Abstract
Background. Enteroaggregative Escherichia coli (EAEC) is a cause of epidemic and sporadic diarrhea, yet its role as an enteric pathogen is not fully understood.
Methods. We characterized 121 EAEC strains isolated in 2008 as part of a case-control study of moderate to severe acute diarrhea among children 0–59 months of age in Bamako, Mali. We applied multiplex polymerase chain reaction and comparative genome hybridization to identify potential virulence factors among the EAEC strains, coupled with classification and regression tree modeling to reveal combinations of factors most strongly associated with illness.
Results. The gene encoding the autotransporter protease SepA, originally described in Shigella species, was most strongly associated with diarrhea among the EAEC strains tested (odds ratio, 5.6 [95% confidence interval, 1.92–16.17]; P = .0006). In addition, we identified 3 gene combinations correlated with diarrhea: (1) a clonal group positive for sepA and a putative hemolysin; (2) a group harboring the EAST-1 enterotoxin and the flagellar type H33 but no other previously identified EAEC virulence factor; and (3) a group carrying several of the typical EAEC virulence genes.
Conclusion. Our data suggest that only a subset of EAEC strains are pathogenic in Mali and suggest that sepA may serve as a valuable marker for the most virulent isolates.
It is estimated that diarrhea causes at least 1.5 million deaths annually, mostly in children <5 years of age [1]. Although in aggregate the diarrheagenic Escherichia coli (DEC) pathotypes comprise the most common bacterial pathogens worldwide [2], each DEC pathotype is clinically, epidemiologically, and pathogenetically distinct. For some pathotypes, the key virulence factors are known, at least in part, whereas for other pathotypes, the key virulence genes and how they coordinately function in the setting of enteric disease remain elusive.
The enteroaggregative E. coli (EAEC) pathotype has been implicated in travelers’ diarrhea [3], in endemic diarrhea among children in both industrialized [4] and resource-poor countries [5], and in persistent diarrhea among individuals infected with human immunodeficiency virus. A recent outbreak of Shiga toxin–producing EAEC highlights its pathogenic potential [6]. Despite this, the molecular epidemiology of EAEC infection remains unclear, largely due to imperfect recognition of the true pathogenic factors within the broadly defined pathotype.
Most EAEC strains colonize the intestinal mucosa via the aggregative adherence fimbriae (AAFs), which include at least 4 major antigenic variants [7–10]. AAFs are transcriptionally regulated by an AraC/XylS family activator called AggR [7, 11]. AggR is also required for expression of genes encoding dispersin (the aap gene), the Aat dispersin translocator [12], and the chromosomal cluster termed Aai, encoding a type VI secretion system [13]. Factors not under AggR control include the Air adhesin, a regulator termed EilA, the EAEC heat-stable toxin EAST-1 (encoded by the astA gene), and a set of toxins termed the serine protease autotransporters of Enterobacteriaceae (SPATEs).
SPATEs have been organized phylogenetically into 2 classes. Members of class 1 are cytotoxic to epithelial cells [14]; class 1 SPATEs found in EAEC strains include the plasmid-encoded toxin (Pet) and its 2 homologs, Sat [15] and SigA [16]. The class 2, or noncytotoxic, SPATEs include Pic, a mucinase that promotes intestinal colonization [17, 18]. As with cytotoxic SPATEs and Pic, we have recently reported that the class 2 SPATE SepA is found commonly among EAEC strains [19]. SepA is a cryptic protease originally described in Shigella species, and is reported to contribute to intestinal inflammation [20]. Importantly, none of these factors are found in all EAEC isolates, and no single factor has ever been consistently implicated in EAEC virulence.
Here, we characterize 121 EAEC strains isolated as part of a case-control study of acute moderate to severe diarrhea among children aged 0–59 months in Mali. We report that the sepA gene and flagellar type H33 are strongly associated with illness, and we define additional sets of virulence genes and factors that are important in this population.
MATERIALS AND METHODS
Study Design
The strains utilized were isolated in the course of a prospective multicenter case-control study (Global Enteric Multi-Center Study, GEMS) of moderate to severe diarrhea among children <5 years of age. Full details of the GEMS design will be published elsewhere. In brief, children ≤59 months presenting to health centers for care with a complaint of diarrhea within the previous 7 days were considered eligible. Cases were enrolled upon parental consent if they met criteria for moderate to severe diarrhea comprising signs of moderate to severe dehydration (sunken eyes, decreased skin turgor), dysentery (blood in stool), or if they were deemed to require hospitalization or intravenous rehydration. Diarrhea was defined as the passage of ≥3 or more unformed stools within a 24-hour period. A stool sample was obtained at enrollment and analyzed comprehensively for bacterial, viral, and protozoal agents. An age-matched asymptomatic control from the same neighborhood was enrolled for each case; a stool sample was obtained from the control child and analyzed similarly.
Specimen Processing and Microbiological Analysis
A single, fresh, whole stool specimen was collected from cases and controls at enrollment for the recovery of potential enteropathogens. Various specific growth media were used for detecting the bacterial pathogens. Up to 3 colonies with the appearance of E. coli on MacConkey agar were selected from each sample and tested using multiplex polymerase chain reaction (PCR) for enterotoxigenic E coli (ETEC) (heat-labile [LT] and heat-stable [ST] enterotoxins), enteropathogenic E. coli (EPEC) (eae and bfpA), and EAEC (aaiC and aatA). Any colonies that were positive for either aaiC (chromosomally encoded) or aatA (encoded on the pAA plasmid) were considered EAEC for the purposes of this analysis.
Serotyping
Somatic (O) and flagella (H) antigens were identified as described elsewhere [21]; the following designations were included: “O rough,” the boiled culture auto-agglutinated, suggesting absence of O antigen; “O?,” it could not be determined whether the strain produces an O antigen (precipitation with Cetavlon indicates an acidic polysaccharide that could represent capsular K antigen); and “O+,” the O antigen is present but could not be typed. Serotyping was performed at the International Escherichia and Klebsiella Centre (World Health Organization), Department of Microbiological Surveillance and Research, Statens Serum Institut, Copenhagen, Denmark.
Polymerase Chain Reaction
Primers and conditions for detecting sequences encoding 21 putative virulence genes, which are described in Table 1, were used in 4 multiplex reactions. Multiplex PCR 1 was performed as previously described [19], with the addition of primers targeting astA. Multiplexes 2–4 were performed using PCR mastermix (2X) according to the manufacturer’s instructions (Fermentas International), with the addition of 1 μL 50 mM magnesium chloride per 50 μL reaction. A DNA template was prepared by boiling a suspension of 10 isolated colonies in 200 μL distilled water. PCR reaction cycles were as follows: (1) 2 minutes denaturation at 95°C, (2) 50 seconds denaturation at 94°C, (3) annealing for 1.5 minutes, and (4) extension for 1.5 minutes at 72°C with 35 cycles returning to step 2. The final extension was 10 minutes at 72°C. Products were amplified using an Eppendorf Mastercycler Gradient thermal cycler (Eppendorf North America) and separated in 2% agarose gels.
Table 1.
Primers Used for the 4 Multiplex Polymerase Chain Reactions (PCRs) and 3 Monoplex PCRs, Description of Target Gene, Product Size in Base Pairs, Annealing Temperature, and Concentration of the Primers
| Multiplex PCR | Gene/Target | Description of Target | Primer Sequence (5′- 3′) | PCR Product, bp | Annealing Temperature Primer Concentration (°C), pmol/μL | GenBank Accession No. |
| 1 | astA | EAST-1 heat-stable toxin | ATGCCATCAACACAGTATAT [22] | 110 | 58/20 | L11241 |
| GCGAGTGACGGCTTTGTAGT [22] | ||||||
| pet | Plasmid-encoded toxin | GGCACAGAATAAAGGGGTGTTT [23] | 302 | 58/25 | AF056581 | |
| CCTCTTGTTTCCACGACATAC [23] | ||||||
| sigA | IgA protease-like homolog | CCGACTTCTCACTTTCTCCCG [19] | 430 | 58/30 | NC_004337 | |
| CCATCCAGCTGCATAGTGTTTG [19] | ||||||
| pic | Serine protease precursor | ACTGGATCTTAAGGCTCAGGAT [23] | 572 | 58/25 | AF097644 | |
| GACTTAATGTCACTGTTCAGCG [23] | ||||||
| sepA | Shigella extracellular protease | GCAGTGGAAATATGATGCGGC [23] | 794 | 58/25 | Z48219 | |
| TTGTTCAGATCGGAGAAGAACG [23] | ||||||
| sat | Secreted autotransporter toxin [15] | TCAGAAGCTCAGCGAATCATTG [19] | 932 | 58/25 | AE014075 | |
| CCATTATCACCAGTAAAACGCACC [19] | ||||||
| 2 | ORF3 | Cryptic proteina | CAGCAACCATCGCATTTCTA | 121 | 57/35 | … |
| CGCATCTTTCAATACCTCCA | ||||||
| aap | Dispersin, antiaggregation protein [12] | GGACCCGTCCCAATGTATAAb | 250 | 57/25 | Z32523 | |
| CCATTCGGTTAGAGCACGATb | ||||||
| aaiC | AaiC, secreted protein | TGGTGACTACTTTGATGGACATTGTb | 313 | 57/25 | … | |
| GACACTCTCTTCTGGGGTAAACGAb | ||||||
| aggR | Transcriptional activator | GCAATCAGATTAARCAGCGATACAb | 426 | 57/25 | Z18751 | |
| CATTCTTGATTGCATAAGGATCTGGb | ||||||
| aatA | Dispersin transporter protein | CAGACTCTGGCRAAAGACTGTATCATb | 642 | 57/35 | AY351860 | |
| CAGCTAATAATGTATAGAAATCCGCTGTb | ||||||
| 3 | agg4A | AAF/IV fimbrial subunit | TGAGTTGTGGGGCTAYCTGGAb | 169 | 57/25 | EU637023 |
| CACCATAAGCCGCCAAATAAGCb | ||||||
| aggA | AAF/I fimbrial subunit | TCTATCTRGGGGGGCTAACGCTb | 220 | 57/20 | Y18149 AY344586 | |
| ACCTGTTCCCCATAACCAGACCb | ||||||
| aafA | AAF/II fimbrial subunit | CTACTTTATTATCAAGTGGAGCCGCTAb | 289 | 57/25 | AF012835 | |
| GGAGAGGCCAGAGTGAATCCTGb | ||||||
| agg3A | AAF/III fimbrial subunit | CCAGTTATTACAGGGTAACAAGGGAAb | 370 | 57/25 | AF411067 | |
| TTGGTCTGGAATAACAACTTGAACGb | ||||||
| agg3/4C c | Usher, AAF/III-IV assembly unit | TTCTCAGTTAACTGGACACGCAATb | 409 | 57/35 | AF411067 AB255435 EU637023 | |
| TTAATTGGTTACGCAATCGCAATb | ||||||
| TCTGACCAAATGTTATACCTTCAYTATGb | ||||||
| aafC | Usher, AAF/II assembly unit | ACAGCCTGCGGTCAAAAGCb | 491 | 57/25 | AF114828 | |
| GCTTACGGGTACGAGTTTTACGGb | ||||||
| 4 | ORF61 | Plasmid-encoded hemolysina | AGCTCTGGAAACTGGCCTCT | 108 | 57/10 | … |
| AACCGTCCTGATTTCTGCTT | ||||||
| eilA | Salmonella HilA homolog | AGGTCTGGAGCGCGAGTGTTb | 248 | 57/30 | … | |
| GTAAAACGGTATCCACGACCb | ||||||
| capU | Hexosyltransferase homolog | CAGGCTGTTGCTCAAATGAAb | 395 | 57/25 | AF134403 | |
| GTTCGACATCCTTCCTGCTCb | ||||||
| air | Enteroaggregative immunoglobulin repeat protein [24] | TTATCCTGGTCTGTCTCAAT | 600 | 57/25 | ||
| GGTTAAATCGCTGGTTTCTT | ||||||
| Singleplex PCR | espY2 | Non-LEE-encoded type III secreted effector | CGCAAAAGATCCGGAAAATAb | 216 | 59/25 | ECSP_0073 |
| TCAGCATTGCTCAGGTCAACb | ||||||
| rmoA | Putative hemolysin expression-modulating protein | TTACCTTACATATTTCCATATCb | 210 | 60/25 | ECUMN_0072 | |
| CGAAAACAAAACAGGAATGGb | ||||||
| shiAd | shiA-like inflammation suppressord | CAGAATGCCCCGCGTAAGGC [25] | 292 | 57/25 | ECB_03517 | |
| CACTGAAGGCTCGCTCATGATCGCCG [25] |
Abbreviations: bp, base pair; PCR, polymerase chain reaction.
Unpublished.
Designed for this study.
Two forward primers and 1 reverse primer were used for the amplification of agg3/4C. This primer set was designed to amplify the usher gene from both AAF/III and IV, hence the name.
Primers used to amplify the shiA gene were forward primer from sisA gene and reverse primer from sisB gene, as described by Lloyd et al [25].
Individual amplification reactions to detect genes designated rmoA, espY2, and shiA were done in 25 μL reaction volumes using crude bacterial cell lysates; PCR reactions were performed as multiplex 2–4. The final extension was 10 minutes at 72°C.
The phylogenetic groups A, B1, B2, and D were determined using triplex PCR methods employing phylogenetic group-specific primers for 2 genes, chuA and yjaA, and a cryptic DNA fragment, TspE4C2. The grouping was coupled to a dichotomous decision tree according to Clermont et al [26].
The following strains where used as controls for detection of target genes: JM221 (aggA, sat) [27], 042 (aatA, aggR, aaiC, aap, ORF3, pic, pet, astA, aafA, aafC, air, capU, eilA) [28], 55989 (agg3A, agg3/4C) [8], H223-1 (sigA) [29], C1010-00 (agg4A, agg3/4C, sat, sepA) [30], MC1061 (negative control), J96 (chuA, yjaA) [26], CFT073 (chuA, yjaA, TspE4.C2) [31], C452-97 (TspE4.C2) [32], and EDL933 (chuA) [33].
Genomic Hybridization
Comparative genome hybridization (CGH) was performed on all the sepA-positive strains as well as sepA-negative strains C801-09 and C46-10 and reference EAEC isolates as previously described [34]. The pan-genome microarrays used in this study were designed by FDA-ECSG Array Probe Set Design and represent the genomes of 32 diverse E. coli and Shigella species, as well as 46 enteric plasmid sequences [35]. Initial data analysis was performed with the Gene Chip Operating System suite of tools provided by Affymetrix. Additional analysis was performed using the Affymetrix power tools software. The MAS5 algorithm was utilized with the perfect match and mismatch calculations and a Tau of 0.150 to detect which probes were present or absent. Features that were present or absent in all samples were removed from further analysis. The resulting features, known as the variable gene set, were analyzed using Multiple Experiment Viewer, version 4.5. The cladogram was constructed using the 12 673 variable features in this dataset, which contained hybridization data from 36 strains. The relationship was determined using hierarchical clustering with Pearson correlation, using the absolute distance and complete linkage run with 500 bootstrap calculations.
Statistics
We utilized classification and regression tree (CART) Pro Version 6.0 (Salford Systems) software inputting 21 or 24 factors of interest as binary (present/absent) independent predictive variables along with a continuous “factor total” that was a sum of all factors including flagellum type H33. Case/control status was the binary dependent outcome variable.
RESULTS
Initial Characterization of EAEC Strains
After 1 year of surveillance, EAEC strains were isolated as the sole DEC pathogen from 60 children with diarrhea and 61 asymptomatic controls. The lack of association of EAEC with diarrhea among the cases persisted even when the presence of other potential pathogens, stratifying for age, was considered or when either aatA or aaiC alone or in combination were considered.
One EAEC isolate was selected from each stool sample that yielded EAEC by multiplex PCR. The 121 EAEC strains belonged to diverse serotypes (Table 2; cases listed in Table 2A and controls listed in Table 2B). Examination of the correlation between serotype and case/control status revealed that only flagellum type H33 was significantly more common among cases than controls (12 cases, 2 controls; odds ratio [OR], 5.9; P = .0138) (Table 3). EAEC cases and control strains were localized to similar positions within a previously published general E. coli phylogenetic tree (Tables 2 and 3).
Table 2a.
Characteristics of 60 Enteroaggregative Escherichia coli Strains Isolated From Children With Diarrhea
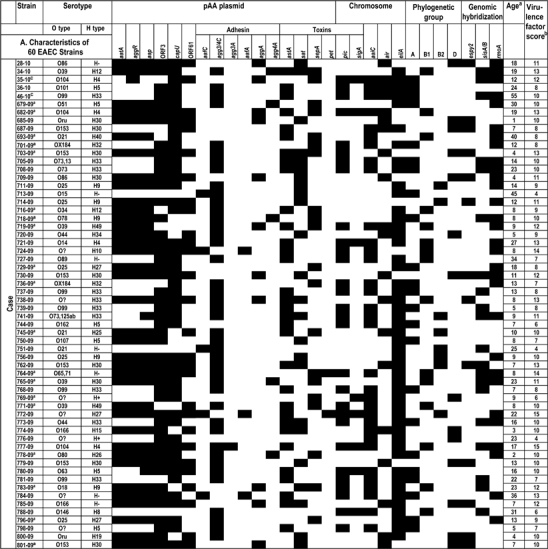 |
Table 2b.
Characteristics of 61 EAEC Strains From Asymptomatic Control Children.
 |
Table 3.
Distribution of Enteroaggregative Escherichia coli Virulence Factors in Cases and Controls
| EAEC |
Cases (n = 60) |
Controls (n = 61) |
Total (N = 121) |
Risk Estimate |
||||||
| Virulence Gene | No. (%) | No. (%) | No. (%) | Odds Ratio | [95% CI] | χ2 | P Value | |||
| aatA | 37 | (61.7) | 46 | (75.4) | 83 | (68.6) | 0.5 | [.24–1.15] | 2.7 | .10 |
| aggR | 38 | (63.3) | 46 | (75.4) | 84 | (69.4) | 0.6 | [.26–1.23] | 2.1 | .15 |
| aaiC | 32 | (53.3) | 26 | (42.6) | 58 | (47.9) | 1.5 | [.75–3.15] | 1.4 | .24 |
| aap | 39 | (65.0) | 48 | (78.7) | 87 | (71.9) | 0.5 | [.22–1.19] | 2.8 | .09 |
| ORF3 | 49 | (81.7) | 55 | (90.2) | 104 | (86.0) | 0.5 | [.17–1.39] | 1.8 | .18 |
| sat | 24 | (40.0) | 33 | (54.1) | 57 | (47.1) | 0.6 | [.28–1.16] | 2.4 | .12 |
| sepA | 20 | (33.3) | 5 | (8.2) | 25 | (20.7) | 5.6 | [1.92–16.17] | 11.7 | .0006 |
| pic | 29 | (48.3) | 27 | (44.3) | 56 | (46.3) | 1.2 | [.58–2.41] | 0.2 | .66 |
| sigA | 8 | (13.3) | 7 | (11.5) | 15 | (12.4) | 1.2 | [.40–3.51] | 0.1 | .76 |
| pet | 4 | (6.7) | 6 | (9.8) | 10 | (8.3) | 0.7 | … | … | .74 |
| astA | 32 | (53.3) | 30 | (49.2) | 62 | (51.2) | 1.2 | [.58–2.41] | 0.2 | .65 |
| aafC | 5 | (8.3) | 5 | (8.2) | 10 | (8.3) | 1.0 | … | … | >.999 |
| agg3/4C | 42 | (70.0) | 40 | (65.6) | 82 | (67.8) | 1.2 | [.57–2.63] | 0.3 | .60 |
| agg3A | 1 | (1.7) | 5 | (8.2) | 6 | (5.0) | 0.2 | … | … | .21 |
| aafA | 3 | (5.0) | 3 | (4.9) | 6 | (5.0) | 1.0 | … | … | >.999 |
| aggA | 11 | (18.3) | 21 | (34.4) | 32 | (26.4) | 0.4 | [.18–.99] | 4.0 | .04 |
| agg4A | 5 | (8.3) | 1 | (1.6) | 6 | (5.0) | 5.5 | … | … | .11 |
| air | 20 | (33.3) | 29 | (47.5) | 49 | (40.5) | 0.6 | [.26–1.15] | 2.5 | .11 |
| capU | 48 | (80.0) | 51 | (83.6) | 99 | (81.8) | 0.8 | [.31–1.99] | 0.3 | .61 |
| eilA | 50 | (83.3) | 53 | (86.9) | 103 | (85.1) | 0.8 | [.28–2.09] | 0.3 | .58 |
| ORF61 | 28 | (46.7) | 44 | (72.1) | 72 | (59.5) | 0.3 | [.16–0.72] | 8.1 | .004 |
| espY2 | 13 | (21.6) | 20 | (32.8) | 33 | (27.3) | 0.6 | [.25–1.28] | 1.9 | .17 |
| rmoA | 30 | (50.0) | 23 | (37.7) | 53 | (43.8) | 1.7 | [.80–3.39] | 1.9 | .17 |
| shiA | 21 | (35) | 22 | (36.1) | 43 | (35.5) | 0.5 | [.45–2.01] | 0.01 | .92 |
| EAEC Serogroup | ||||||||||
| O99 | 5 | (8.3) | 2 | (3.3) | 7 | (5.8) | 2.7 | … | … | .27 |
| O153 | 6 | (10.0) | 1 | (1.6) | 7 | (5.8) | 6.7 | … | … | .61 |
| H- | 7 | (11.7) | 17 | (27.9) | 24 | (19.9) | 0.3 | [.12–.98] | 4.9 | .04 |
| H5 | 6 | (10.0) | 2 | (3.3) | 8 | (6.6) | 3.3 | … | … | .16 |
| H9 | 5 | (8.3) | 1 | (1.6) | 6 | (5.0) | 5.5 | … | … | .21 |
| H30 | 9 | (15.0) | 4 | (6.6) | 13 | (10.7) | 2.5 | [.73–8.66] | 2.2 | .13 |
| H33 | 10 | (16.7) | 2 | (3.3) | 12 | (9.9) | 5.9 | [1.23–28.19] | 6.7 | .01 |
| Phylogenetic Group | ||||||||||
| A | 23 | (38.3) | 14 | (22.9) | 37 | (32.2) | 2.1 | [.95–4.61] | 3.4 | .07 |
| B1 | 15 | (25) | 13 | (21.3) | 28 | (23.1) | 1.2 | [.53–2.78] | 0.2 | .63 |
| B2 | 8 | (13.3) | 9 | (14.7) | 17 | (14) | 0.9 | [.32–2.48] | 0.05 | .82 |
| D | 14 | (23.3) | 25 | (40.9) | 39 | (32.2) | 0.4 | [.19–.96] | 4.3 | .04 |
P < .05 is significant. Fisher exact test was applied when the comparisons between cases and controls were <5 observations.
Abbreviation: EAEC, enteroaggregative Escherichia coli.
Frequencies of Virulence-Related Genes
In order to assess the roles of putative virulence factors in EAEC epidemiology, we developed 4 multiplex PCR assays for the characterization of 21 genes previously found in EAEC strains. The results of the PCR assays for all strains are listed in Table 2. Of the 21 genes scored, hypothetical ORF3 was the most frequently detected (86%) followed by eilA (85.1%), capU (81.8%), aap (71.9%), aggR (69.4%), and aatA (68.6%) (Table 3). There was a high degree of concordance of these genes, which has been demonstrated previously for the plasmid-encoded aap, aggR, and aatA genes [7, 12, 36–38]. Sixty-eight percent of the strains were positive for the usher-encoding gene agg3/4C (the ushers for AAF/III and AAF/IV variants are closely related). The most frequent AAF pilin gene was that of AAF/I, encoded by aggA (26.4%), followed by those of AAF/II (aafA), AAF/III (agg3A), and AAF/IV (agg4A) at 5% each (Table 3). The agg4A gene was found more frequently among cases than controls (5 cases and 1 control), although this difference did not reach statistical significance. A total of 71 strains (58.7%) were negative for a known AAF variant.
Of the 5 genes encoding SPATEs, the most frequent were sat (47.1%) and pic (46.3%). The least common SPATEs were pet (8.3%) and sigA (12.4%). The sepA gene was found in 25 strains (20.7%): 20 from cases and 5 from controls, yielding an OR of 5.6 (P = .0006) (Table 3). Among all the putative virulence factors scored, sepA was the only one significantly associated with moderate to severe diarrheal illness.
Significance of Combinations of EAEC Genes
In addition to considering each factor individually, we pursued a number of approaches to consider the importance of combinations of potential EAEC virulence factors. When crudely considering the collective number of virulence loci present (generating a virulence factor score, VFS), the average number of virulence genes from cases was 8.75 versus 9.5 from control isolates.
To consider combinations of factors, we employed CART analysis, which builds a model in stepwise fashion to yield the combination of factors most strongly associated with the queried outcome. Each branch of a CART output tree ends in a terminal “node”; each observation falls into exactly 1 terminal node; and each terminal node is uniquely defined by a set of rules, such as having or not having a certain factor.
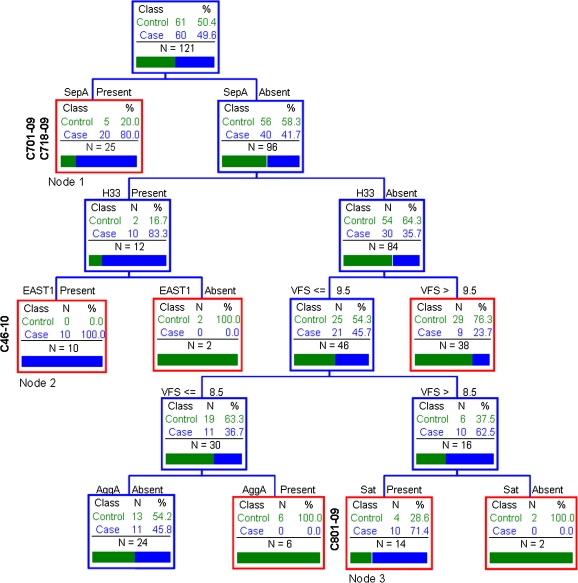
We considered all genotypic and phenotypic assays performed and interrogated the association with case status. Figure 1 illustrates the best CART fit for the dataset. The analysis demonstrates that the presence of sepA, regardless of the presence or absence of any other scored genotype or phenotype among the sepA-positive strains, provides a strong association with diarrhea.
Figure 1.
Classification and regression tree (CART) classification tree topology reveals combinations of factors most strongly associated with moderate to severe diarrhea. We considered all genotypic and phenotypic assays performed: aatA, aggR, aaiC, aap, ORF3, sat, sepA, pic, sigA, pet, astA, aafC, agg3/4C, aafA, agg3A, aggA, agg4A, air, capU, eilA, ORF61, virulence factor score (VSF), and flagellum type H33. Each branch of the CART tree ends in a terminal “node” (red boxes), and each terminal node is uniquely defined by the presence or absence of a predictive factor such as a gene or VFS. The tree is hierarchical in nature. C701-09, C718-09, C801-09, and C46-10 are also shown on the dendrogram.
Among the sepA-negative strains, CART analysis suggested 2 additional trait clusters that were associated with moderate to severe diarrheal illness: 1 cluster included those strains harboring the flagellum H33 and the toxin EAST-1, whereas a second cluster lacked H33 but featured a VFS of 9, suggesting a combination of typical EAEC factors in addition to the Sat toxin.
Genomic Analyses
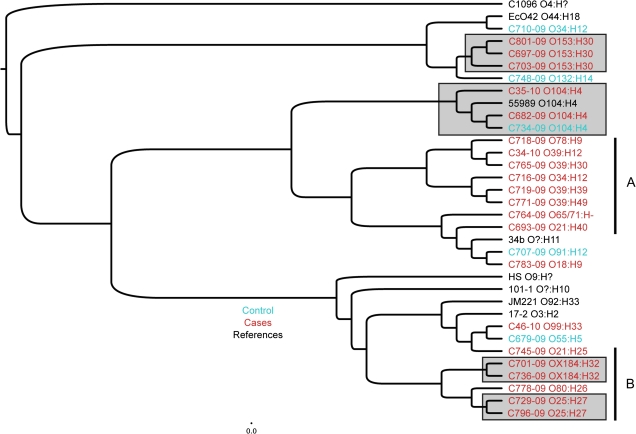
We hypothesized that the strain sets belonging to the nodes most strongly associated with diarrhea would reveal the presence of additional virulence determinants, which themselves might explain the observed clinical correlations. We therefore performed CGH analysis using a previously described microarray containing the full genomes of 32 E. coli and Shigella strains and the genes of an additional 46 E. coli plasmids [35]. For this analysis, we chose all 25 sepA-positive strains, 2 additional strains (C46-10 and C801-09) representing CART (Figure 1) nodes 2 (SepA absent, H33 present, EAST-1 present) and 3 (SepA absent, H33 absent, >8.5VFS, Sat present), and a set of archetype EAEC reference strains. Standard cluster analysis was performed on the microarray data (Figure 2). All isolates belonging to a common serotype clustered together in this analysis. Although cluster analysis did not suggest genomic differences discriminating cases and controls, the analysis did suggest that sepA-positive strains segregated into 2 major clusters (indicated as A and B in Figure 2). We chose for further genomic examination archetypal strains representing sepA-positive clusters A and B, as well as the 2 additional nodes (2 and 3) indicated by CART analysis (Figure 1).
Figure 2.
Cladogram of comparative genomic hybridization data of sepA-positive isolates (C34-10, C35-10, C679-09C, C682-09, C693-09, C701-09, C703-09, C716-09, C718-09, C719-09, C729-09, C736-09, C745-09, C764-09, C765-09, C769-09, C771-09, C778-09, C783-09, C796-09, C697-09, C707-09, C710-09, C734-09, and C748-09), sepA-negative isolates (C46-10 and C801-09), and reference isolates (C1096, 042, 55989, JM221, 17-2, 34b, 101-1, and HS). Notably, C46-10 was most closely related to Mexican enteroaggregative Escherichia coli (EAEC) strain JM221 (isolated from an adult traveler to Guadalajara [27]), and strain C801-09 was most closely related to EAEC strain 042, isolated from a child with diarrhea in Lima, Peru [28]. The phylogenetic comparison was performed using the 12 673 variable features of the 36 hybridizations included. The tree is built using a hierarchical clustering with Pearson correlation using both the absolute distance and complete linkage and viewed in FigTree (http://tree.bio.ed.ac.uk/software/figtree/). Isolates represented in black are reference isolates, controls are indicated in blue, and cases in red. The serotypes of the strains are indicated to the right. The gray boxes identify clusters of serotypes within the context of the larger tree, indicating that those serogroups are genomically similar.
Genome Analysis of sepA-Positive Strains
To represent the sepA-positive strain cluster A (Figure 2), we chose strain C718-09 for further genome analysis; to represent cluster B, we chose strain C701-09. Results are presented in Table 4. Genome analysis of strain C-718-09 did not reveal the presence of additional genes that were not also carried by nonpathogenic E. coli strains. C701-09 hybridized to an open reading frame that was 99% identical to the rmoA gene encoded on plasmid R100 (GenBank accession number Y13856.1); rmoA encodes a predicted 69 amino acid putative hemolysin expression-modulation protein that is 100% identical to protein RmoA found on plasmid R100 from E. coli [39]. The protein sequence of R100 RmoA exhibits 52% identity and 75% amino acid similarity with Hha protein from E. coli K12 (GenBank accession number NP_414993).
Table 4.
Comparative Genomic Hybridization of Strains C701-09, C718-09, C801-09, and C46-10 Against a Microarray That Comprises the Full Genomes of 32 Escherichia coli and Shigella Strains and the Genes of Additional 46 E. coli Plasmids
| Hybridization by Genome |
||||||
| Putative Virulence Genea | Accession No. | Nonpathogenicb | C701-09 | C718-09 | C801-09 | C46 -10 |
| Adhesins | ||||||
| csgA; cryptic curlin major subunita | SBO_2026 | + | + | + | + | – |
| csgA; major curlin subunitb | LF82_0360 | + | + | – | + | – |
| csgC; putative autoagglutination proteinb | ECUMN_1217 | + | + | + | + | – |
| ecpD; putative chaperone protein EcpDb | SBO_0126 | + | – | + | + | – |
| Fimbrial usher family proteinb | SbBS512_E2717 | + | + | + | – | + |
| Flu; antigen 43 (Ag43)b | ECUMN_3400 | + | + | – | – | + |
| Hemagglutinin familyc | SbBS512_E4026 | + | – | + | + | – |
| Putative AidA-I adhesin-like protein | ECO26_3415 | – | – | – | + | + |
| Putative AidA-I adhesin-like proteind | ECO26_1353 | + | – | – | + | – |
| Putative chaperone protein EcpD | ECUMN_0137 | – | + | – | + | – |
| Putative fimbrial biogenesis outer membrane usher protein | ECUMN_0019 | – | + | – | + | – |
| Putative fimbrial proteinb | SbBS512_E2376 | + | + | + | + | – |
| Putative fimbrial-like proteinb | SD.Y_0915 | + | + | + | + | – |
| Putative invasinb | EcSMS35_1146 | + | + | + | + | – |
| Putative type 1 fimbrial protein | ECSP_0022 | – | – | – | + | – |
| sfmD; putative outer membrane export usher protein SfmDb | ECO26_0565 | + | – | + | + | – |
| sfmF; putative fimbrial-like adhesin protein SfmFb | ECO26_0567 | + | – | + | + | – |
| sfmH; putative fimbrial-like adhesin proteinb | ECUMN_0573 | + | – | + | + | – |
| siiEA; adhesin for cattle intestine colonization | ECUMN_0527 | – | – | – | + | – |
| yfaL; adhesin YfaLb | ECO26_3226 | + | + | + | + | – |
| yfcP; putative fimbrial-like adhesin proteinb | BWG_2107 | + | + | – | – | – |
| yfcQ; putative fimbrial-like adhesin proteinb | BWG_2108 | + | + | – | – | – |
| yfcR; putative fimbrial-like adhesin proteinb | BWG_2109 | + | + | – | – | + |
| yfcS; putative periplasmic pilus chaperoneb | BWG_2110 | + | + | – | + | + |
| yfcS; putative periplasmic pilus exported chaperoneb | ECUMN_2676 | + | + | – | + | + |
| yfcT; outer membrane export usher proteinb | ECDH10B_2499 | + | + | – | + | + |
| yfcU; export usher proteinb | ECDH10B_2500 | + | + | – | + | + |
| yfcU; outer membrane usher protein | E2348C_2477 | – | + | – | + | + |
| yfcV; predicted fimbrialprotein–like protein | E2348C_2478 | – | + | – | + | – |
| Toxins | ||||||
| Hcp-like proteinb | SSON_0233 | + | – | + | + | – |
| hlyE; hemolysin Eb | ECO26_1695 | + | + | + | + | – |
| Secretion Systems | ||||||
| espY2; Non-LEE-encoded Type III Secreted Effector | ECSP_0073 | – | – | – | + | – |
| Hypothetical protein; type VI secretion system secreted protein VgrGb | ECSP_0240 | + | + | – | + | + |
| Putative type II secretion protein (GspI-like)b | ECIAI1_3105 | + | + | – | + | – |
| Putative type III secretion protein EpaRb | ECUMN_3195 | + | – | – | + | – |
| T3SS effector-like protein EspL-homologb | ECO111_4829 | + | – | + | + | – |
| tolC; outer membrane channel proteinb | SDY_3205 | + | + | + | – | + |
| Type III secretion protein EpaQb | ECO26_3940 | + | – | + | + | – |
| Type III secretion protein EpaRb | ECO103_3428 | + | – | + | + | – |
| Type III secretion protein EprJb | ECO26_3933 | + | – | + | + | – |
| Other | ||||||
| Hemolysin expression-modulating protein | EC55989_3351 | – | + | + | + | – |
| Putative hemolysin expression-modulating protein RmoA | ECUMN_0072 | – | + | + | – | + |
| Putative hemolysin co-regulated proteinb | SSON_0255 | + | – | – | + | + |
| ShiA-like protein | ECB_03517 | – | – | – | – | + |
Genome Analysis of sepA-Negative Strains
Strains C46-10 and C801-09 were representative of the 2 sepA-negative nodes that were associated with diarrhea in Figure 1.
C46-10 Genome Analysis
Strain C46-10 best represented the confluence of factors identified in node 2 (Figure 1), characterized as sepA absent, H33 present, and EAST-1 present. By CGH, strain C46-10 hybridized to a large number of genes found among DEC pathotypes and Shigella speices (Table 4) and encodes a complete yfc gene cluster, which has been proposed to encode a novel usher-chaperone fimbrial adhesin [40]. C46-10 harbored elements of a type VI secretion system homologous to VgrG of Agrobacterium. However, this component of the newly described type VI systems is also found among nonvirulent isolates and has not yet been assigned any virulence function among DEC or Shigella strains.
C46-10 DNA was found to hybridize with the 347 amino acid ShiA-like protein from E. coli strain REL606 (GenBank accession number YP_003046696). The latter protein exhibited 97% identity with the ShiA protein initially described in Shigella flexneri 5a strain M90T (GenBank accession number AF141323) [41]. ShiA and related proteins identified in uropathogenic E coli and Shigella strains have been found to suppress the inflammatory response in animal models [42].
C801-09 Genome Analysis
C801-09 is closely related to the virulent archetype EAEC strain 042 and harbors many of the same virulence genes, including a near-complete plasmid-borne AggR regulon. It represents the most common serotype found in our study (O153:H30). Like C701-09, C801-09 harbored homologs of a large number of adhesins, including the siiEA locus that is associated with colonization of cattle by E. coli strain UMN026. C801-09 also harbored EspY2, a non-LEE-encoded type III secreted effector from E. coli O157:H7 strain TW14359 (GeneID: 8214639). Five proteins, EspY1-5 from the E. coli O157:H7 Sakai strain, possess an N-terminal WEX5F domain, which has been linked to type III secretion and is conserved in several well-characterized Salmonella effectors and in putative effectors from Edwarsiella and Sodalis [43].
Screening of the EAEC Collection for Presence of espY2, rmoA, and shiA Genes
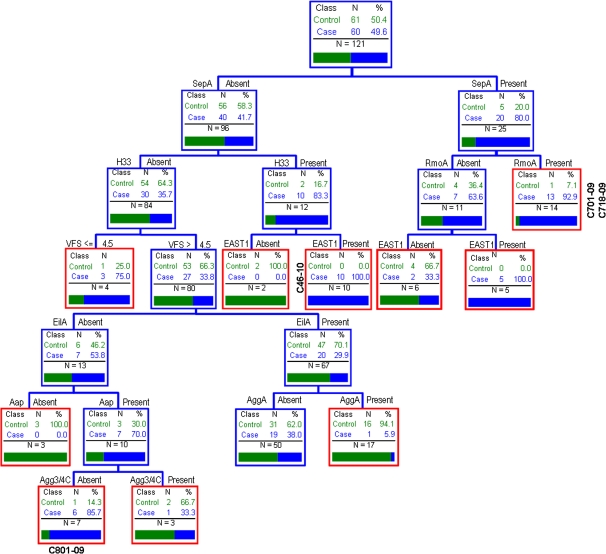
Based on the CGH analysis from strains C46-10, C701-09, C718-09, and C801-09, we inferred that espY2, rmoA, and shiA were the factors most plausibly associated with virulence. Using PCR, we found that 35.5% of the EAEC strains from cases and a similar percentage from controls harbored the shiA gene and 27.3% of each group harbored espY2. The rmoA gene was found in 43.8% of the EAEC strains (50% from cases and 37.7% from controls). None of the 3 genes were independently associated with diarrheal illness (Table 3). However, when we repeated the CART analysis including the espY2, rmoA, and shiA genes (Figure 3), sepA once again exhibited a strong association, yet strains that were both sepA- and rmoA-positive were most strongly associated with disease (13 out of 14 strains positive for this combination were present among cases).
Figure 3.
Classification and regression tree analysis described in Figure 1, adding the genes shiA, espy2, rmoA (hemolysin expression-modulation protein). See Figure 1 legend for details of analysis
DISCUSSION
EAEC is a common diarrheal isolate, yet apart from those outbreak-associated, identification of truly pathogenic strains remains difficult. A large number of virulence factors and combinations have been associated with clinical illness in epidemiologic studies, and it is possible that either the principal determinants of pathogenicity vary by site and population or that the true determinants have not yet been identified.
We report the most detailed genomic characterization of EAEC performed to date, targeting a collection of 121 EAEC strains isolated from children in Mali with or without moderate to severe diarrhea. In agreement with previous reports [44–46], our strains belonged to a diverse range and combination of O:H and phylogenetic types. Although no specific O:H combination was associated with diarrhea, strains expressing the H33 flagellar antigen were found significantly more often in cases than in controls. This association may signify the existence of a specific set of virulence genes in strains of this H type.
To profile the virulence genes of our strain set, we developed and applied 4 multiplex PCR assays targeting 21 putative virulence genes. We found our EAEC strains to be astonishingly diverse. The only factor associated individually with diarrhea in these analyses was the Shigella SPATE toxin SepA. Recognizing that pathogenicity represents the concerted action of multiple virulence factors, which can sort independently throughout the E. coli population, we assessed combinations of virulence factors using CART analysis. This analysis reinforced the association of sepA with diarrhea, independent of any of the other 20 genes scored. We then performed comprehensive genomic analyses on the sepA-positive strains using CGH against a reference set of E. coli genomes. These studies identified the hemolysin expression-modulating protein RmoA as commonly present in combination with SepA and served to strengthen the association of SepA with clinical illness (Figure 3). Our data demonstrate the importance of strains encoding a combination of virulence factors (here SepA and RmoA), although additional factors may colocalize with these genes.
Among the sepA-negative strains, CART analysis suggested 2 combinations of factors that indicate virulent strains (Figure 1). CGH analysis of strains representative of these combinations (strains C801-09 and C46-10) revealed 2 additional factors: T3SS effector EspY2 and ShiA, the latter being associated with modulation of the inflammatory response. However, screening the complete strain set for the presence of these 2 factors, followed by revised CART analysis, did not suggest that these 2 genes strengthened the association with moderate to severe diarrhea.
The association of the toxin EAST-1 with diarrhea only occurred among strains that lacked the majority of the AggR regulon, suggesting that they may require virulence factors not yet apparent; these may occur predominantly in strains harboring flagellar type H33 (Figure 1). EAST-1-positive strains have previously been implicated in pediatric diarrhea [47], so these strains may warrant continued investigation.
The terms typical EAEC and atypical EAEC have been suggested to refer to EAEC strains harboring or lacking AggR, respectively. Some studies have demonstrated an association of typical EAEC with diarrhea [5, 48]. We did not observe any correlation of AggR regulon genes with moderate to severe illness in this study. It is possible that our focus on moderate to severe diarrhea overlooks mild illness due to EAEC and that true determinants of pathogenicity are not recognized. Alternatively, illness may be obscured by epidemiologic factors, such as previous exposure. Also, we note that our EAEC definition included two AggR-related genes, potentially introducing strain selection bias.
This study is notable for the association of the Shigella virulence factor SepA with clinical illness, an association that persisted when the effects of other pathogens were considered (OR, 5.6; P = .0006; data not shown). SepA was first described by Benjelloun-Touimi et al [20] and is a prominent extracellular protein secreted by S. flexneri strains. SepA is produced during infection [20] and has been shown to confer increased epithelial cell exfoliation from human intestinal explants infected with S. flexneri [49]. We also note that SepA is produced by the Shiga toxin–producing outbreak strain from Germany in 2011. The previously unsuspected role of SepA in EAEC warrants further investigation.
Leveraging a large epidemiologic study and powerful genomic techniques, our study sheds additional light on the complex nature of diarrheagenic E. coli genomes and their association with human disease.
Notes
Acknowledgments.
We thank Susanne Jespersen and Pernille Gymoese, Statens Serum Institut, for technical support regarding serotyping and Dr Steen Ethelberg for statistical advice.
This paper is dedicated to the memory of Dr Bernadette Baudry, whose seminal description of the EAEC probe provided the first, and still best, molecular signature for EAEC strains.
Financial support.
This study was supported by the Bill & Melinda Gates Foundation, US Public Health Service grant AI33096 to J. P. N. and Danish Council for Strategic Research grant 2101-07-0023 to K. A. K. N. B. was supported in part by Statens Serum Institut, Copenhagen, Denmark.
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Black RE, Cousens S, Johnson HL, et al. Child Health Epidemiology Reference Group of WHO and UNICEF. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969–87. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- 2.Wanke CA. To know Escherichia coli is to know bacterial diarrheal disease. Clin Infect Dis. 2001;32:1710–2. doi: 10.1086/320763. [DOI] [PubMed] [Google Scholar]
- 3.Adachi JA, Jiang ZD, Mathewson JJ, et al. Enteroaggregative Escherichia coli as a major etiologic agent in traveler’s diarrhea in 3 regions of the world. Clin Infect Dis. 2001;32:1706–9. doi: 10.1086/320756. [DOI] [PubMed] [Google Scholar]
- 4.Tompkins DS, Hudson MJ, Smith HR, et al. A study of infectious intestinal disease in England: microbiological findings in cases and controls [see comments] Commun Dis Public Health. 1999;2:108–13. [PubMed] [Google Scholar]
- 5.Okeke IN, Lamikanra A, Czeczulin J, Dubovsky F, Kaper JB, Nataro JP. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J Infect Dis. 2000;181:252–60. doi: 10.1086/315204. [DOI] [PubMed] [Google Scholar]
- 6.Rasko DA, Webster DR, Sahl JW, et al. Origins of the E. coli strain causing an outbreak of hemolytic-uremic syndrome in Germany. N Engl J Med. 2011;365:709–17. doi: 10.1056/NEJMoa1106920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boisen N, Struve C, Scheutz F, Krogfelt KA, Nataro JP. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect Immun. 2008;76:3281–92. doi: 10.1128/IAI.01646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bernier C, Gounon P, Le Bouguenec C. Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun. 2002;70:4302–11. doi: 10.1128/IAI.70.8.4302-4311.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nataro JP, Yikang D, Giron JA, Savarino SJ, Kothary MH, Hall R. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect Immun. 1993;61:1126–31. doi: 10.1128/iai.61.3.1126-1131.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nataro JP, Deng Y, Maneval DR, German AL, Martin WC, Levine MM. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect Immun. 1992;60:2297–304. doi: 10.1128/iai.60.6.2297-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias WP, Jr, Czeczulin JR, Henderson IR, Trabulsi LR, Nataro JP. Organization of biogenesis genes for aggregative adherence fimbria II defines a virulence gene cluster in enteroaggregative Escherichia coli. J Bacteriol. 1999;181:1779–85. doi: 10.1128/jb.181.6.1779-1785.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheikh J, Czeczulin JR, Harrington S, et al. A novel dispersin protein in enteroaggregative Escherichia coli. J Clin Invest. 2002;110:1329–37. doi: 10.1172/JCI16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:1267–82. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 14.Henderson IR, Hicks S, Navarro-Garcia F, Elias WP, Philips AD, Nataro JP. Involvement of the enteroaggregative Escherichia coli plasmid-encoded toxin in causing human intestinal damage. Infect Immun. 1999;67:5338–44. doi: 10.1128/iai.67.10.5338-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guyer DM, Henderson IR, Nataro JP, Mobley HL. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol Microbiol. 2000;38:53–66. doi: 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 16.Rajakumar K, Sasakawa C, Adler B. Use of a novel approach, termed island probing, identifies the Shigella flexneri she pathogenicity island which encodes a homolog of the immunoglobulin A protease-like family of proteins. Infect Immun. 1997;65:4606–14. doi: 10.1128/iai.65.11.4606-4614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrington SM, Sheikh J, Henderson IR, Ruiz-Perez F, Cohen PS, Nataro JP. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect Immun. 2009;77:2465–73. doi: 10.1128/IAI.01494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun. 1999;67:5587–96. doi: 10.1128/iai.67.11.5587-5596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boisen N, Ruiz-Perez F, Scheutz F, Krogfelt KA, Nataro JP. Short report: high prevalence of serine protease autotransporter cytotoxins among strains of enteroaggregative Escherichia coli. Am J Trop Med Hyg. 2009;80:294–301. [PMC free article] [PubMed] [Google Scholar]
- 20.Benjelloun-Touimi Z, Sansonetti PJ, Parsot C. SepA, the major extracellular protein of Shigella flexneri: autonomous secretion and involvement in tissue invasion. Mol Microbiol. 1995;17:123–35. doi: 10.1111/j.1365-2958.1995.mmi_17010123.x. [DOI] [PubMed] [Google Scholar]
- 21.Orskov F, Orskov I. Escherichia coli serotyping and disease in man and animals. Can J Microbiol. 1992;38:699–704. [PubMed] [Google Scholar]
- 22.Mohamed JA, Huang DB, Jiang ZD, et al. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J Clin Microbiol. 2007;45:121–6. doi: 10.1128/JCM.01128-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Restieri C, Garriss G, Locas MC, Dozois CM. Autotransporter-encoding sequences are phylogenetically distributed among Escherichia coli clinical isolates and reference strains. Appl Environ Microbiol. 2007;73:1553–62. doi: 10.1128/AEM.01542-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheikh J, Dudley EG, Sui B, Tamboura B, Suleman A, Nataro JP. EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol Microbiol. 2006;61:338–50. doi: 10.1111/j.1365-2958.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- 25.Lloyd AL, Smith SN, Eaton KA, Mobley HL. Uropathogenic Escherichia coli suppresses the host inflammatory response via pathogenicity island genes sisA and sisB. Infect Immun. 2009;77:5322–33. doi: 10.1128/IAI.00779-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–8. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathewson JJ, Oberhelman RA, Dupont HL, Javier de la Cabada F, Garibay EV. Enteroadherent Escherichia coli as a cause of diarrhea among children in Mexico. J Clin Microbiol. 1987;25:1917–9. doi: 10.1128/jcm.25.10.1917-1919.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nataro JP, Baldini MM, Kaper JB, Black RE, Bravo N, Levine MM. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J Infect Dis. 1985;152:560–5. doi: 10.1093/infdis/152.3.560. [DOI] [PubMed] [Google Scholar]
- 29.Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun. 1999;67:2692–9. doi: 10.1128/iai.67.6.2692-2699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olesen B, Neimann J, Bottiger B, et al. Etiology of diarrhea in young children in Denmark: a case-control study. J Clin Microbiol. 2005;43:3636–41. doi: 10.1128/JCM.43.8.3636-3641.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welch RA, Burland V, Plunkett G, 3rd, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:17020–4. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson JR, Scheutz F, Ulleryd P, Kuskowski MA, O'Bryan TT, Sandberg T. Phylogenetic and pathotypic comparison of concurrent urine and rectal Escherichia coli isolates from men with febrile urinary tract infection. J Clin Microbiol. 2005;43:3895–900. doi: 10.1128/JCM.43.8.3895-3900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perna NT, Plunkett G, 3rd, Burland V, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–33. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 34.Sahl JW, Lloyd AL, Redman JC, et al. Genomic characterization of asymptomatic Escherichia coli isolated from the neobladder. Microbiology. 2011 doi: 10.1099/mic.0.043018-0. 157(Pt 4):1088–102. [DOI] [PubMed] [Google Scholar]
- 35.Fang H, Xu J, Ding D, et al. An FDA bioinformatics tool for microbial genomics research on molecular characterization of bacterial foodborne pathogens using microarrays. BMC Bioinformatics. 2010;11(Suppl 6):S4. doi: 10.1186/1471-2105-11-S6-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kahali S, Sarkar B, Rajendran K, et al. Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2004;42:4111–20. doi: 10.1128/JCM.42.9.4111-4120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang ZD, Greenberg D, Nataro JP, Steffen R, DuPont HL. Rate of occurrence and pathogenic effect of enteroaggregative Escherichia coli virulence factors in international travelers. J Clin Microbiol. 2002;40:4185–90. doi: 10.1128/JCM.40.11.4185-4190.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerna JF, Nataro JP, Estrada-Garcia T. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J Clin Microbiol. 2003;41:2138–40. doi: 10.1128/JCM.41.5.2138-2140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nieto JM, Prenafeta A, Miquelay E, Torrades S, Juarez A. Sequence, identification and effect on conjugation of the rmoA gene of plasmid R100-1. FEMS Microbiol Lett. 1998;169:59–66. doi: 10.1111/j.1574-6968.1998.tb13299.x. [DOI] [PubMed] [Google Scholar]
- 40.Korea CG, Badouraly R, Prevost MC, Ghigo JM, Beloin C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ Microbiol. 2010;12:1957–77. doi: 10.1111/j.1462-2920.2010.02202.x. [DOI] [PubMed] [Google Scholar]
- 41.Ingersoll MA, Moss JE, Weinrauch Y, Fisher PE, Groisman EA, Zychlinsky A. The ShiA protein encoded by the Shigella flexneri SHI-2 pathogenicity island attenuates inflammation. Cell Microbiol. 2003;5:797–807. doi: 10.1046/j.1462-5822.2003.00320.x. [DOI] [PubMed] [Google Scholar]
- 42.Ingersoll MA, Zychlinsky A. ShiA abrogates the innate T-cell response to Shigella flexneri infection. Infect Immun. 2006;74:2317–27. doi: 10.1128/IAI.74.4.2317-2327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tobe T, Beatson SA, Taniguchi H, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci U S A. 2006;103:14941–6. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vial PA, Robins-Browne R, Lior H, et al. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis. 1988;158:70–9. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]
- 45.Qadri F, Haque A, Faruque SM, Bettelheim KA, Robins-Browne R, Albert MJ. Hemagglutinating properties of enteroaggregative Escherichia coli. J Clin Microbiol. 1994;32:510–4. doi: 10.1128/jcm.32.2.510-514.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto T, Echeverria P, Yokota T. Drug resistance and adherence to human intestines of enteroaggregative Escherichia coli. J Infect Dis. 1992;165:744–9. doi: 10.1093/infdis/165.4.744. [DOI] [PubMed] [Google Scholar]
- 47.Vila J, Gene A, Vargas M, Gascon J, Latorre C, Jimenez de Anta MT. A case-control study of diarrhoea in children caused by Escherichia coli producing heat-stable enterotoxin (EAST-1) J Med Microbiol. 1998;47:889–91. doi: 10.1099/00222615-47-10-889. [DOI] [PubMed] [Google Scholar]
- 48.Sarantuya J, Nishi J, Wakimoto N, et al. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J Clin Microbiol. 2004;42:133–9. doi: 10.1128/JCM.42.1.133-139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Coron E, Flamant M, Aubert P, et al. Characterisation of early mucosal and neuronal lesions following Shigella flexneri infection in human colon. PLoS One. 2009;4:e4713. doi: 10.1371/journal.pone.0004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baudry B, Savarino SJ, Vial P, Kaper JB, Levine MM. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J Infect Dis. 1990;161:1249–51. doi: 10.1093/infdis/161.6.1249. [DOI] [PubMed] [Google Scholar]