibrant and tended to bleach slower as compared to the corresponding control strain indicating that the FtsZ-GFP rings are stable in the the absence of ChiZ (Figure 2). Furthermore, addition of the inducer anhydrotetracycline was not required for visualization of FtsZ-GFP rings in the ΔchiZ strain whereas anhydrotetracycline was required for visualization of FtsZ-GFP rings in wild type strain (Figure 2, compare panels ii and iv). Visualization of FtsZ-YFP structures in M. smegmatis and ΔchiZ strain required the addition of 0.2% acetamide for 2 h. Compared to the wild type strain, a modest 24% increase in cell length but no change in the number of FtsZ-YFP rings was noted in the M. smegmatis ΔchiZ strain. We also found aberrant localization of FtsZ-YFP in the M. smegmatis ΔchiZ strain (see arrowheads in Figure 2, panel viii). These data indicated that the stability of FtsZ –eGFP structures was altered in the absence of ChiZ. Presumably, ChiZ, owing to its cell wall hydrolase activity prevents aberrant FtsZ localizations, thereby confines FtsZ-ring to the midcell sites.
Figure 2.
Loss of ChiZ stabilizes FtsZ rings. Wild type and ΔchiZ strains of M. smegmatis Pami∷ftsZsmeg-yfp and M. tuberculosis Ptet∷ftsZTB-gfp were visualized by brightfield (i, iii, v and vii) and fluorescence (ii, iv, vi and viii) microscopy. Vibrant FtsZ-GFP rings were visible without the addition of inducer anhydrotetracycline in the ΔchiZ (iii, iv) but not in the WT M. tuberculosis strain (i, ii). Addition of 0.2% acetamide led to visualization of vibrant FtsZsmeg-YFP rings in M. smegmatis WT (v, vi) strain but produced extraneous localization in the corresponding ΔchiZ (vii, viii) strain. Arrow – midcell localization. Arrowhead - aberrant localization.
3.3. Overproduction of FtsZ does not reverse the cell viability defect associated with the elevated levels of ChiZ
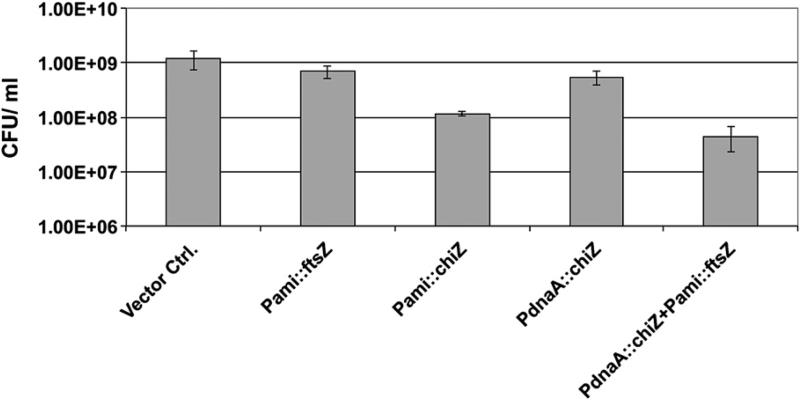
Although overproduction of ChiZ interfers with midcell FtsZ assembly and decreases the viability, purified ChiZ protein did not inhibit in vitro GTP induced FtsZ assembly15 implying that ChiZ does not have a direct effect on FtsZ assembly. To examine if the overproduction of FtsZ reverses the phenotypes associated with ChiZ overproduction, we determined the viability of a M. smegmatis Pami‷ftsZsmeg (pJFR78), PdnaA‷chiZsmeg (pSAR58) strain. Overproduction of ChiZ from the inducible amidase promoter or the constitutive dnaA promoter decreased the viability of M. smegmatis strain (Figure 3). FtsZ overproduction in the WT background caused a small decrease in viability (Figure 3). Increased levels of FtsZ have previously been shown to inhibit cell division and decrease cell viability of M. smegmatis and M. tuberculosis, presumably due to sequestration of cell division proteins and/or formation of non-productive FtsZ structures.26 The combined overproduction of ChiZ and FtsZ additively decreased viability of the recombinant strain (Figure 3). These observations further support our earlier data that ChiZ does not directly affect the FtsZ assembly and imply that the two proteins act at different steps of cytokinesis.
Figure 3.
Reduction of viability caused by ChiZ overproduction is not reversed by increasing the FtsZ levels. Indicated M. smegmatis strains were grown with acetamide for 6 h and plated for colony-forming units as described in materials and methods.
3.4. ChiZ interacts with FtsI and FtsQ
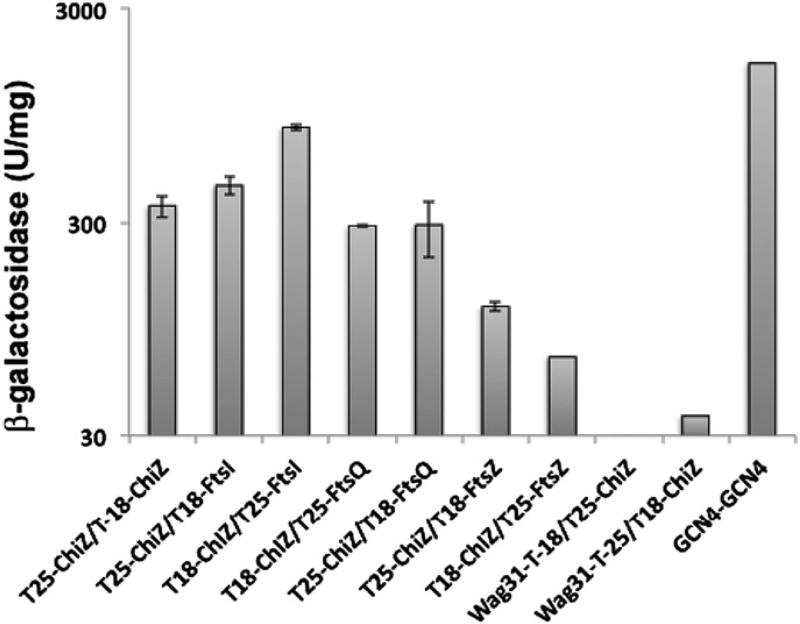
We have previously shown that a GFP-ChiZ fusion protein localizes to midcell sites and poles in M. smegmatis. These sites correspond to sites of nascent PG synthesis in mycobacteria and accordingly stain with Van-FL, a fluoresceinated vancomycin conjugate that binds to the terminal d-ala-d-ala residues in the nascent PG.15,29,30 Given the presence of PG binding LysM domain in ChiZ, its localization to nascent PG synthesis sites is not surprising. However, the localization data tend to suggest that ChiZ interacts with other proteins of PG synthesis and cell division machineries. To identify ChiZ interacting proteins, we used the bacterial two-hybrid assay described by Karimova et al. (2005). This bacterial adenylate cyclase based two-hybrid assay (BACTH) is suitable for membrane proteins and involves cloning of the tested proteins as fusions to the T25 and T18 fragments of the catalytic domain of B. pertussis adenylate cyclase. Interaction between the tested proteins results in functional complementation of adenylate cyclase activity and production of cAMP that triggers the transcriptional activation of lacI and mal operons. Using this system, we examined interactions of ChiZ with several septasomal proteins, viz. FtsZ, FtsI, FtsQ, FtsW and Wag31 (Figure 4). In all cases, the proteins were fused to T18 and T25 domains of adenylate cyclase and as needed membrane topology was taken into consideration such that fusion was always made to the soluble end of the protein. Blue colonies of E. coli BTH101 strain expressing indicated combinations of fusion proteins were tested by broth assay for beta–egalactosidase activity (Figure 4). Our data indicated that ChiZ interacted with FtsI, an essential transpeptidase belonging to the class B penicillin binding protein family, but not with FtsZ and Wag31 (Figure 4). These assays also indicated interactions with FtsQ, another septasomal protein implicated in septal cell wall synthesis (Figure 4). We also found that ChiZ did not interact with FtsW (data not shown). Measurement of beta–egalactosidase activity indicated that the ChiZ/FtsI interaction was stronger as compared to ChiZ/FtsQ interaction. These results were reproducibly obtained with multiple co-transformations of BACTH plasmids. Lack of ChiZ interaction with FtsZ supported the earlier data that ChiZ does not directly affect FtsZ assembly in vitro.15 ChiZ interactions with FtsI and FtsQ suggest a role for ChiZ in septal PG synthesis.
Figure 4.
ChiZ interacts with FtsI and FtsQ. BACTH assays showing ChiZ interactions with other cell division proteins. chiZ, ftsI, ftsQ, ftsZ and wag31 fusions to T18 or T25 fragments of Bordetella pertussis adenylate cyclase were cloned in various BACTH vectors (Table 1) and interactions examined as described.18E. coli BTH101 recombinants bearing indicated combinations of plasmids were plated on indicator plates and incubated at 28 °C. Blue colonies were tested for beta galactosidase activity in broth-grown cultures. Transformants co-expressing gcn4 fusions to T18 and T25 served as positive control.
3.5. ChiZ is a late recruit to the septum
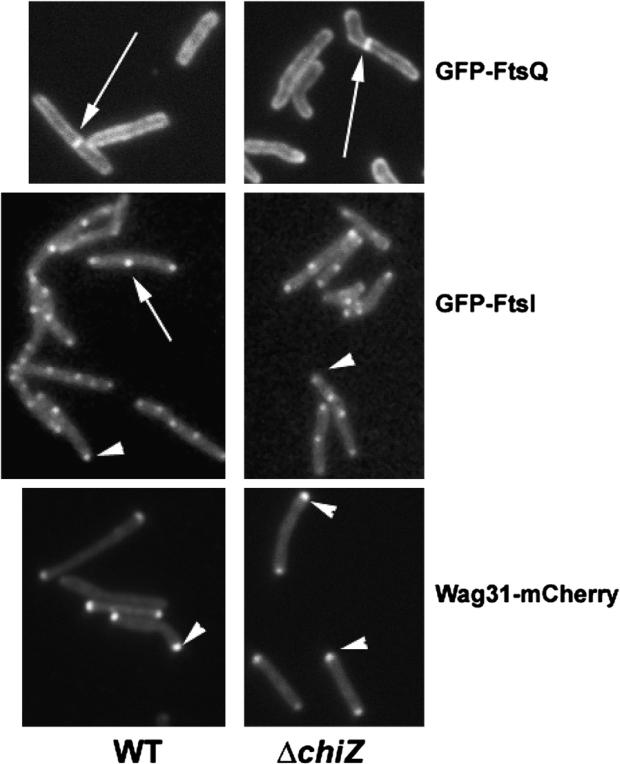
Midcell and polar localization of ChiZ15 combined with its interactions with FtsI and FtsQ proteins prompted an inquiry into the localizations of various septal proteins in the absence of ChiZ. Accordingly, M. smegmatis chiZ deletion strain was transformed with plasmids expressing gfp-ftsQ (pJFR84), gfp-ftsI (pPP79) or wag31-mCherry (pEB16) and the localization of the corresponding fluorescent proteins was examined by fluorescent microscopy. Absence of ChiZ had no significant effect on the localization of GFP-FtsQ, GFP-FtsI and Wag31-mCherry (Figure 5) indicating that ChiZ is not required for localization of FtsI, FtsQ and Wag31 proteins. It is likely that ChiZ is recruited to the septal sites after the assembly of these proteins. Alternatively, its localization could be due to its direct interactions with nascent PG.
Figure 5.
Loss of ChiZ does not affect the localization of FtsQ, FtsI and Wag31. Localization of late septasomal proteins like FtsI, FtsQ and Wag31 was examined in wild type and ΔchiZM. smegmatis strains expressing Pami::gfp-ftsIsmeg, Pami::gfp-ftsQsmeg or Pami::wag31smeg-mCherry. Exponential cultures of the respective recombinant strains were induced with 0.2% acetamide for 3 h and cells examined by fluorescence microscopy. Compare wild type panels with ΔchiZ panels for the respective fusion constructs. Arrow – midcell localization. Arrowhead – polar localization.
3.6. ChiZ is not required for virulence of M. tuberculosis in vivo
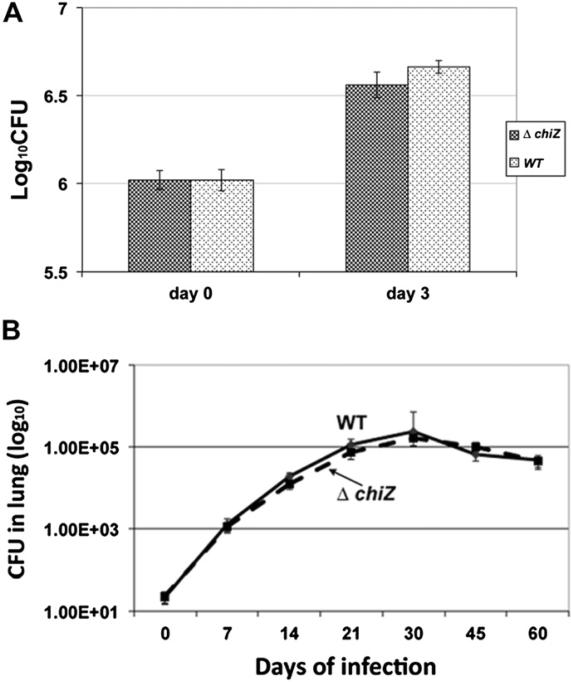
We showed that ChiZ is induced upon DNA damage and during intra-macrophage growth; and modulates FtsZ-ring assembly.15 Since M. tuberculosis is exposed to reactive oxygen and nitrogen species during intracellular growth,31,32 we considered a possibility that ChiZ activity might contribute to optimal M. tuberculosis survival upon infection. To evaluate this possibility, we infected mouse macrophage cell line, J774A.1 with M. tuberculosis ΔchiZ strain as described.28 Macrophages were lysed 3 days post-infection and lysates plated for colony-forming units on Middlebrook-7H11 agar plates. No differences in viabilities were noted between the wild type and mutant strains (Figure 6A). To evaluate the effect of chiZ deletion on growth in vivo, C57BL/6 mice were aerosol infected with about 100 cfu per lung, and growth monitored for 60 days. Bacteria were isolated from lung (Figure 6B) and spleen (not shown) and plated for colony-forming units as described before. The wild type M. tuberculosis and ΔchiZ strains continued to grow and reached a peak level by day 30 and were restricted to the same extent following the induction of adaptive immune response around week 4. These data indicated that loss of chiZ had no effect on the growth of M. tuberculosis in mice lungs and spleen.
Figure 6.
Loss of ChiZ does not influence the growth of M. tuberculosis in murine macrophages and mice. (A) Growth of M. tuberculosis ΔchiZ in murine macrophages. Murine macrophage cell line J774A.1 was infected with actively growing cultures of M. tuberculosis WT or ΔchiZ strain. Macrophages were lysed 3 days post-infection and cfu determined. (B) Growth of M. tuberculosis WT and ΔchiZ strains in mice lungs. C57BL/6 mice were aerosol infected with WT, and ΔchiZ strains at cfu of 100 per mice. Five mice per strain were sacrificed at the time points shown, and cfu counts were determined by plating organ homogenates on 7H11 agar.
4. Conclusions
Septation is thought to require the concerted action of murein biosynthetic and hydrolytic enzymes and it is speculated that septum synthesis complexes contain murein hydrolases and synthases.33,34 We propose that ChiZ partners with PG synthase FtsI to regulate/coordinate cell wall synthesis at septal and polar sites. Interaction of ChiZ with FtsQ, a protein essential for cell division and an interaction partner for multiple cell division proteins, reaffirms a role for ChiZ in septal PG synthesis during normal growth. Our data show that ChiZ protein is not essential for survival under all growth conditions tested, i.e. growth in nutrient broth, ex vivo and in mice lungs (Figure 4, also15). However, chiZ deletion leads to a modest 10% increase in cell length15 indicating that ChiZ activity contributes to cell division. It is known that bacteria contain multiple cell wall hydrolases and deletion of multiple hydrolases in E. coli has minimal effect on cell morphology and cell division.35M. tuberculosis genome appears to contain several cell wall hydrolases.36 Therefore, ChiZ function in M. tuberculosis, like other bacterial cell wall hydrolases, could be redundant. Nonetheless, results shown in this study tend to suggest that ChiZ activities could contribute to cell wall hydrolysis during septum formation and to help focus the FtsZ assembly to the midcell sites. Since chiZ expression is downregulated during hypoxia and ChiZ loss leads to an enhanced cell division defect during intra-macrophage growth,15 we speculate that ChiZ activity is dispensable during persistent growth, but could be important for recovery of growth from the persistent phase. Further studies are needed to resolve the exact role of ChiZ in M. tuberculosis.
Acknowledgements
We thank Drs. Marek Fol and Ashwini Chauhan for help with some experiments.
Funding: This work was supported by NIH grants, RO1AI48417 (MR), RO1AI84734 (MM) and RO1AI49534 (JC).
Footnotes
Competing interests: The authors have no conflicts of interest to declare.
Ethical approval: Not required.
References
- 1.Dye C, Williams BG. The population dynamics and control of tuberculosis. Science. 2010;328:856–61. doi: 10.1126/science.1185449. [DOI] [PubMed] [Google Scholar]
- 2.Rachman H, Strong M, Ulrichs T, Grode L, Schuchhardt J, Mollenkopf H, Kosmiadi GA, Eisenberg D, Kaufmann SH. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immunity. 2006;74:1233–42. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–64. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 4.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiology. 2002;43:717–31. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 6.Rhee KY, Carvalho LP, Bryk R, Ehrt S, Marrero J, Park SW, Schnappinger D, Venugopal A, Nathan C. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol. 2011;19:307–14. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farhana A, Guidry L, Srivastava A, Singh A, Hondalus MK, Steyn AJ. Reductive stress in microbes: implications for understanding Mycobacterium tuberculosis disease and persistence. Adv Microb Physiol. 2010;57:43–117. doi: 10.1016/B978-0-12-381045-8.00002-3. [DOI] [PubMed] [Google Scholar]
- 8.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med. 2010;16:466–9. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hugonnet JE, Tremblay LW, Boshoff HI, Barry CE, 3rd, Blanchard JS. Meropenem-clavulanate is effective against extensively drug-resistant Mycobacterium tuberculosis. Science. 2009;323:1215–8. doi: 10.1126/science.1167498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, Mainardi JL. The peptidoglycan of stationary-phase Mycobacterium tuberculosis predominantly contains cross-links generated by L, D-transpeptidation. J Bacteriol. 2008;190:4360–6. doi: 10.1128/JB.00239-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Payne DJ. Microbiology. Desperately seeking new antibiotics. Science. 2008;321:1644–5. doi: 10.1126/science.1164586. [DOI] [PubMed] [Google Scholar]
- 12.Chauhan A, Madiraju MV, Fol M, Lofton H, Maloney E, Reynolds R, Rajagopalan M. Mycobacterium tuberculosis cells growing in macrophages are filamentous and deficient in FtsZ rings. J Bacteriol. 2006;188:1856–65. doi: 10.1128/JB.188.5.1856-1865.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajagopalan M, Atkinson MA, Lofton H, Chauhan A, Madiraju MV. Mutations in the GTP-binding and synergy loop domains of Mycobacterium tuberculosis ftsZ compromise its function in vitro and in vivo. Biochem Biophys Res Commun. 2005;331:1171–7. doi: 10.1016/j.bbrc.2005.03.239. [DOI] [PubMed] [Google Scholar]
- 14.White EL, Ross LJ, Reynolds RC, Seitz LE, Moore GD, Borhani DW. Slow polymerization of Mycobacterium tuberculosis FtsZ. J Bacteriol. 2000;182:4028–34. doi: 10.1128/jb.182.14.4028-4034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan A, Lofton H, Maloney E, Moore J, Fol M, Madiraju MV, Rajagopalan M. Interference of Mycobacterium tuberculosis cell division by Rv2719c, a cell wall hydrolase. Mol Microbiol. 2006;62:132–47. doi: 10.1111/j.1365-2958.2006.05333.x. [DOI] [PubMed] [Google Scholar]
- 16.Datta P, Dasgupta A, Bhakta S, Basu J. Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J Biol Chem. 2002;277:24983–7. doi: 10.1074/jbc.M203847200. [DOI] [PubMed] [Google Scholar]
- 17.Datta P, Dasgupta A, Singh AK, Mukherjee P, Kundu M, Basu J. Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol Microbiol. 2006;62:1655–73. doi: 10.1111/j.1365-2958.2006.05491.x. [DOI] [PubMed] [Google Scholar]
- 18.Dziedzic R, Kiran M, Plocinski P, Ziolkiewicz M, Brzostek A, Moomey M, Vadrevu IS, Dziadek J, Madiraju M, Rajagopalan M. Mycobacterium tuberculosis ClpX interacts with FtsZ and interferes with FtsZ assembly. PloS One. 2010;5:e11058. doi: 10.1371/journal.pone.0011058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plocinski P, Ziolkiewicz M, Kiran M, Vadrevu SI, Nguyen HB, Hugonnet J, Veckerle C, Arthur M, Dziadek J, Cross TA, Madiraju M, Rajagopalan M. Characterization of CrgA, a New partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J Bacteriology. 2011;193:3246–56. doi: 10.1128/JB.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sureka K, Hossain T, Mukherjee P, Chatterjee P, Datta P, Kundu M, Basu J. Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One. 2010;5:e8590. doi: 10.1371/journal.pone.0008590. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Thakur M, Chakraborti PK. GTPase activity of mycobacterial FtsZ is impaired due to its transphosphorylation by the eukaryotic-type Ser/Thr kinase, PknA. J Biol Chem. 2006;281:40107–13. doi: 10.1074/jbc.M607216200. [DOI] [PubMed] [Google Scholar]
- 22.England K, Crew R, Slayden RA. Mycobacterium tuberculosis septum site determining protein, Ssd encoded by rv3660c, promotes filamentation and elicits an alternative metabolic and dormancy stress response. BMC Microbiol. 2011;11:79. doi: 10.1186/1471-2180-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, Lamichhane G. The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Medicine. 2010;16:466–9. doi: 10.1038/nm.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hett EC, Chao MC, Deng LL, Rubin EJ. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog. 2008;4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol. 2007;66:658–68. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 26.Dziadek J, Madiraju MV, Rutherford SA, Atkinson MA, Rajagopalan M. Physiological consequences associated with overproduction of Mycobacterium tuberculosis FtsZ in mycobacterial hosts. Microbiology. 2002;148:961–71. doi: 10.1099/00221287-148-4-961. [DOI] [PubMed] [Google Scholar]
- 27.Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–43. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fol M, Chauhan A, Nair NK, Maloney E, Moomey M, Jagannath C, Madiraju MV, Rajagopalan M. Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol Microbiol. 2006;60:643–57. doi: 10.1111/j.1365-2958.2006.05137.x. [DOI] [PubMed] [Google Scholar]
- 29.Kang CM, Nyayapathy S, Lee JY, Suh JW, Husson RN. Wag31, a homologue of the cell division protein DivIVA, regulates growth, morphology and polar cell wall synthesis in mycobacteria. Microbiology. 2008;154:725–35. doi: 10.1099/mic.0.2007/014076-0. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen L, Scherr N, Gatfield J, Walburger A, Pieters J, Thompson CJ. Antigen 84, an effector of pleiomorphism in Mycobacterium smegmatis. J Bacteriol. 2007;189:7896–910. doi: 10.1128/JB.00726-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenberger CM, Finlay BB. Macrophages inhibit Salmonella typhimurium replication through MEK/ERK kinase and phagocyte NADPH oxidase activities. J Biological Chemistry. 2002;277:18753–62. doi: 10.1074/jbc.M110649200. [DOI] [PubMed] [Google Scholar]
- 32.Woodmansee DB. Kinetics of the initial rounds of cell division of Toxoplasma gondii. J Parasitol. 2003;89:895–8. doi: 10.1645/GE-112R. [DOI] [PubMed] [Google Scholar]
- 33.Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holtje JV, Heidrich C. Enzymology of elongation and constriction of the murein sacculus of Escherichia coli. Biochimie. 2001;83:103–8. doi: 10.1016/s0300-9084(00)01226-8. [DOI] [PubMed] [Google Scholar]
- 35.Heidrich C, Ursinus A, Berger J, Schwarz H, Holtje JV. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J Bacteriol. 2002;184:6093–9. doi: 10.1128/JB.184.22.6093-6099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 37.Triccas JA, Parish T, Britton WJ, Gicquel B. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol Lett. 1998;167:151–6. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]