Abstract
Early detection (localized stage) of colon cancer is associated with a five-year survival rate of 91%. Only 39% of colon cancers, however, are diagnosed at that early stage. Early and accurate diagnosis, therefore, constitutes a critical need and a decisive factor in the clinical treatment of colon cancer and its success. In this study, using supervised linear discriminant analysis, we have developed three diagnostic biomarker models that—based on global micro-RNA expression analysis of colonic tissue collected during surgery—can discriminate with a perfect accuracy between subjects with colon cancer (stages II–IV) and normal healthy subjects. We developed our three diagnostic biomarker models with 57 subjects [40 with colon cancer (stages II–IV) and 17 normal], and we validated them with 39 unknown (new and different) subjects [28 with colon cancer (stages II–IV) and 11 normal]. For all three diagnostic models, both the overall sensitivity and specificity were 100%. The nine most significant micro-RNAs identified, which comprise the input variables to the three linear discriminant functions, are associated with genes that regulate oncogenesis, and they play a paramount role in the development of colon cancer, as evidenced in the tumor tissue itself. This could have a significant impact in the fight against this disease, in that it may lead to the development of an early serum or blood diagnostic test based on the detection of those nine key micro-RNAs.
Keywords: colon cancer, ROC-supervised linear discriminant analysis, biomarkers, diagnostic biomarker models, global micro-RNA expression analysis, systems biology
Introduction
Colorectal cancer is the third most common type of cancer for both males and females. In 2010, there were an estimated 102,900 new cases of colon and 39,670 cases of rectal cancer, and an estimated 51,370 deaths from colorectal cancer occurred.1,2 The five-year survival rate for those patients who are diagnosed at an early stage (localized stage—stage I or II) is 91%; however, only 39% of colorectal cancer patients are diagnosed at an early stage.1 If the colorectal cancer has spread to adjacent organs or lymph nodes, the five-year survival drops to 70%, and if it has spread to distant organs, the five-year survival is 11%.1 It follows, therefore, that early and accurate diagnostic tests would have a significant impact in the fight against this disease by saving thousands of lives every year. Furthermore, if an early and accurate diagnostic test were based on serum or blood, then it would have additional significant advantages: it would be considerably less invasive and expensive than colonoscopy, the current standard diagnostic procedure for colon cancer (CCA).
In this study, we analyzed the global micro-RNA (miRNA) expression data of colonic tumor and healthy tissue obtained during surgery from 96 subjects [68 with CCA (stages II–IV) and 28 normal]. We developed three different and independent diagnostic biomarker models using 57 subjects [40 with CCA (stages II–IV) and 17 normal], and we validated all three of them with 39 unknown subjects [28 with CCA (stages II–IV) and 11 normal] that were new and different from those 57 subjects used in the development of the models. Our three diagnostic biomarker models were able to identify with a perfect accuracy (overall sensitivity: 100.00% and overall specificity: 100.00%) all 68 subjects with CCA and all 28 normal subjects.
Each of our three diagnostic biomarker models is a linear discriminant function of a number of miRNAs. Altogether, nine miRNAs constitute the input variables to all three diagnostic biomarker models, and they are deemed highly significant in the discrimination between healthy normal tissue and tumor tissue, as well as, therefore, in the development of colon cancer.
Materials and Methods
Data acquisition
We used the normalized miRNA data for 68 subjects with CCA (stages II–IV) (labeled ‘pMMR’) and for 28 normal subjects by Sarver et al3 posted at the GEO (Gene Expression Omnibus) of the NCBI (National Center for Biotechnology Information) [ID: GSE18392].
Discovery and validation studies
Of the total 96 subjects, we randomly selected 57 of them [40 with CCA (stages II–IV) and 17 normal (NRM)] for the development and training of the diagnostic biomarker models. The remaining 39 subjects [28 with CCA (stages II–IV) and 11 NRM] constituted the unknown subjects with which all diagnostic biomarker models were tested. This validation method provided us with the means to test our diagnostic biomarker models with 39 new and real unknowns that were different from the subjects used for—and, therefore, completely extraneous to—the development and training of the models. The proportions of the stages (II–IV) in the total set of 68 CCA subjects were maintained in both the discovery and validation subsets of CCA subjects.
Statistical methods
In order to reduce the dimensionality of the data and zero in on those variables (miRNAs) that are most significant in the process that differentiates between normal healthy tissue and CCA tissue, we applied our bioinformatic methods that we have developed, presented, and explained in a great detail in our previous studies.4–7 Briefly, we performed ROC curve analysis on the entire data matrix, i.e., on all variables (735 miRNAs × 96 subjects) in order to assess the discriminating capability of all variables with respect to our two groups, namely, CCA and NRM. In the final round, we selected only those variables with an AUC ≥ 0.97. Twelve variables (miRNAs) fulfilled this criterion, and they constituted the final pool of the most significant variables. We should point out that our method used in this study constitutes a novel linear discriminant analysis method, i.e., one that is carefully supervised by ROC curve analysis.
Generation of linear discriminant functions
From the aforementioned 12 most significant variables, 9 became the input variables to the three linear discriminant functions (D1, D2, and D3), which we were able to generate in the discovery study {57 subjects [40 with CCA (stages II–IV) and 17 NRM]}. Those three different and independent linear discriminant functions are the final diagnostic biomarker models. The D1 is a function of the following 4 of the 9 aforementioned significant variables (miRNAs):
| (1.1) |
The letter ‘T’ preceding the name of a miRNA indicates that that miRNA variable was transformed in order to meet normality, equality of variance, and/or equality of covariance requirements.
The D2 is a function of the following 3 of the 9 aforementioned significant variables (mi RNAs):
| (1.2) |
The D3 is a function of the following 4 of the 9 aforementioned significant variables (miRNAs):
| (1.3) |
As can be seen from Equations (1.1), (1.2), and (1.3), the three linear discriminant functions D1, D2, and D3 are three different and independent functions. Table S1 (Supplementary Data) shows the exact D1, D2, and D3 functions. Table 1 shows the top 12 most significant miRNAs, including the 9 miRNAs that constitute the input variables to D1, D2, and D3 functions, along with their ROC AUC rank, relative expression, and other properties.
Table 1.
The 12 miRNAs (constituent variables) of the three diagnostic biomarker models (D1, D2, and D3), ranked according to their ROC AUC value.
| Rank | ROC AUC | miRNA symbol | miRNA Signif. Diff. Expr. (CCA) | Known gene interactions | Observed processes | Known drugs/Chemicals/Hormones |
|---|---|---|---|---|---|---|
| 1 | 0.99813 | miR-182* | ↑ | colon cancer | ||
| 2 | 0.99440 | miR-182 | ↑ | RASA1, RAC1, TP53I13, EGR3, BCL10 | endometrial ovarian cancer, endometrioid carcinoma, lung cancer, lung squamous cell carcinoma, metastasis, colon cancer | 5-fluorouracil, vorinostat, trichostatin A, 25-hydroxy-vitamin D3, decitabine |
| 3 | 0.99360 | miR-30a-3p | ↓ | AQP4, CDK6, CYR61, FMR1, SLC7A6, THBS1, TMEM2, TUBA1A, VEZT, WDR82 | head and neck cancer, hypopharyngeal squamous cell carcinoma, uterine cancer, cervical carcinoma | vorinostat, docetaxel, Insulin |
| 4 | 0.99307 | miR-147 | ↓ | BCL7B, TP53RK, VEGFA, PNP, FAM123C, SLC25A34, C17orf55, ONECUT2, PDLIM2, SLC8A3, HPCAL4, AHCYL1, BCOR, MAPRE3, RGS17, DGKG, SLC10A7 | hepatocellular carcinoma, liver cancer | 5-fluorouracil |
| 5 | 0.99200 | miR-183 | ↑ | BCL10, BCL11B, BCLAF1, RBM8A, FOXO1, SRSF2, PDCD4, TRIM2, NUFIP2, GREM2, VAMP7, HECTD2, KY, PPP2R2A, CTDSPL, PLA2G2D, CDH9, THEM4, C16orf54, TUB, TUBA8, TUBGCP4 | liver cancer, endometrial ovarian cancer, endometrioid carcinoma, head and neck cancer, hypopharyngeal squamous cell carcinoma, hepatocellular carcinoma, melanoma metastases, melanoma, lung squamous cell carcinoma, metastasis, lung cancer | vorinostat |
| 6 | 0.99147 | miR-378 | ↓ | RASAL1, METTL4, RASAL1, RBBP9, RBM12, TUSC2, SUFU, SOST, TBX3, NR2C2, FAM60A, SH3GLB1, SDCCAG3, BCL2L2, TRAF5, SHANK3, CSNK1G2, SLC7A6OS, WRB, BEND4 | head and neck cancer, hypopharyngeal squamous cell carcinoma | 4-hydroxynonenal, 25-hydroxy-vitamin D3 |
| 7 | 0.98694 | HS-94 | ↑ | |||
| 8 | 0.98507 | miR-135b | ↑ | RASAL2, RASSF2, TP53TG3/TP53TG3B, BCL11A, BCL11B, BCL2L2, BCL9L, SMAD5, APC, JAK2, ALOX5AP, SUCLG2, ZNF28, OSCP1, PDCD6IP, C1QBP, GULP1, KCTD1, YBX2, OTUD6B, BHLHB9, DEPTOR, NR3C2, RUNX2 | liver cancer, hepatocellular carcinoma, melanoma metastases, melanoma, clear-cell adenocarcinoma, renal cancer, uterine cancer, cervical carcinoma | perchlorate, methimazole, tretinoin |
| 9 | 0.98028 | miR-224 | ↑ | RASA4/RASA4B, RASD1, RASEF, RASGRP1, TP53INP1, METAP2, METTL10, BCL2L11, BEND2, BEND6, API5, ZC3HC1, OSBPL11, NPY1R, ATG2B, BNIP3L, GPR137C, ZNF585A, TSPYL5, USP6NL, PLCD3, METAP2, C3orf64, ZNF140, ANKS1A, PAK4, MMP9 | papillary thyroid cancer, papillary thyroid carcinoma, endometrial cancer, pancreatic cancer, pancreatic ductal adenocarcinoma, lung squamous cell carcinoma, lung cancer | docetaxel, lipopolysaccharide, fluorouracil |
| 10 | 0.97735 | miR-30a-5p | ↓ | RASA1, RASAL2, RASD1, RASGEF1A, RASGRP3, RASL12, RASSF4, TP53, TP53INP1, TP63, BCL10, BCL11A, BCL11B, BCL2, BCL2L11, BCL2L15, BCL6, BCL9, BCLAF1, EED, ACTBL2, ACTC1, ACTN1, NEFL, NEFM, GNAI2, NEUROD1, TMED2, TMED10, CHD1, CBFB, RAD23B, AP2A1, SLC7A11, SLC4A7, MBNL1, TNRC6A, NUFIP2, P4HA2, NT5E, BDNF, RUNX2 | uterine cancer, liver cancer, uterine leiomyoma, papillary thyroid cancer, papillary thyroid carcinoma, head and neck cancer, hypopharyngeal squamous cell carcinoma, prostate cancer, cervical carcinoma, lung cancer, brain cancer, medulloblastoma, colorectal cancer, hepatocellular carcinoma, early-onset breast cancer, breast cancer, hormonedependent breast cancer, breast carcinoma | acetaminophen, 5-fluorouracil, docetaxel, oxaliplatin, 25-hydroxy-vitamin D3, Gulo, Hcg (chorionic gonadotropin complex), trichostatin A, ethanol, androgen, valproic acid |
| 11 | 0.97601 | miR-137 | ↓ | RASGRP3, RASIP1, TP53TG3/TP53TG3B, TP63, BCL11A, BCL11B, BCL2L11, BCL2L13, MAF, MAFK, MED1, MED11, MED14, MED27, MEF2A, MEGF11, MEGF9, METAP1, METTL8, METTL9, ACTBL2, ACTC1, ACTN2, BCL11A, BCL11B, BCL2L11, BCL2L13, TNFAIP1, TNFAIP6, TNFAIP8, TNFSF10, TUBB1, EGR2, RB1, CDK2, CDK6, MET, MITF, CDK6, E2F6, NCOA2, SNAPC1, PLA2G15, STC2, DNAJB12, SSR1, RELL1, SLC6A17, C7orf28B/CCZ1, ASH1L, TMEM229B, C18orf1 | hepatocellular carcinoma, liver cancer | decitabine, trichostatin A, phorbol myristate acetate |
| 12 | 0.97015 | miR-493–5p | ↑ |
Note: Their significant differential expression [over-expression (↑) or under-expression (↓)] as observed in the CCA group relative to the NRM group is shown, along with their symbol, known gene interactions, the processes wherein they have been observed to be involved, and known drug/chemical/hormone interactions.
As was mentioned above, 9 of the 12 most significant miRNA variables were employed to develop the D1, D2, and D3 functions. The remaining 3 miRNA variables were not employed due to high degree of multi collinearity, as well as due to inequality of covariance, with the other miRNA variables. For those same reasons, the D1, D2, and D3 functions with their respective miRNA variables represent the miRNA groups (out of the 12 most significant miRNA variables) that fulfilled all conditions required by discriminant analysis. Table S2 ( Supplementary Data) shows the test results for equality of covariance and variance among the constituent miRNA variables of the D1, D2, and D3 functions. Table S3 ( Supplementary Data) shows the test results for normality for the D1, D2, and D3 functions. We should point out here that, having lowered the criterion of significance (ROC AUC ≥ 0.90), we were able to generate several other discriminant functions, whose constituent miRNA variables were less significant than those employed by the D1, D2, and D3 functions; but, following final assessment, they proved to be not as robust as the D1, D2, and D3, and they are consequently not presented here.
The D1, D2, and D3 functions that we generated are canonical linear discriminant functions; this means that all three of them, by definition, are centered at zero, i.e., the mean D1, D2, and D3 scores of the 57 subjects used in the discovery study are all zero. In order to avoid having to deal with negative scores, especially in the case of the graphs, we centered all three discriminant functions at +20.
Computer programs
Computer programs were written using MATLAB R2011b by The MathWorks, Inc., Natick, MA, USA.
Results
Discovery study
As was mentioned earlier, from the total number of 96 subjects [68 with CCA (stages II–IV) and 28 NRM] used in this study, we randomly selected 57 subjects [40 with CCA (stages II–IV) and 17 NRM] for the development and training of the three diagnostic biomarker models (D1, D2, and D3); and we will henceforward refer to those 57 subjects as the 57 original subjects. After the development of the three diagnostic biomarker models, we assessed their diagnostic accuracy using the aforementioned 57 original subjects, which were employed for their development. This constitutes an important first step in the assessment of a diagnostic test.
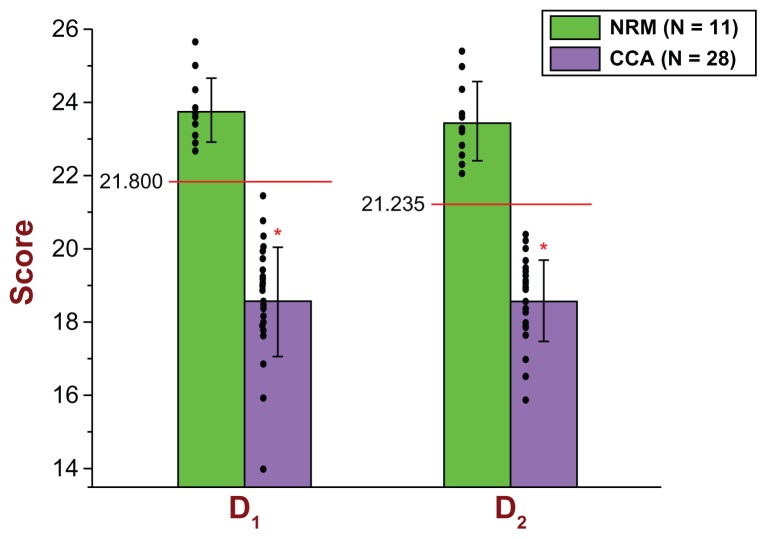
The cut-off score of the D1 diagnostic biomarker model, as well as those of the other two models, was determined by taking into account the results of the following two analyses: (1) calculation of the optimal point on the ROC curve based on the 57 scores of the 57 original subjects used in the discovery study [ optimal point is defined as the point with the highest sensitivity and the lowest false positive rate ( 1-specificity)] and (2) calculation of the 99.99% confidence intervals for the mean D1 scores of the two groups (CCA and NRM) and their respective standard deviations. Based on that, the cut-off score of the D1 model was determined to be 21.800. If a subject has a D1 score less than 21.800, then that subject is classified as a CCA; otherwise (≥21.800), that subject is classified as an NRM. As can be seen from Figure 1, the D1 model correctly identified all (40/40) CCA subjects and all (17/17) NRM subjects. Since our target group is the CCA group, and since our reference group is the NRM group, it follows that, for the discovery study, the D1 model exhibited a sensitivity = 40/40 = 1.000 and a specificity = 17/17 = 1.000. Figure 1 and Table 2A show all pertinent statistical results of the D1 diagnostic biomarker model in connection with the discovery study in great detail.
Figure 1.
Scatter plot and bar graph of all 57 original subjects (40 CCA and 17 NRM) used in the Discovery Study in connection with the D1 and D2 diagnostic biomarker models.
Notes: As can be seen, 40/40 CCA subjects (purple color) had D1 and D2 scores lower than the determined cut-off scores of 21.800 and 21.235, respectively; therefore, 40/40 CCA subjects were identified correctly by both D1 and D2 diagnostic biomarker models [sensitivity = 40/40 = 1.000 for both D1 and D2]. Regarding the NRM group (green color), all 17 subjects had D1 and D2 scores greater than the determined cut-off scores of 21.800 and 21.235, respectively; therefore, 17/17 NRM subjects were identified correctly by both D1 and D2 diagnostic biomarker models [specificity = 17/17 = 1.000 for both D1 and D2]. For the Discovery Study, the mean D1 and D2 scores of the 40 CCA subjects were18.4054 and 18.3266 respectively (top of the D1 and D2 purple bars) and their respective standard deviations (whiskers above or below the top of the D1 and D2 purple bars) were 1.0899 and 1.0703. The mean D1 and D2 scores of the 17 NRM subjects were 23.7523 and 23.9373 respectively (top of the D1 and D2 green bars) and their respective standard deviations (whiskers above or below the top of the D1 and D2 green bars) were 0.7363 and 0.8029. The significance level was set at α = 0.001 (two-tailed), and the probability of significance for the D1 was P = 3.05 × 10−25 (independent t-Test with T-value = 18.4664), whereas the probability of significance for the D2 was P = 3.01 × 10−26 (independent t-Test with T-value = 19.3834). Both the D1 and the D2 are parametrically distributed with respect to both groups.
Table 2.
Statistical results of the three diagnostic biomarker models (D1, D2, and D3) in the Discovery Study (identification of the 57 original subjects) and in the Validation Study (identification of the 39 unknown subjects, which were new and different from the 57 original subjects).
| Diagnostic Test | ROC AUC | T-Value | P | CCA Group | NRM Group |
|---|---|---|---|---|---|
|
|
|
|
|||
| (2-tailed) | [99.99% CI of mean] | [99.99% CI of mean] | |||
|
|
|
|
|||
| α = 0.001 | (SD) | (SD) | |||
| A (Discovery study) | |||||
| D1 | 1.000 | 18.4664 | 3.05 × 10−25 | [17.8457, 18.9607] (1.0899) | [23.2097, 24.3414] (0.7363) |
| D2 | 1.000 | 19.3834 | 3.01 × 10−26 | [17.8040, 18.9029] (1.0703) | [23.3861, 24.5940] (0.8029) |
| D3 | 1.000 | 23.1476 | 4.96 × 10−30 | [17.4960, 18.4864] (0.9684) | [23.7995, 25.4473] (1.0730) |
| CCA Group | NRM Group | ||||
|
|
|
||||
| Mean ± SD | Mean ± SD | ||||
|
| |||||
| B (Validation study) | |||||
| D1 | 1.000 | 10.8991 | 4.17 × 10−13 | 18.5568 ± 1.4817 | 23.7912 ± 0.9013 |
| D2 | 1.000 | 12.4374 | 8.76 × 10−15 | 18.5869 ± 1.1167 | 23.4817 ± 1.0766 |
| D3 | 1.000 | 12.9987 | 2.30 × 10−15 | 18.1475 ± 1.2818 | 24.5298 ± 1.6149 |
Notes: (A) The ROC AUC value, the T value and probability of significance (P) of the independent t-Test, the 99.99% confidence interval for the mean score of the CCA group and that of the NRM group, along with their respective standard deviations, of the D1, D2, and D3 diagnostic biomarker models in the Discovery Study are shown. (B) The ROC AUC value, the T value and probability of significance (P) of the independent t-Test, and the mean score of the CCA group and that of the NRM group, along with their respective standard deviations, of the D1, D2, and D3 diagnostic biomarker models in the Validation Study are shown. As can be seen, all six of those group mean scores, as observed in the validation study with the 39 unknown subjects, fall within the 99.99% confidence intervals of the respective group mean scores as predicted in the discovery study (A).
The cut-off score of the D2 diagnostic biomarker model was determined to be 21.235. If a subject has a D2 score less than 21.235, then that subject is classified as a CCA; otherwise (≥21.235), that subject is classified as an NRM. As can be seen from Figure 1, the D2 model correctly identified all (40/40) CCA subjects and all (17/17) NRM subjects. Therefore, for the discovery study, the D2 model exhibited a sensitivity = 40/40 = 1.000 and a specificity = 17/17 = 1.000. Figure 1 and Table 2A show all pertinent statistical results of the D2 diagnostic biomarker model in connection with the discovery study in great detail.
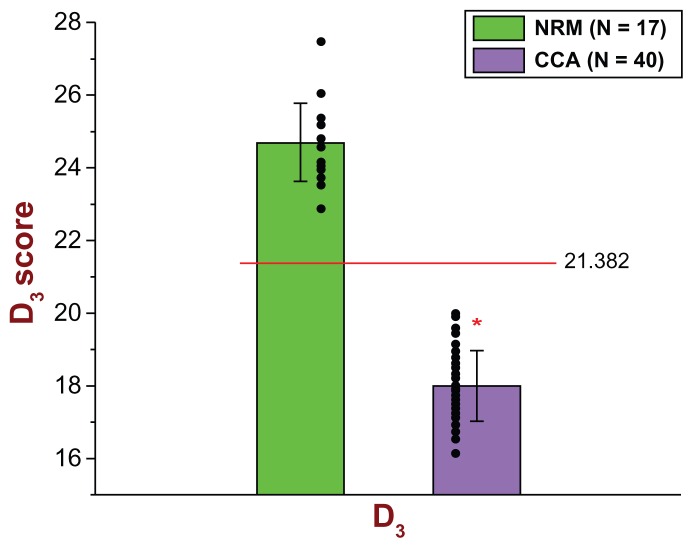
Regarding the D3 diagnostic biomarker model, the cut-off score was determined to be 21.382. If a subject has a D3 score less than 21.382, then that subject is classified as a CCA; otherwise (≥21.382), that subject is classified as an NRM. As can be seen from Figure 2, the D3 model correctly identified all (40/40) CCA subjects and all (17/17) NRM subjects. Therefore, for the discovery study, the D3 model exhibited a sensitivity = 40/40 = 1.000 and a specificity = 17/17 = 1.000. Figure 2 and Table 2A show all pertinent statistical results of the D3 diagnostic biomarker model in connection with the discovery study in great detail.
Figure 2.
Scatter plot and bar graph of all 57 original subjects (40 CCA and 17 NRM) used in the Discovery Study in connection with the D3 diagnostic biomarker model.
Notes: As can be seen, 40/40 CCA subjects (purple color) had D3 scores lower than the determined cut-off score of 21.382; therefore, 40/40 CCA subjects were identified correctly by the D3 diagnostic biomarker model [sensitivity = 40/40 = 1.000]. Regarding the NRM group (green color), all 17 subjects had D3 scores greater than the determined cut-off score of 21.382; therefore, 17/17 NRM subjects were identified correctly by the D3 diagnostic biomarker model [specificity = 17/17 = 1.000]. For the Discovery Study, the mean D3 score of the 40 CCA subjects was 18.0010 (top of the purple bar) and the standard deviation (whiskers above or below the top of the purple bar) was 0.9684. The mean D3 score of the 17 NRM subjects was 24.7016 (top of the green bar) and the standard deviation (whiskers above or below the top of the green bar) was 1.0730. The significance level was set at α = 0.001 (two-tailed), and the probability of significance for the D3 was P = 4.96 × 10−30 (independent t-Test with T-value = 23.1476). The D3 is parametrically distributed with respect to both groups.
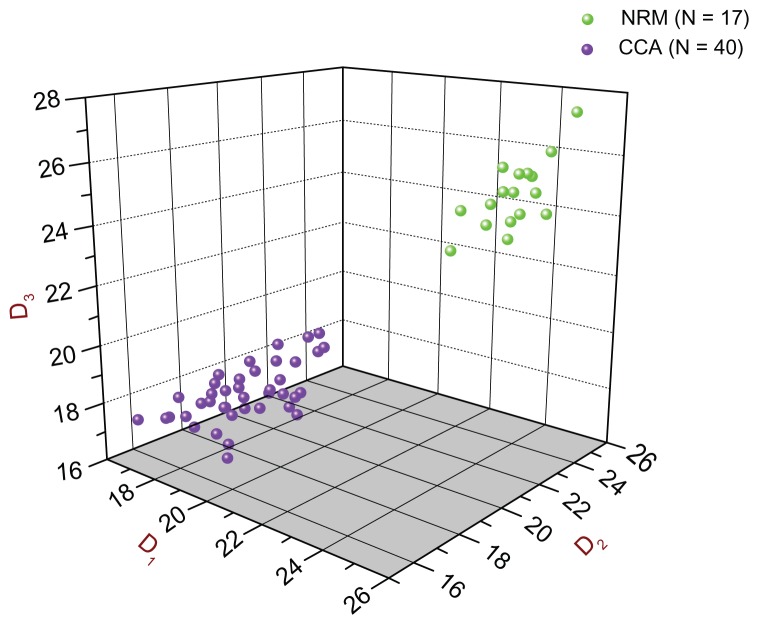
Figure 3 shows the 3D scatter plot of the D1 vs. D2 vs. D3 scores of all 57 original subjects, providing, thus, a visual depiction of the diagnostic accuracy of all three models with respect to the discovery study. As can be seen, the two groups are segregated into two distinct and completely separate clusters: the CCA group (purple spheres) is at the front and lower level, whereas the NRM group (green spheres) is at the back and higher level. It can also be seen that there were no misclassifications by any of the three diagnostic models.
Figure 3.
3D Scatter plot of all 57 original subjects [40 CCA (purple) and 17 NRM (green)] used in the Discovery Study in connection with the D1, D2, and D3 diagnostic biomarker models.
Notes: The D1, D2, and D3 scores of all 57 original subjects are plotted against each other (D1 vs. D2 vs. D3). As can be seen, there are two distinct, separate clusters: the purple one (CCA group) is at the front and at a lower level, whereas the green one (NRM group) is at the back and at a higher level. It can also be seen that there were no misclassifications.
Validation study
As was mentioned earlier, from the total number of 96 subjects [68 with CCA (stages II–IV) and 28 NRM] used in this study, we had randomly segregated 39 subjects [28 with CCA (stages II–IV) and 11 NRM] for the sole and express purpose of testing our three diagnostic biomarker models. Those 39 unknown subjects were completely extraneous to all three models, that is to say they were new and different from the original 57 subjects used for the development of the three models, and they had never before been encountered by any of the three models. This constitutes the most important test in the assessment of a diagnostic test.
As can be seen from Figures 4 and 5 and Table 2B, all three diagnostic biomarker models correctly diagnosed all of the 39 unknown subjects. More specifically, all 28 unknown CCA subjects had D1, D2, and D3 scores that were less than the respective cut-off scores (21.800, 21.235, 21.382); whereas all 11 unknown NRM subjects had D1, D2, and D3 scores that were greater than the respective aforementioned cut-off scores. Therefore, in connection with the validation study, both the sensitivity and the specificity of all three diagnostic biomarker models were 1.000. Figure 6 shows the 3D scatter plot of the D1 vs. D2 vs. D3 scores of all 39 unknown subjects, providing, thus, a visual depiction of the diagnostic accuracy of all three models with respect to the validation study. As can be seen, the 39 unknown subjects are segregated into two distinct and completely separate clusters: the CCA group (purple spheres) is at the front and lower level, whereas the NRM group (green spheres) is at the back and higher level. It can also be seen that there were no misclassifications by any of the three diagnostic models.
Figure 4.
Scatter plot and bar graph of all 39 unknown (new and different) subjects (28 CCA and 11 NRM) used in the Validation Study in connection with the D1 and D2 diagnostic biomarker models.
Notes: As can be seen, 28/28 unknown CCA subjects (purple color) had D1 and D2 scores lower than the determined cut-off scores of 21.800 and 21.235, respectively; therefore, 28/28 unknown CCA subjects were identified correctly by both D1 and D2 diagnostic biomarker models [sensitivity = 28/28 = 1.000 for both D1 and D2]. Regarding the NRM group (green color), all 11 unknown subjects had D1 and D2 scores greater than the determined cut-off scores of 21.800 and 21.235, respectively; therefore, 11/11 unknown NRM subjects were identified correctly by both D1 and D2 diagnostic biomarker models [specificity = 11/11 = 1.000 for both D1 and D2]. For the Validation Study, the mean D1 and D2 scores of the 28 unknown CCA subjects were 18.5568 and 18.5869 respectively (top of the D1 and D2 purple bars) and their respective standard deviations (whiskers above or below the top of the D1 and D2 purple bars) were 1.4817 and 1.1167. The mean D1 and D2 scores of the 11 unknown NRM subjects were 23.7912 and 23.4817 respectively (top of the D1 and D2 green bars) and their respective standard deviations (whiskers above or below the top of the D1 and D2 green bars) were 0.9013 and 1.0766. The significance level was set at α = 0.001 (two-tailed), and the probability of significance for the D1 was P = 4.17 × 10−13 (independent t-Test with T-value = 10.8991), whereas the probability of significance for the D2 was P = 8.76 × 10−15 (independent t-Test with T-value = 12.4374). Both the D1 and the D2 are parametrically distributed with respect to both groups.
Figure 5.
Scatter plot and bar graph of all 39 unknown (new and different) subjects (28 CCA and 11 NRM) used in the Validation Study in connection with the D3 diagnostic biomarker model.
Notes: As can be seen, 28/28 unknown CCA subjects (purple color) had D3 scores lower than the determined cut-off score of 21.382; therefore, 28/28 unknown CCA subjects were identified correctly by the D3 diagnostic biomarker model [ sensitivity = 28/28 = 1.000]. Regarding the NRM group (green color), all 11 unknown subjects had D3 scores greater than the determined cut-off score of 21.382; therefore, 11/11 unknown NRM subjects were identified correctly by the D3 diagnostic biomarker model [ specificity = 11/11 = 1.000]. For the Validation Study, the mean D3 score of the 28 unknown CCA subjects was 18.1475 (top of the purple bar) and the standard deviation (whiskers above or below the top of the purple bar) was 1.2818. The mean D3 score of the 11 unknown NRM subjects was 24.5298 (top of the green bar) and the standard deviation (whiskers above or below the top of the green bar) was 1.6149. The significance level was set at α = 0.001 (two-tailed), and the probability of significance for the D3 was P = 2.30 × 10−15 (independent t-Test with T-value = 12.9987). The D3 is parametrically distributed with respect to both groups.
Figure 6.
3D Scatter plot of all 39 unknown (new and different) subjects [28 CCA (purple) and 11 NRM (green)] used in the Validation Study in connection with the D1, D2, and D3 diagnostic biomarker models.
Notes: The D1, D2, and D3 scores of all 39 unknown subjects are plotted against each other (D1 vs. D2 vs. D3). As can be seen, there are two distinct, separate clusters: the purple one (CCA group) is at the front and at a lower level, whereas the green one (NRM group) is at the back and at a higher level. It can also be seen that there were no misclassifications.
Table 2B, in addition to other pertinent statistical results of our three diagnostic biomarker models, shows the observed mean D1, D2, and D3 scores of the two groups (CCA and NRM) of the 39 unknown subjects. As can be seen, all six of those group mean scores, as observed in the validation study with the 39 unknown subjects, fall within the 99.99% confidence intervals of the respective group mean scores as predicted in the discovery study (Table 2A).
Overall diagnostic biomarker model performance
If we combined the discovery study results with those of the validation study, then the overall performance of our three diagnostic biomarker models would be as follows. All three of them (D1, D2, and D3) exhibited an overall sensitivity = 1.000 (68/68 CCA subjects) and an overall specificity = 1.000 (28/28 NRM subjects).
On the top 12 most significant miRNAs
In connection with the aforementioned 12 most significant miRNAs identified in our study, we conducted an Ingenuity Pathway Analysis (IPA) search. We sought to ascertain information about those 12 miRNAs pertaining to their known interactions with genes; their known interactions with drugs, chemicals, and/or hormones; and their known associations with various types of cancer as derived from the findings of scientific, peer-reviewed studies. The IPA search results are listed in Table 1, along with the direction of the statistically significant differential expression (over-expression or under-expression) of those 12 miRNAs in the CCA group relative to that of the NRM group. As can be seen from Table 1, nearly all of those 12 miRNAs are known to interact with genes, such as RASA1, TP53, CDK6, BCL10, EGR1, and RB1—genes that are involved in the regulation of oncogenesis.
Numerous miRNAs have been observed to be differentially expressed in various types of cancer as compared with the normal healthy state. More specifically, miR-183 and miR-135b have been observed to be over-expressed in colon cancer cells as compared to healthy tissue cells,8,9 and that agrees with our results (Table 1). Also in connection with colon cancer, miR-182* and miR-224 have been observed to be over-expressed, whereas miR-30a- 3p and miR- 137 have been observed to be under-expressed;8 those observations are also in agreement with our findings (Table 1). In connection with colon cancer cell lines, miR-182 and miR-147 have been observed to be over-expressed and under-expressed, respectively;8 and that also accords with the results of our analysis (Table 1). In the cases of hypopharyngeal squamous cell carcinoma and gastric cancer, miR-378 has been observed to be under-expressed,9,10 which is in agreement with our findings. In the cases of prostate cancer and lung cancer, miR-30a-5p has been observed to be under-expressed,11,12 and that is also in agreement with our findings.
The original study by Sarver et al3 was an observational study. Using the criteria of P value and fold change, the authors reported over forty miRNAs that were determined to be differentially expressed between the subjects with colon cancer and the normal subjects. We should point out here that Sarver et al3 did not develop any diagnostic models (tests), much less validate them with unknown subjects that were new and different from the original subjects and report the performance results of such diagnostic models (tests).
Discussion
Having employed 57 subjects [40 with CCA (stages II–IV) and 17 NRM], we were able to generate three different and independent linear discriminant functions, i.e. three different and independent diagnostic tests, that, based on the global miRNA analysis of tissue, can diagnose with perfect accuracy colon cancer. Following validation with 39 unknown (new and different) subjects [28 with CCA (stages II–IV) and 11 NRM], our three diagnostic tests (D1, D2, and D3) exhibited an overall sensitivity = 1.000 (68/68 CCA subjects) and an overall specificity = 1.000 (28/28 NRM subjects). This robust performance should be further tested using a wider pool of subjects in terms of demographics, family history, and syndromic associations.
The clinical significance of our study is as follows. We were able to develop and independently validate three different and independent diagnostic tests that, based on the global miRNA analysis of tumor and healthy tissue, can discriminate with a perfect accuracy between subjects with colon cancer and normal subjects. The nine most significant miRNAs identified, which comprise the input variables to our three diagnostic tests, play, therefore, a key role in the development of colon cancer, as evidenced by the tissue analysis. If an accurate and reliable detection and quantification of those nine key miRNAs were possible in the circulation (plasma or serum), then that would lead to early, accurate, and far less invasive diagnostic tests for colon cancer. Since early detection of colon cancer is associated with 91% survival,1 the results of our study may have a significant impact in the fight against this disease by contributing to the saving of thousands of lives of patients with colon cancer each year.
Detection of miRNAs in the circulation, be it in circulating tumor cells13 or in exosomes,14,15 has been demonstrated by numerous studies over the last several years. Circulating miRNAs have also been detected in connection with various types of cancer, such as breast cancer,15 prostate cancer,16 liver cancer,17 esophageal cancer,18 etc. Therefore, identifying and quantifying accurately and reliably, either in serum or in plasma, the aforementioned nine miRNAs that play a key role in the development of colon cancer constitutes the ultimate goal of this study.
Acknowledgements
We would like to extend our gratitude to Dr. Paul A. Burgio for his comments on clinical and epidemiological matters.
Footnotes
Author Contributions
JBN generated and developed the three linear discriminant functions in this study. JBN conceived, designed, performed the analysis, and executed this project; and he wrote and co-edited the manuscript. WCL participated in the discussions, provided the necessary support and resources for this project, and co-edited the manuscript.
Grant Support
This study was funded by the National Institutes of Health (grant number: T32 DA007097).
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
Supplementary Tables
Table S1.
Canonical linear discriminant functions of D1, D2, and D3 diagnostic biomarker models developed from the original 57 subjects [17 NRM (Group 0) and 40 CCA (Group 1)].
| Discriminant Analysis Report | |||
|---|---|---|---|
| Group | 0 | 1 | Overall |
| Count | 17 | 40 | 57 |
| Variable | Canonical Variate |
|---|---|
| Canonical coefficients (D1) | |
| Constant | −18.363945 |
| miR_182 | −0.146842 |
| miR_30a_5p | 1.612585 |
| miR_183 | −0.609552 |
| TmiR_378 | 0.000264 |
| Canonical coefficients (D2) | |
| Constant | −0.360000 |
| miR_182* | −1.018370 |
| miR_147 | 0.800789 |
| TmiR_30a_3p | 0.0000002 |
| Canonical coefficients (D3) | |
| Constant | −16.476653 |
| miR_182 | −1.216682 |
| miR_137 | 0.566376 |
| TmiR_30a_3p | 0.169121 |
| TmiR_224 | 271.728594 |
Notes: The constituent miRNA variables, their respective coefficients, and the constant of each of the three canonical linear discriminant functions (D1, D2, and D3) are shown. The letter ‘T’ preceding the name of a miRNA indicates that that miRNA variable was transformed in order to meet normality, equality of variance, and/or equality of covariance requirements.
Table S2.
Test results for equality of covariance and variance among the constituent miRNA variables of the D1, D2, and D3 functions developed from the original 57 subjects [17 NRM (Group 0) and 40 CCA (Group 1)].
| Equality of Covariance and Variance Report | |||
|---|---|---|---|
| Group | 0 | 1 | Overall |
| Count | 17 | 40 | 57 |
| Variable | Barlett | F | F | Chi2 | Chi2 | ||
|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||
| Value | DF1 | DF2 | Approx | Prob | Approx | Prob | |
| Bartlett-Box homogeneity tests (D1) | |||||||
| miR_182 | 0.8189 | 1 | 5517 | 0.80 | 0.371063 | 0.80 | 0.371148 |
| miR_30a_5p | 2.3170 | 1 | 5517 | 2.26 | 0.132402 | 2.26 | 0.132496 |
| miR_183 | 0.2023 | 1 | 5517 | 0.20 | 0.656596 | 0.20 | 0.656644 |
| TmiR_378 | 2.1038 | 1 | 5517 | 2.06 | 0.151640 | 2.05 | 0.151736 |
| Box’s M | 8.6511 | 10 | 4538 | 0.78 | 0.651808 | 7.78 | 0.649968 |
| Bartlett-Box homogeneity tests (D2) | |||||||
| miR_182* | 2.2110 | 1 | 5517 | 2.16 | 0.141599 | 2.16 | 0.141693 |
| miR_147 | 0.1519 | 1 | 5517 | 0.15 | 0.700072 | 0.15 | 0.700114 |
| TmiR_30a_3p | 0.0523 | 1 | 5517 | 0.05 | 0.821247 | 0.05 | 0.821272 |
| Box’s M | 6.6653 | 6 | 6101 | 1.03 | 0.406328 | 6.16 | 0.405495 |
| Bartlett-Box homogeneity tests (D3) | |||||||
| miR_182* | 0.8189 | 1 | 5517 | 0.80 | 0.371063 | 0.80 | 0.371148 |
| miR_137 | 0.0281 | 1 | 5517 | 0.03 | 0.868393 | 0.03 | 0.868412 |
| TmiR_30a_3p | 0.0523 | 1 | 5517 | 0.05 | 0.821247 | 0.05 | 0.821272 |
| TmiR_224 | 2.3096 | 1 | 5517 | 2.26 | 0.133022 | 2.26 | 0.133116 |
| Box’s M | 14.9302 | 10 | 4538 | 1.34 | 0.202563 | 13.43 | 0.200457 |
Notes: As can be seen from the probability of significance values of both the F and the χ2 tests for the Box’s M test, there are no statistically significant covariance differences among the constituent miRNA variables of the D1, D2, or D3 function. Likewise, the Bartlett test shows that there are no statistically significant variance differences among the constituent miRNA variables of the D1, D2, or D3 function.
Table S3.
Normality test results for the D1, D2, and D3 linear discriminant functions with respect to both groups of the original 57 subjects [17 NRM (Group 0) and 40 CCA (Group 1)] used for the development of the three functions.
| Normality Tests Report | |||||
|---|---|---|---|---|---|
|
| |||||
| Test name | Test value | Prob level | 10% Critical value | 5% Critical value | Decision (5%) |
| Normality test section of D1 when Group = 0 (Count 17) | |||||
| Shapiro-Wilk W | 0.9679477 | 0.7815679 | Can’t reject normality | ||
| Anderson-Darling | 0.3600979 | 0.4483844 | Can’t reject normality | ||
| Martinez-lglewicz | 1.135777 | 1.252524 | 1.438767 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.1029178 | 0.19 | 0.207 | Can’t reject normality | |
| D’Agostino Skewness | −0.6319371 | 0.527428 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | 0.9578 | 0.338181 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 1.3167 | 0.517716 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D1 when Group = 1 (Count 40) | |||||
| Shapiro-Wilk W | 0.9523966 | 9.170641E-02 | Can’t reject normality | ||
| Anderson-Darling | 0.4800356 | 0.233547 | Can’t reject normality | ||
| Martinez-lglewicz | 0.9609824 | 1.114676 | 1.175041 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.0905983 | 0.126 | 0.139 | Can’t reject normality | |
| D’Agostino Skewness | −0.3980126 | 0.6906209 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | −2.4009 | 0.016356 | 1.645 | 1.960 | Reject normality |
| D’Agostino Omnibus | 5.9226 | 0.051752 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D2 when Group = 0 (Count 17) | |||||
| Shapiro-Wilk W | 0.9018213 | 7.286435E-02 | Can’t reject normality | ||
| Anderson-Darling | 0.6532255 | 8.824592E-02 | Can’t reject normality | ||
| Martinez-lglewicz | 1.067013 | 1.252524 | 1.438767 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.1256069 | 0.19 | 0.207 | Can’t reject normality | |
| D’Agostino Skewness | −1.408385 | 0.159017 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | −0.4372 | 0.661989 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 2.1747 | 0.337114 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D2 when Group = 1 (Count 40) | |||||
| Shapiro-Wilk W | 0.9654804 | 0.2565536 | Can’t reject normality | ||
| Anderson-Darling | 0.4056016 | 0.3517282 | Can’t reject normality | ||
| Martinez-lglewicz | 1.038377 | 1.114676 | 1.175041 | Can’t reject normality | |
| Kolmogorov-Smirnov | 7.907125E-02 | 0.126 | 0.139 | Can’t reject normality | |
| D’Agostino Skewness | −1.585528 | 0.1128464 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | 0.4021 | 0.687630 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 2.6756 | 0.262427 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D3 when Group = 0 (Count 17) | |||||
| Shapiro-Wilk W | 0.9496766 | 0.4514251 | Can’t reject normality | ||
| Anderson-Darling | 0.3490809 | 0.4751235 | Can’t reject normality | ||
| Martinez-lglewicz | 1.136325 | 1.252524 | 1.438767 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.1442362 | 0.19 | 0.207 | Can’t reject normality | |
| D’Agostino Skewness | 1.580456 | 0.1140025 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | 1.5018 | 0.133142 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 4.7533 | 0.092860 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D3 when Group = 1 (Count 40) | |||||
| Shapiro-Wilk W | 0.9784388 | 0.6317195 | Can’t reject normality | ||
| Anderson-Darling | 0.2572377 | 0.7206884 | Can’t reject normality | ||
| Martinez-lglewicz | 0.9622557 | 1.114676 | 1.175041 | Can’t reject normality | |
| Kolmogorov-Smirnov | 7.959955E-02 | 0.126 | 0.136 | Can’t reject normality | |
| D’Agostino Skewness | 0.802801 | 0.4220898 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | −0.6426 | 0.520487 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 1.0574 | 0.589366 | 4.605 | 5.991 | Can’t reject normality |
Note: As can be seen, D1, D2, and D3 are normally distributed with respect to both groups.
References
- 1.American Cancer Society. Cancer Facts & Figures 2010. Atlanta: American Cancer Society; 2010. [Google Scholar]
- 2.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KAT, et al. Human colon cancer profiles show differential micro RNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer. 2009;9:401. doi: 10.1186/1471-2407-9-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nikas JB, Keene CD, Low WC. Comparison of analytical mathematical approaches for identifying key nuclear magnetic resonance spectroscopy biomarkers in the diagnosis and assessment of clinical change of diseases. Journal of Comparative Neurology. 2010;518:4091–112. doi: 10.1002/cne.22365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikas JB, Low WC. ROC-supervised principal component analysis in connection with the diagnosis of diseases. American Journal of Translational Research. 2011;3(2):180–96. [PMC free article] [PubMed] [Google Scholar]
- 6.Nikas JB, Low WC. Application of clustering analyses to the diagnosis of Huntington disease in mice and other diseases with well-defined group boundaries. Computer Methods and Programs in Biomedicine. 2011;104(3):e133–e147. doi: 10.1016/j.cmpb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nikas JB, Boylan KLM, Skubitz APN, Low WC. Mathematical Prognostic Biomarker Models for Treatment Response and Survival in Epithelial Ovarian Cancer. Cancer Informatics. 2011;10:233–47. doi: 10.4137/CIN.S8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bandres E, Cubedo E, Agirre X, Malumbres R, Zarate R, Ramirez N, et al. Identification by Real-time PCR of 13 mature micro RNAs differentially expressed in colorectal cancer and non-tumoraltissues. Molecular Cancer. 2006;5:29. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paranjape T, Slack FJ, Weidhaas JB. MicroRNAs: tools for cancer diagnostics. Gut. 2009;58:1546–54. doi: 10.1136/gut.2009.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, et al. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypo-pharyngeal squamous cell carcinoma (HSCC) British Journal of Cancer. 2010;103:877–84. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–93. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 12.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci. 2009;106(29):12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mostert B, Sieuwerts AM, Martens JW, Sleijfer S. Diagnostic applications of cell-free and circulating tumor cell-associated miRNAs in cancer patients. Expert Rev Mol Diagn. 2011;11(3):259–75. doi: 10.1586/erm.11.11. [DOI] [PubMed] [Google Scholar]
- 14.Rani S, O’Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, et al. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods Mol Biol. 2011;784:181–95. doi: 10.1007/978-1-61779-289-2_13. [DOI] [PubMed] [Google Scholar]
- 15.Rupp AK, Rupp C, Keller S, Brase JC, Ehehalt R, Fogel M, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol. 2011;122(2):437–46. doi: 10.1016/j.ygyno.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 16.Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Müller SC, Ellinger J. Circulating micro RNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011;77(5):1265.e9–16. doi: 10.1016/j.urology.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Qu KZ, Zhang K, Li H, Afdhal NH, Albitar M. Circulating microRNAs as biomarkers for hepatocellular carcinoma. J Clin Gastroenterol. 2011;45(4):355–60. doi: 10.1097/MCG.0b013e3181f18ac2. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105(1):104–1. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Canonical linear discriminant functions of D1, D2, and D3 diagnostic biomarker models developed from the original 57 subjects [17 NRM (Group 0) and 40 CCA (Group 1)].
| Discriminant Analysis Report | |||
|---|---|---|---|
| Group | 0 | 1 | Overall |
| Count | 17 | 40 | 57 |
| Variable | Canonical Variate |
|---|---|
| Canonical coefficients (D1) | |
| Constant | −18.363945 |
| miR_182 | −0.146842 |
| miR_30a_5p | 1.612585 |
| miR_183 | −0.609552 |
| TmiR_378 | 0.000264 |
| Canonical coefficients (D2) | |
| Constant | −0.360000 |
| miR_182* | −1.018370 |
| miR_147 | 0.800789 |
| TmiR_30a_3p | 0.0000002 |
| Canonical coefficients (D3) | |
| Constant | −16.476653 |
| miR_182 | −1.216682 |
| miR_137 | 0.566376 |
| TmiR_30a_3p | 0.169121 |
| TmiR_224 | 271.728594 |
Notes: The constituent miRNA variables, their respective coefficients, and the constant of each of the three canonical linear discriminant functions (D1, D2, and D3) are shown. The letter ‘T’ preceding the name of a miRNA indicates that that miRNA variable was transformed in order to meet normality, equality of variance, and/or equality of covariance requirements.
Table S2.
Test results for equality of covariance and variance among the constituent miRNA variables of the D1, D2, and D3 functions developed from the original 57 subjects [17 NRM (Group 0) and 40 CCA (Group 1)].
| Equality of Covariance and Variance Report | |||
|---|---|---|---|
| Group | 0 | 1 | Overall |
| Count | 17 | 40 | 57 |
| Variable | Barlett | F | F | Chi2 | Chi2 | ||
|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||
| Value | DF1 | DF2 | Approx | Prob | Approx | Prob | |
| Bartlett-Box homogeneity tests (D1) | |||||||
| miR_182 | 0.8189 | 1 | 5517 | 0.80 | 0.371063 | 0.80 | 0.371148 |
| miR_30a_5p | 2.3170 | 1 | 5517 | 2.26 | 0.132402 | 2.26 | 0.132496 |
| miR_183 | 0.2023 | 1 | 5517 | 0.20 | 0.656596 | 0.20 | 0.656644 |
| TmiR_378 | 2.1038 | 1 | 5517 | 2.06 | 0.151640 | 2.05 | 0.151736 |
| Box’s M | 8.6511 | 10 | 4538 | 0.78 | 0.651808 | 7.78 | 0.649968 |
| Bartlett-Box homogeneity tests (D2) | |||||||
| miR_182* | 2.2110 | 1 | 5517 | 2.16 | 0.141599 | 2.16 | 0.141693 |
| miR_147 | 0.1519 | 1 | 5517 | 0.15 | 0.700072 | 0.15 | 0.700114 |
| TmiR_30a_3p | 0.0523 | 1 | 5517 | 0.05 | 0.821247 | 0.05 | 0.821272 |
| Box’s M | 6.6653 | 6 | 6101 | 1.03 | 0.406328 | 6.16 | 0.405495 |
| Bartlett-Box homogeneity tests (D3) | |||||||
| miR_182* | 0.8189 | 1 | 5517 | 0.80 | 0.371063 | 0.80 | 0.371148 |
| miR_137 | 0.0281 | 1 | 5517 | 0.03 | 0.868393 | 0.03 | 0.868412 |
| TmiR_30a_3p | 0.0523 | 1 | 5517 | 0.05 | 0.821247 | 0.05 | 0.821272 |
| TmiR_224 | 2.3096 | 1 | 5517 | 2.26 | 0.133022 | 2.26 | 0.133116 |
| Box’s M | 14.9302 | 10 | 4538 | 1.34 | 0.202563 | 13.43 | 0.200457 |
Notes: As can be seen from the probability of significance values of both the F and the χ2 tests for the Box’s M test, there are no statistically significant covariance differences among the constituent miRNA variables of the D1, D2, or D3 function. Likewise, the Bartlett test shows that there are no statistically significant variance differences among the constituent miRNA variables of the D1, D2, or D3 function.
Table S3.
Normality test results for the D1, D2, and D3 linear discriminant functions with respect to both groups of the original 57 subjects [17 NRM (Group 0) and 40 CCA (Group 1)] used for the development of the three functions.
| Normality Tests Report | |||||
|---|---|---|---|---|---|
|
| |||||
| Test name | Test value | Prob level | 10% Critical value | 5% Critical value | Decision (5%) |
| Normality test section of D1 when Group = 0 (Count 17) | |||||
| Shapiro-Wilk W | 0.9679477 | 0.7815679 | Can’t reject normality | ||
| Anderson-Darling | 0.3600979 | 0.4483844 | Can’t reject normality | ||
| Martinez-lglewicz | 1.135777 | 1.252524 | 1.438767 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.1029178 | 0.19 | 0.207 | Can’t reject normality | |
| D’Agostino Skewness | −0.6319371 | 0.527428 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | 0.9578 | 0.338181 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 1.3167 | 0.517716 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D1 when Group = 1 (Count 40) | |||||
| Shapiro-Wilk W | 0.9523966 | 9.170641E-02 | Can’t reject normality | ||
| Anderson-Darling | 0.4800356 | 0.233547 | Can’t reject normality | ||
| Martinez-lglewicz | 0.9609824 | 1.114676 | 1.175041 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.0905983 | 0.126 | 0.139 | Can’t reject normality | |
| D’Agostino Skewness | −0.3980126 | 0.6906209 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | −2.4009 | 0.016356 | 1.645 | 1.960 | Reject normality |
| D’Agostino Omnibus | 5.9226 | 0.051752 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D2 when Group = 0 (Count 17) | |||||
| Shapiro-Wilk W | 0.9018213 | 7.286435E-02 | Can’t reject normality | ||
| Anderson-Darling | 0.6532255 | 8.824592E-02 | Can’t reject normality | ||
| Martinez-lglewicz | 1.067013 | 1.252524 | 1.438767 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.1256069 | 0.19 | 0.207 | Can’t reject normality | |
| D’Agostino Skewness | −1.408385 | 0.159017 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | −0.4372 | 0.661989 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 2.1747 | 0.337114 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D2 when Group = 1 (Count 40) | |||||
| Shapiro-Wilk W | 0.9654804 | 0.2565536 | Can’t reject normality | ||
| Anderson-Darling | 0.4056016 | 0.3517282 | Can’t reject normality | ||
| Martinez-lglewicz | 1.038377 | 1.114676 | 1.175041 | Can’t reject normality | |
| Kolmogorov-Smirnov | 7.907125E-02 | 0.126 | 0.139 | Can’t reject normality | |
| D’Agostino Skewness | −1.585528 | 0.1128464 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | 0.4021 | 0.687630 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 2.6756 | 0.262427 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D3 when Group = 0 (Count 17) | |||||
| Shapiro-Wilk W | 0.9496766 | 0.4514251 | Can’t reject normality | ||
| Anderson-Darling | 0.3490809 | 0.4751235 | Can’t reject normality | ||
| Martinez-lglewicz | 1.136325 | 1.252524 | 1.438767 | Can’t reject normality | |
| Kolmogorov-Smirnov | 0.1442362 | 0.19 | 0.207 | Can’t reject normality | |
| D’Agostino Skewness | 1.580456 | 0.1140025 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | 1.5018 | 0.133142 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 4.7533 | 0.092860 | 4.605 | 5.991 | Can’t reject normality |
| Normality test section of D3 when Group = 1 (Count 40) | |||||
| Shapiro-Wilk W | 0.9784388 | 0.6317195 | Can’t reject normality | ||
| Anderson-Darling | 0.2572377 | 0.7206884 | Can’t reject normality | ||
| Martinez-lglewicz | 0.9622557 | 1.114676 | 1.175041 | Can’t reject normality | |
| Kolmogorov-Smirnov | 7.959955E-02 | 0.126 | 0.136 | Can’t reject normality | |
| D’Agostino Skewness | 0.802801 | 0.4220898 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Kurtosis | −0.6426 | 0.520487 | 1.645 | 1.960 | Can’t reject normality |
| D’Agostino Omnibus | 1.0574 | 0.589366 | 4.605 | 5.991 | Can’t reject normality |
Note: As can be seen, D1, D2, and D3 are normally distributed with respect to both groups.