CCR5-binding chemokines produced in the draining lymph node after vaccinia virus infection guide naive CD8+ T cells toward DCs and away from the macrophage-rich zone, thereby facilitating optimal CD8+ T cell activation and cytokine production.
Abstract
Naive antiviral CD8+ T cells are activated in the draining LN (DLN) by dendritic cells (DCs) presenting viral antigens. However, many viruses infect LN macrophages, which participate in initiation of innate immunity and B cell activation. To better understand how and why T cells select infected DCs rather than macrophages, we performed intravital microscopy and ex vivo analyses after infecting mice with vaccinia virus (VV), a large DNA virus that infects both LN macrophages and DCs. Although CD8+ T cells interact with both infected macrophages and DCs in the LN peripheral interfollicular region (PIR), DCs generate more frequent and stable interactions with T cells. VV infection induces rapid release of CCR5-binding chemokines in the LN, and administration of chemokine-neutralizing antibodies diminishes T cell activation by increasing T cell localization to macrophages in the macrophage-rich region (MRR) at the expense of PIR DCs. Similarly, DC ablation increases both T cell localization to the MRR and the duration of T cell–macrophage contacts, resulting in suboptimal T cell activation. Thus, virus-induced chemokines in DLNs enable antiviral CD8+ T cells to distinguish DCs from macrophages to optimize T cell priming.
DCs and macrophages are highly heterogeneous cells (Vremec and Shortman, 1997; Shortman and Liu, 2002; Gordon and Taylor, 2005; Ziegler-Heitbrock et al., 2010). Their myriad subtypes exhibit a blend of shared and unique functions that tailor their abilities to regulate innate and adaptive immune responses. In mouse LNs, conventional DCs include the CD8α+ and CD11b+ subgroups, the latter subdivided based on CD4 expression (Vremec et al., 2000; Allan et al., 2003; Belz et al., 2004; Hildner et al., 2008). Recent DC immigrants populating skin draining LNs (DLNs) include Langerhans cells and CD8α− CD103+ dermal DCs (Randolph et al., 2008; Bedoui et al., 2009). An additional subset, plasmacytoid DCs (pDCs) are copious producers of type I IFNs (Swiecki and Colonna, 2010). Microbial infections typically increase DC numbers in the DLN, and induce the differentiation of inflammatory DCs (Serbina et al., 2003).
Macrophage subtypes are less clearly established than DC subtypes, but can be delineated based on anatomical location (e.g., marginal zone splenic macrophages) or expression of cell surface receptors (e.g., CD169, mannose receptor, MARCO, and dectin-1; Taylor et al., 2005). Within the LN, both subcapsular sinus (SCS) and medullary macrophages express sialoadhesin (CD169), with the latter distinguished by coexpression of F4/80 (Phan et al., 2009). The inflammatory milieu heavily influences the differentiation state of macrophages (Serbina et al., 2008; Liu et al., 2009).
LN macrophages have recently been the subject of intense investigation because of their rapid and efficient uptake of lymph-borne particles deposited into the LN SCS. After antigen capture, SCS macrophages can relay antigen to follicular B cells (Carrasco and Batista, 2007; Junt et al., 2007; Phan et al., 2007), a step that promotes the affinity maturation of antibodies (Abs; Phan et al., 2009). As SCS macrophages express lower levels of several proteases involved in proteolytic degradation of phagocytosed antigen, Phan et al. (2007) suggested that their function may be to maintain a reservoir of antigen for relay to LN B cells. However, many SCS macrophages are not located above B cell follicles, but rather above the T cell zone in the peripheral interfollicular regions (PIRs) of the node (Hickman et al., 2008), suggesting their participation in T cell responses.
As with other particulate antigens, virions trafficking to the LNs via the lymphatics are internalized by SCS macrophages (Norbury et al., 2002; Junt et al., 2007; Hickman et al., 2008; Iannacone et al., 2010). Both vaccinia virus (VV) and vesicular stomatitis virus readily infect nodal macrophages, which account for up to 85% of virus-infected LN cells within a few hours of infection (Norbury et al., 2002; Hickman et al., 2008). Functionally, SCS macrophages limit vesicular stomatitis viremia and dissemination (Junt et al., 2007), and are a critical source of type I IFNs (Iannacone et al., 2010). Although macrophages express the appropriate machinery for T cell activation, numerous studies have shown through in vivo ablation or ex vivo isolation that DCs prime CD8+ T cells after viral infection (Allan et al., 2003; Probst et al., 2005; Ciavarra et al., 2006; Kassim et al., 2006). It is currently unknown whether CD8+ T cells interact with infected LN macrophages. Additionally, because infected macrophages and DCs are intimately intermingled in the PIRs, it is unclear how T cells select DCs for priming over the more numerous macrophages.
To better understand the priming of antiviral CD8+ T cells in vivo, we have combined intravital multiphoton microscopy (MPM) with other approaches to dissect mechanisms controlling T cell activation during viral infection. We show that CD8+ T cells interact with both virally infected macrophages and DCs in vivo in the DLN. Macrophage interactions, however, cannot fully prime antiviral CD8+ T cells. To optimize priming, CD8+ T cells use chemokines to guide them to DCs in vivo, and neutralizing these chemokines diminishes the antiviral CD8+ T cell response.
RESULTS
Definition of a macrophage-rich region (MRR) of the inguinal LN
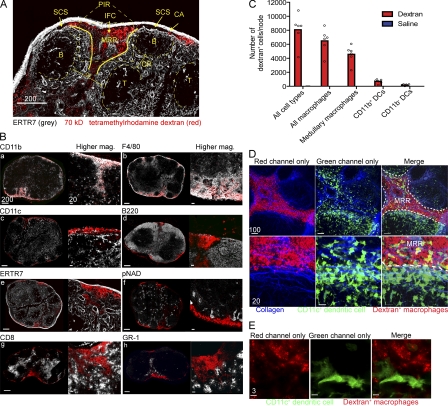
Visualizing macrophages and DCs in vivo using MPM necessitates the unambiguous, simultaneous fluorescent detection of both cell populations. Although CD11c-eYFP reporter mice allow DC analysis via MPM (Lindquist et al., 2004), no red fluorescent counterpart exists for macrophages. Gretz et al. (2000) showed that s.c. injected dextran ≥70 kD was initially excluded from cortical areas of the LN but flowed into the subcapsular and medullary sinuses, where it accumulated in unidentified phagocytic cells. Subsequently, Carrasco and Batista (2007) identified this region as a macrophage-rich area. Therefore, to visualize macrophages, we injected mice s.c. with 70 kD tetramethylrhodamine dextran (red fluorescent) and examined excised nodes at 30–45 min after injection by immunohistochemistry and flow cytometry (Fig. 1).
Figure 1.
Histological characterization of the MRR of the LN. (A and B) Frozen LN sections from WT mice given 70-kD tetramethylrhodamine-labeled dextran (red) 30 min before excision. (A) ER-TR7 staining (grey) identifies the following nodal regions: MRR, macrophage-rich region; PIR, peripheral interfollicular region; CA, capsule; SCS, subcapsular sinus; CR, cortical ridge; B, B cell follicle; T, T cell zone. (B) Mosaic-tiled confocal images of whole LNs stained with different Abs (grey, left images), and higher magnification images of staining of the MRR (right images). (C) Number of dextran+ cells per node per cell type as determined by node dissociation followed by flow cytometry. Cells were gated on FITC-dextran positivity and divided into CD11b+ DCs (CD11c+CD11b+ cells), CD11b− DCs (CD11c+CD11b− cells), and macrophages (CD11c−CD11b+ cells). Macrophages were further gated on F4/80 to identify medullary macrophages (F4/80+). Dextran+ cells belonging to each population were quantified using flow cytometric percentages and total node counts. (D) MPM images from CD11c-eYFP mice given dextran. Bottom panels show higher magnification. CD11c+ cells (green), collagen (second harmonic generation, blue), dextran+ cells (red). (E) High-magnification confocal image of whole-mounted CD11c-eYFP LNs after dextran administration. CD11c+ cells (green), dextran+ cells (red). Scale bars are shown in micrometers. We made similar observations in at least three additional experiments per image.
Dextran+ cells were clearly visible in frozen sections at the periphery of the LN, both in the SC and medullary sinuses, as well as in the interfollicular channels identified via staining of the LN stroma with ER-TR7 (Fig. 1 A). Dextran+ cells composed a distinct area of the cortical ridge (CR) region of the LN (Katakai et al., 2004) between the T cell zone and B cell follicles. Additional staining clearly identified the dextran+ cells as SCS and medullary macrophages (Fig. 1, B and C), indicated by expression of CD11b and CD169 (SCS macrophages) and F4/80 on medullary macrophages (Phan et al., 2009). Although the overwhelming majority of conventional (CD11c+) DCs do not endocytose dextran (visualized by MPM; Fig. 1, D and E), we detected a minor population of dextran+ CD11b+ CD11c+ DCs using flow cytometry (Fig. 1 C). Conventional DCs are positioned on either side of the dextran+ cells in the CR, clearly distinct from macrophages using MPM (Fig. 1, D and E; and Videos 1 and 2). This region of dense dextran+ macrophages overlaps but is not identical to either the PIR or CR and represents the MRR.
Together, these data support the use of dextran endocytosis for the in vivo labeling of LN macrophages in the sinuses and MRR.
SCS and medullary macrophages become infected by lymph-borne virus
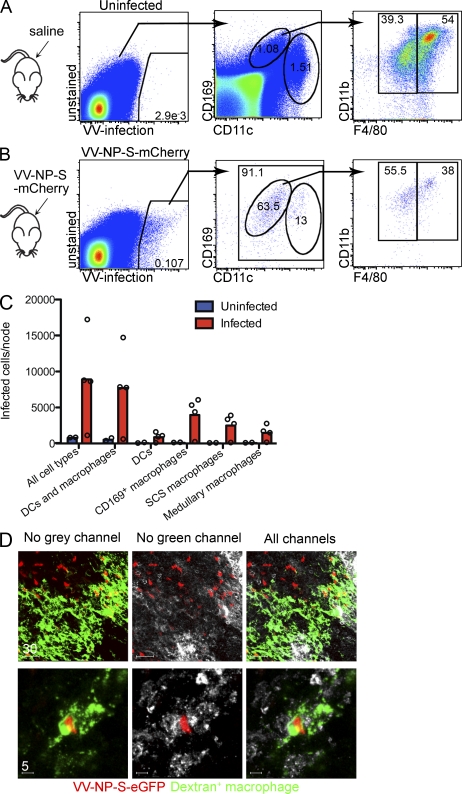
We previously reported that SCS macrophages are the predominant infected cell in the DLN after s.c. VV injection (Norbury et al., 2002; Hickman et al., 2008). Now other lymph-borne viruses have been shown to infect or accumulate in SCS macrophages (Hsu et al., 2009; Gonzalez et al., 2010). Although these studies relied exclusively on CD169 to identify SCS macrophages, it is now clear that medullary macrophages also express this marker (Phan et al., 2009).
Thus, we reexamined LNs (Fig. 2) using CD169 and F4/80 in flow cytometry to distinguish macrophage populations 8–10 h after infecting mice s.c. with a recombinant VV (rVV) expressing a red fluorescent protein (VV-NP-S-mCherry; Table I). This revealed that >90% of VV-infected LN cells were CD169+ or CD11chigh/intermediate (Fig. 2, A–C). Approximately 15% of infected cells were DCs, with the remainder being CD169high/intermediate. As expected, almost all of the CD169+ cells were also CD11b+, with 56% being F4/80dim SCS macrophages (Fig. 2 B, far right) and 38% being F4/80+ medullary macrophages.
Figure 2.
LN SCS and medullary macrophages are infected by VV. (A) Flow cytometric analysis of single-cell LN suspensions from saline-injected mice. CD169+CD11clo macrophages can be further divided into CD11b+F4/80− SCS or CD11b+F4/80+ medullary macrophages. (B) LNs from VV-infected mice 10 hpi. Cells were gated based on expression of a vaccinia-expressed fluorescent protein, into DCs and CD169+ macrophages, and finally on F4/80+ or F4/80− cells. (C) Numbers of vaccinia-infected cells in different nodal populations. Circles indicate individual LNs, and bars show mean. (D) Whole-mount confocal images of LNs 10 hpi, with VV-NP-S-mCherry (red). Macrophages, green (FITC-labeled dextran); F4/80, grey. Scale bars are shown in micrometers. Data are representative of three experiments.
Table I.
Viruses used in this study
| Abbreviation | Full name | Antigen | Fluorescent protein | Advantage |
| VV-ova | VV-ovalbumin | full-length ovalbumin (secreted) | none | standard construct |
| VV-SIINFEKL | VV-SIINFEKL | MSIINFEKL | none | not cross-primed |
| VV-NP-S-eGFP | VV-Influenza A virus nucleoprotein-SIINFEKL-enhanced green fluorescent protein | SIINFEKL as a fusion protein | eGFP | standard construct |
| VV-NP-S-mCherry | VV-Influenza A virus nucleoprotein-SIINFEKL-mCherry fluorescent protein | SIINFEKL as a fusion protein | mCherry | standard construct; red fluorescence |
| VV-Venus-ub-S | VV-Venus enhanced yellow fluorescent protein-ubiquitin-SIINFEKL | SIINFEKL as a minigene liberated via ubiquitin hydrolases | Venus eYFP | brightest fluorophore |
| VV-Venus-ub-NP | VV-Venus enhanced yellow fluorescent protein-ubiquitin-ASNENMETM | none for OT-I cells | Venus eYFP | control fluorescent virus |
| VV-β-gal | VV-β-galactosidase | none; backbone for recombinant VVs | none | control nonfluorescent virus |
To confirm this finding, we performed whole-mount F4/80 staining of nodes removed 10 h post infection (hpi; Fig. 2 D). We detected a substantial number of F4/80+, dextran+ VV-infected cells in the LN. Thus, both medullary macrophages and SCS macrophages are robustly infected by virus draining to the LN.
CD8+ T cells interact with VV-infected macrophages and DCs in vivo
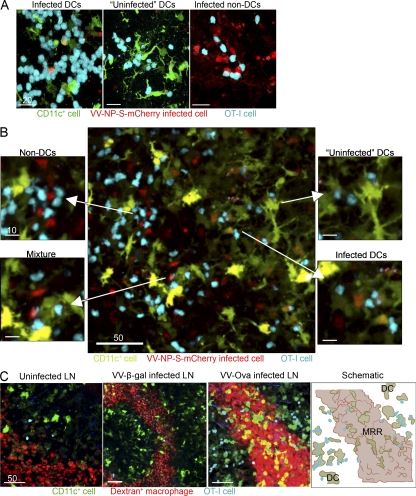
Previously, we showed that CD8+ T cells in virus-DLNs form intimate, enduring antigen-specific interactions with virus-infected cells, leading to T cell activation (Norbury et al., 2002; Hickman et al., 2008). Notably, interactions occurred in a relatively small subregion of the LN, the PIR, and antigen was required for T cell accumulation in the PIR and for sustained DC–T cell interactions (Hickman et al., 2008). However, we were technically unable to simultaneously identify virus-infected cells and DCs by MPM because of spectral overlap of the fluorescent proteins used to visualize viral gene products and DCs. By generating rVVs that express an antigenic protein genetically fused to mCherry (NP-S-mCherry) we could simultaneously image viral antigens, eYFP-expressing DCs, and adoptively transferred, fluorescently labeled, antigen-specific CD8+ T cells (OT-I cells, recognizing Kb-SIINFEKL; Clarke et al., 2000).
At 10 hpi, we imaged excised inguinal LNs via confocal microscopy (Fig. 3 A), or inguinal LNs in living animals using MPM (Fig. 3 B). In uninfected nodes or nodes infected with a control rVV lacking SIINFEKL, few OT-I cells are present in the PIR (Video 3). After VV infection with a SIINFEKL-containing virus (VV-NP-S-mCherry [red]), we commonly observed OT-I cells contacting infected DCs (Fig. 3, A and B; and Video 4). We also identified OT-I cells interacting with likely uninfected DCs (we cannot rigorously exclude expression of fluorescent protein below detection limits) and infected eYFP− cells identified as macrophages in other analyses (Video 4). We routinely identified multiple examples of each interaction occurring in individual LNs (Fig. 3 B).
Figure 3.
CD8+ T cells interact with LN DCs and macrophages after viral infection. (A) LNs from CD11c-eYFP mouse (green DCs) excised 10 hpi with VV-NP-S-mCherry (red) were examined as whole mounts by confocal microscopy. OT-I cells were labeled with Cell Tracker Blue before transfer. (B) MPM images of the inguinal LN, as in A. (C) MPM images from the top 30 µm of the inguinal node of CD11c-eYFP mice given fluorescent-dextran (red) to label macrophages and OT-I cells (blue). Uninfected nodes (left) 10 hpi with nonfluorescent backbone virus (VV-β-gal; control, middle left) or VV-Ova (middle right). (right) A schematic of T cell localization in relation to macrophages after VV infection. DCs (green), MRR (red), OT-I cells (blue; right). Scale bars are shown in micrometers. We made similar observations in two additional experiments.
We next labeled macrophages in CD11c-eYFP mice via s.c. dextran injection. Few OT-I cells were located at the periphery of the uninfected LNs, or in nodes infected with control virus (Fig. 3 C left and center panels and Video 3). In contrast, infection with VV expressing cognate antigen induced OT-I cells to localize to the PIR (Fig. 3 C, Video 3, and Video 5). Most (but not all) OT-I cells were excluded from the MRR (Fig. 3 C, schematic, far right).
Collectively, these data indicate that although most CD8+ T cells interact with infected and uninfected DCs, they can and do interact with VV-infected PIR macrophages under normal circumstances.
VV-infected macrophages stimulate T cell division but not activation
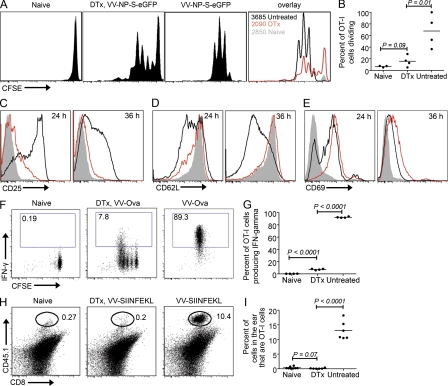
What happens when CD8+ T cells interact exclusively with infected macrophages? We forced T cells to use infected macrophages as APCs, using CD11c-diphtheria toxin (DTx) receptor (DTR)-eGFP mice (Jung et al., 2002) that were previously used for ex vivo analyses of immune responses to other viruses (Ciavarra et al., 2005; Probst et al., 2005; Kassim et al., 2006). In this system, administration of DTx results in DC ablation in vivo, leaving macrophages as the only infected cell type in the DLNs. Although long-term DC depletion requires generation of bone-marrow chimeras, our short-term assays were performed simply with transgenic animals. Thus, to measure T cell activation in the absence of DCs, we adoptively transferred OT-I cells (because of the potential effects of DTx on autologous T cells in DTR mice, which can express CD11c [and DTR] upon activation; Jung et al., 2002), administered DTx, and examined OT-I cell proliferation in rVV-infected LNs (Fig. 4, A and E). DTx treatment greatly reduced (but did not eliminate) OT-I cell proliferation in response to VV-encoded antigen. We next analyzed several common T cell activation markers (Fig. 4, C–E). Macrophage-activated T cells failed to normally up-regulate CD25 or down-regulate CD62L (Fig. 4, C and D). Although CD69 was induced in OT-I cells, it was dysregulated in comparison to cells activated in the presence of DCs (at 36 h there was less down-regulation compared with WT; Fig. 4 E), and CD69 was not up-regulated at all under the same circumstances in different TCR transgenic T cells (unpublished data). Reducing the number of OT-I cells transferred failed to restore their normal activation phenotype (unpublished data), indicating that this phenomenon is unlikely to be an artifact of using a high precursor frequency of TCR-transgenic cells (Badovinac et al., 2007).
Figure 4.
CD8+ T cell interactions with macrophages are largely nonproductive. (A) LN OT-I cells dividing in response to s.c. delivery of VV-NP-S-eGFP. 5 × 106 OT-I CFSE-labeled OT-I cells were transferred into CD11c-DTR-eGFP mice. 12 h later, mice were treated with DTx or PBS (untreated). After an additional 12 h, mice were infected with VV (untreated and DTx) or left uninfected (naive). Cells were analyzed at 48 hpi for division. Far right panel shows overlays. Untreated mice, black lines; DTx-treated mice, red lines; uninfected DTx-treated, filled grey lines. Numbers = OT-I cells recovered. (B) Graph showing percentage of dividing OT-I cells. Dots represent individual nodes. (C–E) Activation marker profile of T cells. Histograms were only gated on dividing cells. Untreated mice, black lines; DTx-treated mice, red lines; uninfected DTx-treated, filled grey lines. Numbers in top right corner indicate time after infection. (F) IFN-γ production (y axes) versus division (indicated by CFSE dilution; x axes) at 2 days after infection with VV-Ova. (G) Graphical representation of data shown in F. Dots represent individual nodes. (H) 2 × 106 OT-I cells were transferred into CD11c-DTR-eGFP mice (expressing CD45.2) that were treated with DTx (middle) or PBS (right) before intradermal infection with VV-SIINFEKL in the ear pinnae. 4 days after infection, ears were removed and analyzed for the percentage of OT-I cells present (CD8+CD45.1+ cells). (I) Graphical representation of data in H. All experiments were performed at least three times with three to six animals/group yielding similar results.
Such macrophage activated-CD8+ T cells demonstrated functional defects as well (Fig. 4, F–I). OT-I cells were transferred into CD11c-DTR-eGFP mice that received DTx 12 h. before infection with VV-Ova (Table I). In untreated mice, ∼90% of OT-I cells produced IFN-γ at 2 d after infection when restimulated with SIINFEKL peptide (compared with <0.2% when stimulated with an irrelevant peptide; Fig. 4, F and G). In CD11c-depleted mice, however, <8% of cells produced IFN-γ in response to SIINFEKL peptide, and even cells that had divided 5 times synthesized less IFN-γ on average. DTx treatment did not modify IFN-γ production in non–DTR transgenic (C57BL/6) mice (unpublished data).
A hallmark of effector CD8+ cells is their ability to traffic from LNs to peripheral infection sites. We transferred OT-I cells into CD11c-DTR-eGFP mice treated with DTx or untreated-mice and infected intradermally in the ear with VV-SIINFEKL. 4 d later, ears were removed and examined for the presence of OT-I cells (Fig. 4, H and I). Remarkably, macrophage-primed OT-I cells from DTx-treated mice completely failed to traffic to the infection site.
Together, these data indicate that macrophage-primed CD8+ T cells exhibit diminished proliferation, altered activation, and greatly reduced functionality, pointing to the importance of DC-mediated activation in this system.
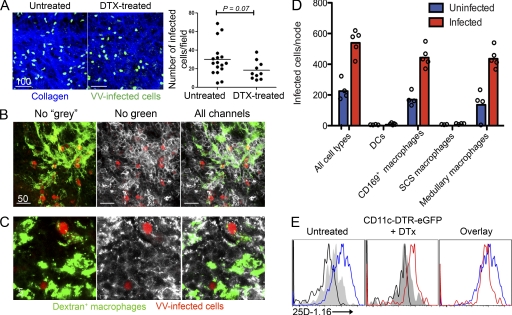
DC-ablated LNs still present viral antigen
Several possible explanations exist for the altered priming in DTx-treated CD11c-DTR-eGFP mice, the simplest of which is decreased viral infection/antigen presentation in the DC-depleted LNs. To analyze this, we infected DTX-treated CD11c-DTR-eGFP mice or nontreated WT mice with VV-NP-S-eGFP and visualized infected cells by MPM (Fig. 5 A). Numerous VV-infected cells were present just beneath the SCS of the DLN in untreated or DTx-treated animals (Fig. 5 A). Most infected cells were macrophages (Video 6), as indicated by in vivo endocytosis of fluorescent dextran administered s.c., and confirmed by CD11b+ staining (Fig. 5 B) or F4/80 staining (Fig. 5 C) of whole-mount LN sections. Using flow cytometry, we found that almost all of the infected LN cells recovered from DTx-treated mice (∼10-fold fewer than recovered from untreated mice) were CD11b+, CD169+, F4/80+ medullary macrophages (Fig. 5 D).
Figure 5.
Infected macrophages present antigen in DC-ablated mice. (A) VV-NP-S-eGFP–infected cells (green) just under the collagenous capsule of the node (second harmonic generation (blue)). Numbers of infected cells in nonablated (untreated) and DC-ablated (DTx-treated) mice (right). (B) Confocal microscopy of a frozen section of a DC-ablated LN 10 hpi with VV-NP-S-mCherry (red) showing only the red and green channels (left), only the red and grey channels (middle), or all channels (right). FITC-dextran (green) and anti-CD11b staining (white) identify macrophages. (C) Same as shown in B, except staining with F4/80 (grey) to identify medullary macrophages. (D) Numbers of infected cells of each cell type in the node. Cells were gated into CD11c+CD169− (DCs) or CD11cdimCD169+ (CD169+ macrophages), then CD169+ macrophages were further gated based on F4/80 expression. Background (blue) is caused by low cell recovery and macrophage autofluorescence. (E) Flow cytometric analysis of LN single-cell suspensions from untreated or DTx-treated animals 6 hpi with VV-venus-ubiquitin-SIINFEKL. Cells were stained with 25D-1.16 recognizing Kb-SIINFEKL complexes. Histograms are gated on infected cells (Venus eYFP+). Infected with virus lacking SIINFEKL (grey shaded histograms), infected without 25D-1.16 stain (black lines), infected untreated mice (blue lines), and infected DTx-treated mice (red lines). Scale bars are shown in micrometers. Results are shown from one experiment of two (A–C) or four (D and E).
To examine the ability of DTx-resistant macrophages to present rVV antigens, we infected mice with a rVV expressing a highly fluorescent fusion protein (Venus-ubiquitin-SIINFEKL) that generates large amounts of cytosolic SIINFEKL because of co-translational liberation of SIINFEKL by cellular ubiquitin hydrolases (or an irrelevant control virus expressing Venus-eYFP but lacking SIINFEKL; Table I; Fruci et al., 2003; Lev et al., 2010). We removed DLN 6 hpi and measured expression of Kb-SIINFEKL on dissociated cells by flow cytometry using a fluorescent conjugate of the TCR-like antibody 25D-1.16 (Fig. 5 E). Cells from both untreated and DTx-treated mice were specifically stained with 25-D1.16, demonstrating that APCs in DTx-treated mice are able to generate class I peptide complexes from endogenous antigens, although at lower levels than in untreated mice.
Collectively, these data demonstrate that medullary macrophages are infected in DTX-treated mice and infected cells still robustly present antigen in the node, albeit at lower numbers and antigen levels than in untreated mice.
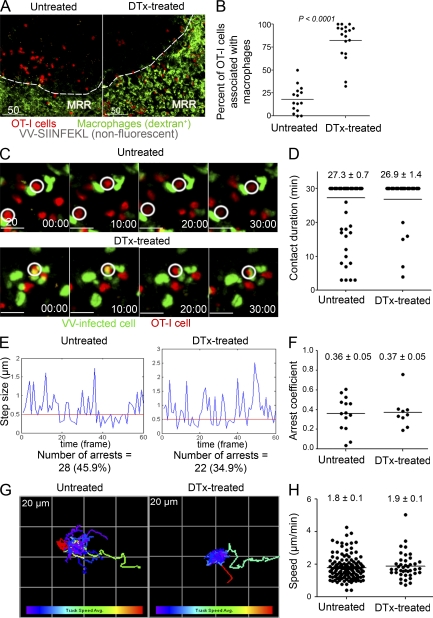
CD8+ T cells establish stable interactions with infected macrophages in the absence of DCs
How do CD8+ T cells behave in the absence of DCs? In DC-competent mice, T cells rapidly transit to the periphery after infection, forming stable contacts with virus-infected cells in the presence of cognate antigen (Hickman et al., 2008). Notably, in the absence of specific antigen, CD8+ T cells in the periphery move at reduced speeds compared with those in the deeper T cell zone, yet fail to form immotile, stable contacts with non–antigen-expressing APCs. To analyze T cell behavior in DC-ablated mice, we transferred 107 OT-I cells into CD11c-DTR-eGFP mice, treated mice with DTx, infected with VV-SIINFEKL 12 h later, and imaged the DLN 6 hpi after injection of fluorescent dextran to identify macrophages (Fig. 6, A and B and Video 6). Remarkably, in DTx-treated mice, 82% of OT-I cells localized to the MRR compared with 18% in untreated mice. Complete T cell activation requires stable prolonged contact between T cells and APCs (Mempel et al., 2004; Henrickson et al., 2008). MPM revealed that most OT-I cells in the MRR of DTx-treated mice were relatively immotile throughout the course of three consecutive 20-min imaging sessions (the first of which is shown in Video 7). To image the interaction between OT-I cells and infected macrophages, we transferred OT-I cells into mice 12 h before DTx treatment, and then infected them with VV-NP-S-eGFP. 6–10 hpi, we imaged OT-I cells interacting with VV-infected cells in the DLN (Fig. 6 C and Video 8). In both control and DTx-treated mice, OT-I cells formed numerous tight contacts with infected cells. DTx treatment had no significant effect on the mean contact time (27 min in each case during a 30-min imaging period), arrest coefficients (the fraction of time OT-I cells paused on infected cells), or mean T cell speeds (Fig. 6, C–H).
Figure 6.
CD8+ T cells stably interact with VV-infected cells in DC-ablated mice. (A) MPM images of untreated or DTx-treated CD11c-DTR-eGFP animals that were given 1.0 × 107 Cell Tracker Red (red) labeled OT-I cells before DC depletion. Images acquired 6 hpi with VV-SIINFEKL (nonfluorescent). The macrophage rich-region (MRR) was identified by in vivo uptake of FITC-dextran (green). (B) Percentage of OT-I cells per 63X field (238 × 238 × 85 µm) located in the MRR in untreated or DTx-treated mice. Results were analyzed with an unpaired t test. (C) Time-lapse MPM images of OT-I cells (red) in the LN of untreated (top panels) or DC-ablated (bottom panels) mice 6–10 hpi with VV-NP-S-eGFP (green). White circles indicate stable contacts. For untreated mice, 106 OT-I cells were transferred; for DTx-treated, 107 (it was necessary to transfer more OT-I cells into DTx-treated mice than untreated mice due to decreased homing to the LN following DTx-mediated ablation). (D) Calculation of contact times between OT-I cells and VV-infected cells over the course of a 30 min imaging session. (E) Plot of OT-I cells’ movement (step size) between individual frames of a 30 min movie in untreated vs. DC-ablated mice. We classified movement under 0.5 µm between frames as a pause step. (F) Calculation of the percentage of T cells arresting in each condition. (G) Color-mapped plot of T cell tracks during a 30-min movie. Each box on grid is 20 × 20 µm. Increasing track speed is colored from purple to red (bottom bar). (H) Calculation of average OT-I cell speed in untreated or DTX-treated mice. Results are shown from one experiment of three to six with similar results. No differences between untreated and DTx-treated mice were statistically significant according to unpaired Student’s t tests. Time is shown in minutes. Scale bars are shown in micrometers.
Collectively, these data demonstrate that in the absence of DCs, OT-I cells migrate into the MRR and form contacts with VV-infected macrophages that are grossly indistinguishable from their contacts with infected DCs.
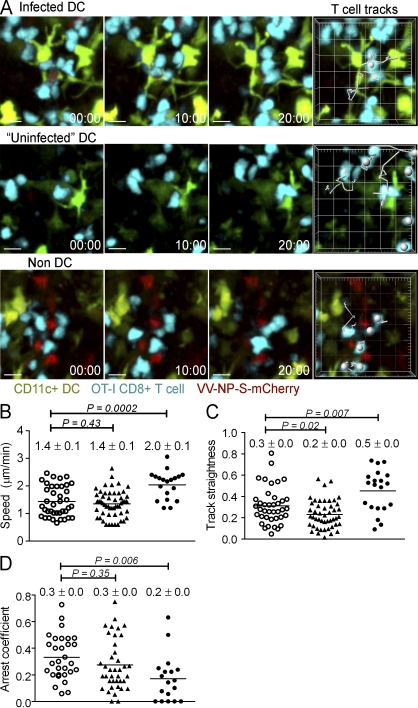
CD8+ T cells rapidly scan infected non-DCs
Although T cells can form stable contacts with macrophages when DCs are absent, we reasoned that the presence of DCs could significantly impact the nature of contacts between T cells and macrophages. Therefore, we next used MPM to analyze contacts in CD11c-eYFP mice between T cells and DCs or macrophages 6 h after VV infection. Over a 20-min imaging period, we tracked T cells moving among infected (direct-priming) DCs, uninfected DCs (which likely consisted of cross-priming DCs and those expressing amounts of fluorescent proteins below detectable levels), and infected non-DCs (macrophages; Fig. 7 A). Although there was no clear difference in the speed of T cells interacting with infected and uninfected DCs (∼1.4 µm/min, similar to values for previously unidentified infected cells; Hickman et al., 2008), T cells interrogated infected non-DCs at a significantly higher speed (Fig. 7 B). Likewise, T cells closely scanned DCs in a curved path, but moved in straighter lines (indicated by higher track linearity) when interacting with infected non-DCs (Fig. 7 C). Finally, we monitored T cell arrest over a 30-min period for T cells interacting with each APC category (Fig. 7 D). T cells arrested for similar periods over infected and uninfected DCs, but paused for shorter amounts of time over non-DCs.
Figure 7.
CD8+ T cells rapidly scan infected non-DCs. (A) MPM time series of T cells interacting with infected DCs, uninfected DCs, or infected non-DCs. CD11c-eYFP mice were given 107 Cell Tracker Blue-labeled OT-I cells (blue) and were infected s.c. with VV-NP-S-mCherry 12 h. later. Six hpi, inguinal LNs were imaged over sequential 20–30 min periods using MPM. The tracks of individual T cells during a 20-min imaging period are plotted (right). (B) T cell speeds, (C) track straightness (T cell displacement/track length), and (D) T cell arrest calculated for each type of APC interaction over 30 min. Open circles, infected DCs; closed triangles, uninfected DCs; closed circles, macrophages. Mean and SEM is shown. Statistics were performed using an unpaired Student’s t test. Shown is one experiment of two analyzed with 10–20 nodes per group.
Overall, these data indicate that T cells preferentially form stable contacts with DCs (whether overtly infected or not) over infected macrophages.
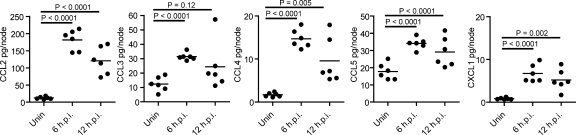
VV induces chemokine production in the DLN
What factors modulate CD8+ T cell interaction with and activation by DCs? The migration of OT-I cells into the MRR in the absence of DCs demonstrates that CD8+ T cells are fully capable of entering the MRR, suggesting chemotactic recruitment of CD8+ T cells to PIR DCs. Chemokines attract naive CD8+ T cells to antigen-bearing LN DCs in noninfectious models (with peptide-pulsed DCs or antigen targeted to DCs via conjugated Abs; Castellino et al., 2006; Hugues et al., 2007). Thus, we performed bioplex analysis for the protein levels of several chemokines on excised nodes. VV infection increased levels of 5 chemokines examined at 6 hpi, with all except CCL3 being significantly greater at 12 hpi (Fig. 8). The greatest increases were seen with CCL2 and CCL4. We confirmed VV induction of CCL3 expression in BMDCs (unpublished data).
Figure 8.
VV infection induces rapid DC chemokine secretion. Mice were infected s.c. with VV-Ova and LN harvested at 6 or 12 hpi Chemokine protein levels were determined from clarified node homogenates via Bioplex assay. Dots represent individual mice. Data are shown from two of four independent experiments.
Chemokine-neutralizing Abs impair the primary CD8+ T cell response to VV
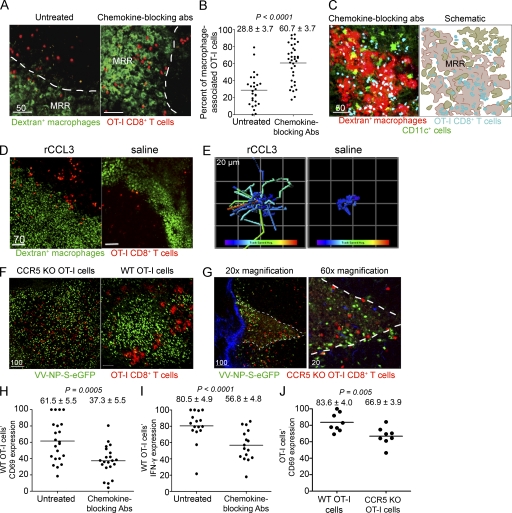
Do VV-induced chemokines play an essential role in attracting CD8+ T cells to DCs during viral infection? Remarkably, administering a cocktail of neutralizing Abs against CCL3, CCL4, and CCL5 dramatically redistributed OT-I cells from DCs into the MRR (Fig. 9), which is visualized clearly using either two-color (red T cells, green macrophages; Fig. 9 A) or three-color MPM (blue CD8+ T cells, green DCs, and red macrophages; Fig. 9 C and Video 9). In untreated mice, ∼30% of OT-I cells were associated with MRR macrophages, but in mice treated with anti-CCL3, -CCL4, and -CCL5 Abs, >60% of OT-I cells localized in the MRR (Fig. 9 B). s.c. injection of recombinant CCL3 (introducing artificial gradients of CCL3 in the uninfected node) also induced T cell scanning of both DCs and the MRR early after VV infection (Fig. 9, D and E and Video 10). Additionally, CCR5KO OT-I cells failed to form large clusters with VV-infected cells at 6 hpi (Fig. 9 F), and many of the cells were located in the MRR (visualized using the natural autofluorescence of the region; Fig. 9 G and Video 11).
Figure 9.
Chemokine-based CD8+ T cell homing to DCs. (A) MPM images of animals that were given 1.0 × 107 Cell Tracker Red (red)–labeled OT-I cells in the presence or absence (untreated) of chemokine-neutralizing Abs against CCL3, CCL4, and CCL5. Images acquired 6–8 hpi with VV-Ova (nonfluorescent). MRR delineated by white lines. (B) Percentage of OT-I cells per 63× field (238 × 238 × 85 µm) located in the MRR in untreated or antibody-treated mice. Results were analyzed with an unpaired Student’s t test. (C) MPM images from the top 30 µm of the inguinal node of CD11c-eYFP mice given fluorescent-dextran (red) to label macrophages and OT-I cells (blue). Mice were given chemokine neutralizing Abs; nodes imaged at 8 hpi with nonfluorescent VV-Ova. (right) A schematic of T cell and macrophage localization after VV infection with CCR5-ligand blockade. Scale bars are shown in micrometers. (D and E) OT-I cells (red) in the MRR (green) after s.c. administration of recombinant CCL3 at 2.25 hpi with VV-SIINFEKL (nonfluorescent; E) tracks of OT-I cells in the presence (left) or absence (right) of rCCL3 2–3 hpi. Tracks are colored according to mean track speed (slowest [purple) to fastest [red]). (F) Distribution of CCR5KO OT-I cells (red, left) or WT OT-I cells (red, right) 6 hpi with VV-NP-S-eGFP (green). (G) CCR5KO OT-I cells (red) in the MRR (visualized using the intrinsic autofluorescence of macrophages) at 6 hpi (virus=green, collagen=blue) (H) CD69 expression by LN OT-I cells 15 hpi with VV-Ova in the presence or absence of chemokine-neutralizing Abs. Data were averaged from three independent experiments normalized using the highest mean fluorescence intensity in an individual experiment as 100. (I) IFN-γ production by OT-I cells in the node 48 hpi with VV-Ova. Data were compiled from two independent experiments and normalized to the highest mean fluorescence intensity per experiment. (J) Same as described in H, but with CCR5KO OT-I cells. All experiments were performed at least twice with three to six mice/group.
We examined the participation of these chemokines in regulating antiviral immune responses via APC selection by s.c. injecting a mixture of CCL3-, CCL4-, and CCL5-blocking Abs at the time of infection. Chemokine neutralization reduced OT-I CD69 induction (an early indicator of activation) 15 hpi with VV-Ova (Fig. 9 H), and decreased IFN-γ synthesis 2 d after infection (Fig. 9 I). In contrast, injection of isotype-control Abs did not impact T cell activation (unpublished data). In untreated, infected LNs, CCR5KO OT-I cells (lacking the CCR5 receptor for these chemokines, but retaining CCR1 which also signals via several CCR5 ligands) up-regulated CD69 to a lesser extent than WT OT-I cells (Fig. 9 J).
Together, these findings demonstrate that the chemokines CCL3, CCL4, and CCL5 play essential roles in enabling T cells to reach a complete effector differentiation state and maximizing primary antiviral CD8+ T cell responses by enhancing CD8+ T cell interactions with DCs in the infected node.
DISCUSSION
After s.c. delivery of infectious virions, the most rapid antigen presentation occurs via direct priming. Within seconds, lymphatics convey virions to LNs, where they can infect macrophages and DCs in the LN parenchyma (Hickman et al., 2008). Because viral peptides are generated primarily from DRiPs (Dolan et al., 2011), the onset of viral protein synthesis is immediately accompanied by the generation of class I peptide complexes, which can rapidly initiate CD8+ T cell priming. For VV, the initial phase of direct priming likely persists for 12–24 h (Hickman et al., 2008), after which infected DCs and macrophages succumb to viral cytopathogenesis or NK-mediated lysis. Because the first 12 hpi are critical for activating antiviral T cells after s.c. VV infection (Hickman et al., 2008), it is imperative that T cells rapidly contact the appropriate infected APC.
The second wave of antigen presentation likely involves transfer or acquisition of antigen by LN-resident APCs. CD8α+ DCs are particularly adept at cross-priming antigens acquired from dead or dying cells (den Haan et al., 2000; Iyoda et al., 2002). For viruses that infect dermal epithelial cells, like herpes simplex virus-1, these cells provide an antigen source for cross-priming by migratory dermal DCs (Bedoui et al., 2009). To date, antiviral cross-priming has been investigated largely by ex vivo analyses of cells recovered from dissociated tissues (Allan et al., 2003; Belz et al., 2004; Bedoui et al., 2009). It will be important in future studies to confirm the central role of CD8α+ DCs in antiviral cross-priming using MPM, although it will be challenging to unambiguously identify APCs actively engaged in cross-priming and not direct priming after synthesis of undetectable levels of viral antigens.
The LN CR, a reticular stromal structure at the border of the T/B zones, likely serves as a staging ground for both direct and cross-priming T cell–APC interactions (Katakai et al., 2004). Migratory, antigen-bearing DCs are thought to accumulate in the CR, positioning themselves to maximize cross-priming interactions with nearby T cells (Bajénoff et al., 2003; Lindquist et al., 2004). We show here that direct priming interactions independent of DC migration also occur in the outermost edges of the CR as virions drain to the LN and are captured by resident APCs. After the first wave of virus-infected cells die or are eliminated in the LN, it will be interesting to determine the location of virus-specific CD8+ T cell–APC interactions in relation to the macrophage-rich and dendritic regions of the CR.
Controversy swirls around the definition of macrophages versus DCs and their putative differences in priming capacity. Hume (2008) has eloquently argued that DCs are simply mononuclear phagocytes that do not comprise a unique cellular subset, and are not especially adept at priming. A key element of Hume’s argument is that the heightened priming ability of DCs relative to macrophages ex vivo is caused by removing suppressive macrophages during the process of isolating DCs. Here, we show that in vivo, T cells scan DCs and macrophages at different rates in infected LNs. Additionally, LN depletion of CD11c+ cells, which eliminates nearly all DCs and SCS macrophages, has profound effects on antiviral priming, despite our direct observations that CD8+ T cells now form contacts with macrophages that are indistinguishable by IVM from their contacts with DCs under nonablative conditions. This argues strongly that DCs have a unique role in direct priming that macrophages cannot replace.
What is the role for the large numbers of infected LN macrophages if not for priming naive CD8+ T cells? Recently, Asano et al. (2011) showed that these macrophages serve as facile APCs via cross-presentation of tumor associated antigens, making their failure to drive antiviral T cell responses even more enigmatic. Several imaging studies have now shown antigen acquisition by SCS sinus macrophages and subsequent antigen donation to LN B cells (Batista and Harwood, 2009). Perhaps virally infected nodal macrophages specialize in presentation to B cells and are largely ignored by T cells (much like CD8α+ DCs preferentially present to CD8+ and not CD4+ T cells; Dudziak et al., 2007). Additionally, these macrophages express the molecule sialoadhesin, a newly identified participant in regulatory T cell function and expansion, raising the possibility that macrophage infection activates suppressive rather than effector T cells (Wu et al., 2009).
We show that macrophages prime CD8+ T cells that are sub-optimally activated by the standard criteria. It remains to be determined, however, whether there is method to this madness and the “partially” activated cells are fully activated for a specific alternative function in primary or memory responses. Alternatively, macrophages may serve to dampen initial CD8+ T cell responses generated via direct priming, favoring instead cross-priming at later time points. In any event, it is a safe assumption that macrophages perform multiple functions in the infected LNs.
In clearly demonstrating the essential role DCs play in direct antiviral priming, our findings emphasize the importance of understanding the basis for CD8+ T cell attraction to infected PIR DCs. This knowledge is likely to be useful in maximizing the ability of vaccines to elicit effective CD8+ T cell responses. We provide the initial evidence that CCR5 and R1-signaling chemokines, known to guide CD8+ T cells to cross-priming DCs in noninfectious models (Castellino et al., 2006; Hugues et al., 2007), also play a critical role in direct priming during viral infection. We show, first, that viruses rapidly induce chemokines in the LN, and second, that an Ab cocktail that neutralizes several of the known CCR5 ligands inhibits both DC attraction of CD8+ T cells and CD8+ T cell IFN-γ responses. Additionally, these experiments likely underestimate the importance of CCR5 agonist chemokines in direct antiviral CD8+ T cell priming, since it is unlikely that the Ab cocktail we used covers all CCR5 agonists or completely neutralizes the agonists covered. Likewise, experiments using CCR5KO OT-I cells will similarly undervalue the effect of these chemokines on CD8+ T cell responses as the cells retain signaling through CCR1–CCL3 and –CCL5 interactions.
Because of its relevance for rational vaccine design, there is tremendous interest in identifying APC types that most effectively prime functional CD8+ T cell responses. We have identified a factor that optimizes CD8+ T cell targeting to appropriate APCs during viral infection. This raises the exciting possibility of engineering vaccines to express optimal antigens as well as a guidance system to attract CD8+ T cells to DCs for maximal priming of effector cells with optimal effector profiles.
MATERIALS AND METHODS
Mice.
Specific pathogen–free CD11c-DTR-eGFP transgenic mice on a C57BL/6 background were acquired from The Jackson Laboratory (stock # 4509) and bred in house. Nontransgenic WT controls were C57BL/6 mice. CD11c-eYFP mice (Lindquist et al., 2004) were acquired through the National Institute for Allergy and Infectious Disease (NIAID) Intramural Research Repository and are now available from The Jackson Laboratory (stock# 8829). CCR5KO mice (The Jackson Laboratory; stock #5427) were crossed to OT-I TCR transgenic mice (acquired through the NIAID Intramural Research Repository) and bred for homozygosity. 6–16-wk-old adult mice were used in all experiments. All mice were housed under specific pathogen–free conditions (including MNV, MPV, and MHV) and maintained on standard rodent chow and water supplied ad libitum. All animal procedures were approved by and performed in accordance with the NIAID Animal Care and Use Committee.
DC depletion and adoptive transfer.
DC depletion was performed as described previously (Jung et al., 2002). Mice were given a single of dose of DTx at 4 ng/g body weight (Sigma-Aldrich) i.p. in PBS unless another dose is noted. CD8+ T cells (TCR transgenic) were transferred 12 h before depletion. Virus infections were always performed 12–24 h after DTx administration, allowing complete DC depletion before infection. Mice received only one dose of DTx.
Dextran and virus injection.
Approximately 25 µg of FITC- or tetramethylrhodamine-conjugated 70 kD dextran (Invitrogen) was injected s.c. 30–45 min before LN analysis where indicated. VV was injected s.c. (3.5 × 107 PFU) or footpad (104 PFU) in sterile saline. Route of infection is indicated for each figure. Previously described VVs used in this study include the following: VV-Ova, VV-NP-S-eGFP, VV-SIINFEKL, VV-Venus-ubiquitin-SIINFEKL, VV-Venus-ubiquitin-NP366-374 (Norbury et al., 2002; Norbury et al., 2004; Lev et al., 2008). VV-NP-S-mCherry was constructed according to established protocols (Earl et al., 2001).
Adoptive transfers.
CD8+ T cells were purified from TCR-transgenic rag−/− mice by negative selection using magnetic-activated cell sorting (MACS) according to the manufacturer’s instruction (Miltenyi Biotec). Cells were 95–99% pure by flow cytometry. Purified cells were labeled with 2 µM CFSE (Invitrogen) for 10 min at room temperature in PBS. For IVM, cells were labeled with 2 µm CMPTX or CMF2HC (Invitrogen). The amount of T cells transferred is indicated in specific figure legends.
Confocal whole-mount microscopy.
Whole LNs were removed at indicated time points after infection and placed in 3.2% paraformaldehyde for 2 h at 4°C. For CD11c-eYFP experiments, mice were given 107 CMF2HC-labeled (blue) OT-I cells 12 h before infection. Where indicated, mice were given lysine-fixable fluorescent dextran s.c. (Invitrogen) 30–45 min before removal. Nodes were washed extensively in PBS, and then stained overnight with anti-CD11b (clone M1/70; eBioscience) conjugated to Alexa Fluor 647, or for 2 h with anti-F4/80 (clone BM8; eBioscience). Nodes were washed in PBS before slicing into smaller pieces, which were mounted on slides in Fluoromount G (Electron Microscopy Sciences). Slides were imaged on an SP5 confocal microscope (Leica).
Ex vivo analyses of dissociated LNs.
For flow cytometric determination of the nature and number of infected cells per node, untreated or CD11c-DTR-eGFP mice treated with DTx were given 3.5 × 107 PFU VV-NP-S-mCherry 8–12 h before removal of inguinal LNs. Single-cell suspensions were generated via digestion in collagenase (Worthington Biochemical Corporation) + dispase (Roche) before staining for CD11c, CD11b, CD169, F4/80, and GR-1. Cells were gated on mCherry+ vaccinia-infected cells before additional analyses of individual cell populations.
For determination of the nature and number of dextran+ cells, mice were given 0.25 µg FITC-conjugated 70 kD dextran 30–45 min before node removal and digestion with collagenase + dispase. Single-cell suspensions were stained for CD11c, CD11b, F4/80, and GR-1. Cells were gated on the FITC+ population before analyzing individual cell types.
Histochemistry on frozen LN sections.
LNs were removed and embedded in OCT medium (Electron Microscopy Sciences) and frozen in dry-ice cooled isopentane. 20 µm sections were cut on a Leica cryostat (Leica). Sections were fixed in ice cold-acetone for 5 min before blocking with 5% goat or donkey serum, then staining with the following Abs: ER-TR7 (clone ER-TR7; Abcam), CD11b (clone M1/70; eBioscience), F4/80 (clone BM8; eBioscience), CD11c (clone N418; eBioscience), B220 (clone RA3-6B2; eBioscience), pNAd (clone Meca-79; BD), CD8 (clone 53–6.7; eBioscience), and GR-1 (clone RB6-8C5; eBioscience).
MPM.
CD8+ T cells labeled with CFSE, CMTPX (C42H40ClN3O4), or CMF2HC (4-chloromethyl-6,8-difluoro-7-hydroxycoumarin; all from Invitrogen) were injected intravenously 12–24 h before injection with virus, unless otherwise specified. When indicated, mice were given 25 µg FITC- or rhodamine-conjugated 70-kD dextran (Invitrogen) s.c. for drainage to the inguinal LN. Two-photon imaging used an inverted TCS-SP2 MP confocal microscope (Leica) equipped with a 20× objective (numerical aperture, 0.7) or 63× objective (numerical aperture, 1.30) and with 80% glycerol as the immersion medium for each objective. Two-photon excitation was provided by a Mai Tai Ti:Sapphire laser (Spectra Physics) with a 10-Watt pump, tuned to 800 nm for imaging of cells labeled with CMTPX, FITC, or CMF2HC; 850 nm for imaging of cells labeled with CMTPX in combination with eGFP; or 900 nm for imaging of eGFP alone or for imaging of second-harmonic generation. Emitted fluorescence was collected with a two-channel nondescanned detector. Wavelength separation was accomplished with a dichroic mirror at 560 nm, followed by emission filters of 525/50 nm bandpass and 610/75 nm bandpass. For imaging of cellular interactions, the 63× objective was used, and stacks of 25 sections obtained with a 2.5-µm z-step for a total depth of 40–60 µm were obtained every 30 s.
For imaging of CD11c-eYFP LNs after infection, a TCS-SP5 MP confocal microscope (Leica) equipped with dual MP lasers was used. One laser was set at 800 nm for imaging dye-labeled cells and CMF2HC-labeled OT-I cells, and the other was set at 900–930 nm for imaging VV-NP-S-mCherry and eYFP. Images were obtained in sequential mode using the same 63× objective as in the previous paragraph. Sections were taken at 2.5-µm intervals for a total depth of 40 µm. Series were acquired every minute.
Data analyses.
Data were analyzed with Imaris 64 version 6.1.2 (Bitplane). Images were processed using the Gaussian filter algorithm. Contact times between VV-infected cells and OT-I cells were calculated manually and compiled from 3–5 separate movies for each condition (DTx-treated or WT). Contacts were defined as being in close proximity (<1 µm) from a nuclear-expressed fluorescent protein. Arrest coefficients were calculated using the Imaris XT function “plot length of selected track,” and the percentage of pause steps for all of the T cells in a given image were plotted using GraphPad Prism 5. Overlays of tracks were generated using Imaris XT function “translate tracks,” and then pseudocolored according to the mean speed of the cell creating the track. Average speeds were calculated using the spot-detection function and the following parameters: autoregressive motion, gapclose 3, 7.5-µm object diameter, 40 µm maximum distance.
The number of infected cells under DTx-treated or untreated conditions was calculated from 3–6 fields 63× fields (238 × 238 × 85 µms) chosen randomly in 3 separate VV-infected LNs. The percentage of cells in the MRR was calculated by determining the number of OT-I cells associated with the MRR compared with the total number of OT-I cells in the 63× field. 3–6 fields per LN were selected for the MRR and the zone just outside it (but otherwise randomly); data from at least four individual LNs were compiled. Results were analyzed using GraphPad Prism 5 software. Means are indicated with a solid line and standard error of the mean is shown. Groups were analyzed using an unpaired Student’s t test.
Bioplex assay.
LN chemokines were analyzed during infection using a bead-based assay (Bioplex; Bio-Rad Laboratories) and results were read out on a Bioplex Suspension Array System (Bio-Rad Laboratories). Experiments were basically performed according to the manufacturer’s instructions. To gain more reproducible virus drainage to the LN, we analyzed cytokines within the popliteal LN instead of the inguinal. Mice were infected for various time points with VV-NP-S-eGFP (104 PFU footpad), and the skin-draining popliteal LN was removed and placed 250 µl buffered saline solution with 0.1% BSA. Two nodes were pooled for a single sample. LNs were homogenized in solution, and samples were centrifuged at 14,000 g for 6 min to remove cellular debris. Exactly 100 µl of clarified supernatant was used per sample, and samples performed in duplicate. Results were analyzed using GraphPad Prism Software Version 5.0 (GraphPad). Error bars represent SEM, and groups compared using unpaired Student’s t tests.
Kb-SIINFEKL complex staining.
CD11c-DTR-eGFP mice received 4 ng/g DTx i.p. or PBS i.p. 12 h before s.c. infection with VV-Venus-ubiquitin-SIINFEKL or VV-Venus-Ub-NP366-374. 6 hpi with VV, draining inguinal LNs were removed and digested with collagenase type II (Worthington Biochemical Corporation). Single-cell suspensions were stained with 25D-1.16-Alexa Fluor 647 (produced in-house but comparable product available from eBioscience) and propidium iodide and analyzed on a LSRII flow cytometer (BD). Plots were gated on live cells and Venus (eYFP)+ cells before analysis of 25D-1.16 staining.
Chemokine-neutralizing Abs and recombinant chemokine injection.
50 µg each of anti-CCL3 (MAB450), anti-CCL4 (MAB451), and anti-CCL5 (MAB478; all from RND Systems) were given i.v. in sterile saline at the same time as infection. Alternatively, 150 µg rat IgG2a (BioXcell) was given in the same manner. 1 µg of rCCL3 (R&D Systems) in sterile saline was given s.c. 45 min to 1 h before imaging.
CFSE proliferation.
Spleens, inguinal, brachial, cervical, and mesenteric LNs were removed and homogenized to produce single-cell suspensions. Red blood cells were lysed, and samples were filtered through a 70-µm nylon filter. Cells were labeled for 10 min at room temperature in 2 µM CFSE (Invitrogen). 5 × 106 OT-I cells were transferred i.v. into CD11c-DTR-eGFP mice, which were treated with DTx or PBS i.p. 12 h later, mice were infected s.c. with 3.5 × 107 PFU VV-NP-S-eGFP. 48 hpi, inguinal nodes were removed and analyzed by flow cytometry. OT-I cells were stained for CD45.1 (clone A20) and CD8 (clone 53–6.7) from eBioscience as well as propidium iodide, and analyzed for CFSE fluorescence on a BD LSRII flow cytometer (BD). Proliferation was then examined using FlowJo (Tree Star).
OT-I cellular activation assays.
12 h before infection s.c. with 3.5 × 107 PFU VV-ovalbumin, 5 × 106 OT-I cells were transferred i.v. into WT mice. For activation marker analysis, OT-I cells were removed from the draining ILN at indicated time points after infection using homogenization. Red bloods cells were lysed, and samples were filtered through a 70 µm nylon filter. Single-cell suspensions were stained for CD69 (clone H1.2F3), CD25 (clone PC61), CD62L (clone Mel-14), CD45.1 (clone A20), and CD8 (clone 53–6.7; eBioscience). Results were compiled from four independent experiments by normalizing the maximum CD69 MFI per experiment to 100. Where indicated, mice received a single injection of chemokine-neutralizing Abs at the same time as infection.
For analysis of IFN-γ production, WT or CD11c-DTR-eGFP mice (indicated in figure legends) were infected s.c. with 2.5 × 107 PFU VV-ovalbumin or VV-NP-S-eGFP. Where indicated, mice received a single injection of chemokine-neutralizing Abs i.v. concomitant with infection. Inguinal LNs were harvested 48 hpi and homogenized, and cells were resuspended in RPMI-10 + 10 mM Hepes buffer and plated at 2 × 106 cells/well in U-bottom, 96-well plates along with SIINFEKL or an irrelevant control peptide (SSIEFARL) at a final concentration of 100 nM. Cells and peptide were incubated for 3 h. at 37°C in the presence of 10 µg/ml brefeldin A (Sigma-Aldrich) to allow IFN-γ accumulation. For dead cell exclusion, cells were treated with ethidium monoazide (Invitrogen) before washing, incubating with Fc block (antibody 2.4G2 produced in house), and staining with anti-CD8 (clone 53–6.7) and anti-CD45.1 (clone A20; eBioscience). After staining, cells were washed and fixed at room temperature for 20 min with 1% paraformaldehyde. Cells were next incubated overnight at 4°C with Alexa Fluor 647 anti–IFN-γ (clone XMG1.2, eBioscience) diluted in PBS containing 0.5% saponin (EMD Biosciences). Cells were analyzed on a BD LSR II and the MFI of IFN-γ signal normalized to the maximum MFI in a given experiment. Results were compiled from three individual experiments.
Online supplemental material.
Video 1 shows the MRR CD11cYFP mice. Video 2 shows macrophages and DCs in the inguinal LN. Video 3 shows OT-I cells interacting with DCs outside the MRR. Video 4 shows OT-I cells interacting with different DCs. Video 5 shows T cells in relation to the MRR after infection. Video 6 shows infected macrophages in the LN. Video 7 shows T cells in the MRR in the absence of DCs. Video 8 shows T cell contacts in DC ablated nodes. Video 9 shows T cell location in the presence of chemokine neutralizing Abs. Video 10 shows T cell behavior in the presence of rCCL3. Video 11 shows CCR5 KO OT-I T cell location after VV infection. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20102545/DC1.
Acknowledgments
We thank the NIAID Bldg. 33 Comparative Medicine Branch for their support in care for the mice used in this study.
This work was funded by the Division of Intramural Research, NIAID, National Institutes of Health
The authors have no conflicting financial interests.
Author contributions: H.D. Hickman, J.W. Yewdell, and J.R. Bennink designed experiments and wrote the manuscript. H.D. Hickman, L. Li, G.V. Reynoso, E.J. Rubin, C.N. Skon, and J.W. Mays performed experiments. H.D. Hickman and O. Schwartz devised MPM methods. H.D. Hickman analyzed the data. J. Gibbs created VV-NP-S-mCherry.
Footnotes
Abbreviations used:
- Ab
- antibody
- CR
- cortical ridge
- DLN
- draining LN
- DTR
- DTx receptor
- DTx
- diphtheria toxin
- eGFP
- enhanced GFP
- eYFP
- enhanced yellow fluorescent protein
- hpi
- hours post infection
- MPM
- multiphoton microscopy
- MRR
- macrophage-rich region
- PIR
- peripheral interfollicular region
- SCS
- subcapsular sinus
- VV
- vaccinia virus
References
- Allan R.S., Smith C.M., Belz G.T., van Lint A.L., Wakim L.M., Heath W.R., Carbone F.R. 2003. Epidermal viral immunity induced by CD8alpha+ dendritic cells but not by Langerhans cells. Science. 301:1925–1928 10.1126/science.1087576 [DOI] [PubMed] [Google Scholar]
- Asano K., Nabeyama A., Miyake Y., Qiu C.H., Kurita A., Tomura M., Kanagawa O., Fujii S., Tanaka M. 2011. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity. 34:85–95 10.1016/j.immuni.2010.12.011 [DOI] [PubMed] [Google Scholar]
- Badovinac V.P., Haring J.S., Harty J.T. 2007. Initial T cell receptor transgenic cell precursor frequency dictates critical aspects of the CD8(+) T cell response to infection. Immunity. 26:827–841 10.1016/j.immuni.2007.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajénoff M., Granjeaud S., Guerder S. 2003. The strategy of T cell antigen-presenting cell encounter in antigen-draining lymph nodes revealed by imaging of initial T cell activation. J. Exp. Med. 198:715–724 10.1084/jem.20030167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista F.D., Harwood N.E. 2009. The who, how and where of antigen presentation to B cells. Nat. Rev. Immunol. 9:15–27 10.1038/nri2454 [DOI] [PubMed] [Google Scholar]
- Bedoui S., Prato S., Mintern J., Gebhardt T., Zhan Y., Lew A.M., Heath W.R., Villadangos J.A., Segura E. 2009. Characterization of an immediate splenic precursor of CD8+ dendritic cells capable of inducing antiviral T cell responses. J. Immunol. 182:4200–4207 10.4049/jimmunol.0802286 [DOI] [PubMed] [Google Scholar]
- Belz G.T., Smith C.M., Eichner D., Shortman K., Karupiah G., Carbone F.R., Heath W.R. 2004. Cutting edge: conventional CD8 alpha+ dendritic cells are generally involved in priming CTL immunity to viruses. J. Immunol. 172:1996–2000 [DOI] [PubMed] [Google Scholar]
- Carrasco Y.R., Batista F.D. 2007. B cells acquire particulate antigen in a macrophage-rich area at the boundary between the follicle and the subcapsular sinus of the lymph node. Immunity. 27:160–171 10.1016/j.immuni.2007.06.007 [DOI] [PubMed] [Google Scholar]
- Castellino F., Huang A.Y., Altan-Bonnet G., Stoll S., Scheinecker C., Germain R.N. 2006. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 440:890–895 10.1038/nature04651 [DOI] [PubMed] [Google Scholar]
- Ciavarra R.P., Taylor L., Greene A.R., Yousefieh N., Horeth D., van Rooijen N., Steel C., Gregory B., Birkenbach M., Sekellick M. 2005. Impact of macrophage and dendritic cell subset elimination on antiviral immunity, viral clearance and production of type 1 interferon. Virology. 342:177–189 10.1016/j.virol.2005.07.031 [DOI] [PubMed] [Google Scholar]
- Ciavarra R.P., Stephens A., Nagy S., Sekellick M., Steel C. 2006. Evaluation of immunological paradigms in a virus model: are dendritic cells critical for antiviral immunity and viral clearance? J. Immunol. 177:492–500 [DOI] [PubMed] [Google Scholar]
- Clarke S.R., Barnden M., Kurts C., Carbone F.R., Miller J.F., Heath W.R. 2000. Characterization of the ovalbumin-specific TCR transgenic line OT-I: MHC elements for positive and negative selection. Immunol. Cell Biol. 78:110–117 10.1046/j.1440-1711.2000.00889.x [DOI] [PubMed] [Google Scholar]
- den Haan J.M., Lehar S.M., Bevan M.J. 2000. CD8(+) but not CD8(-) dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192:1685–1696 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan B.P., Bennink J.R., Yewdell J.W. 2011. Translating DRiPs: progress in understanding viral and cellular sources of MHC class I peptide ligands. Cell. Mol. Life Sci. 68:1481–1489 10.1007/s00018-011-0656-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.W., Park C.G., et al. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science. 315:107–111 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- Earl P.L., Moss B., Wyatt L.S., Carroll M.W. 2001. Generation of recombinant vaccinia viruses. Curr Protoc Protein Sci. Chapter 5:Unit5 13 [DOI] [PubMed] [Google Scholar]
- Fruci D., Lauvau G., Saveanu L., Amicosante M., Butler R.H., Polack A., Ginhoux F., Lemonnier F., Firat H., van Endert P.M. 2003. Quantifying recruitment of cytosolic peptides for HLA class I presentation: impact of TAP transport. J. Immunol. 170:2977–2984 [DOI] [PubMed] [Google Scholar]
- Gonzalez S.F., Lukacs-Kornek V., Kuligowski M.P., Pitcher L.A., Degn S.E., Kim Y.A., Cloninger M.J., Martinez-Pomares L., Gordon S., Turley S.J., Carroll M.C. 2010. Capture of influenza by medullary dendritic cells via SIGN-R1 is essential for humoral immunity in draining lymph nodes. Nat. Immunol. 11:427–434 10.1038/ni.1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S., Taylor P.R. 2005. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 5:953–964 10.1038/nri1733 [DOI] [PubMed] [Google Scholar]
- Gretz J.E., Norbury C.C., Anderson A.O., Proudfoot A.E., Shaw S. 2000. Lymph-borne chemokines and other low molecular weight molecules reach high endothelial venules via specialized conduits while a functional barrier limits access to the lymphocyte microenvironments in lymph node cortex. J. Exp. Med. 192:1425–1440 10.1084/jem.192.10.1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrickson S.E., Mempel T.R., Mazo I.B., Liu B., Artyomov M.N., Zheng H., Peixoto A., Flynn M.P., Senman B., Junt T., et al. 2008. T cell sensing of antigen dose governs interactive behavior with dendritic cells and sets a threshold for T cell activation. Nat. Immunol. 9:282–291 10.1038/ni1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman H.D., Takeda K., Skon C.N., Murray F.R., Hensley S.E., Loomis J., Barber G.N., Bennink J.R., Yewdell J.W. 2008. Direct priming of antiviral CD8+ T cells in the peripheral interfollicular region of lymph nodes. Nat. Immunol. 9:155–165 10.1038/ni1557 [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.M., Pratt J.R., Akers W.J., Achilefu S.I., Yokoyama W.M. 2009. Murine cytomegalovirus displays selective infection of cells within hours after systemic administration. J. Gen. Virol. 90:33–43 10.1099/vir.0.006668-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugues S., Scholer A., Boissonnas A., Nussbaum A., Combadière C., Amigorena S., Fetler L. 2007. Dynamic imaging of chemokine-dependent CD8+ T cell help for CD8+ T cell responses. Nat. Immunol. 8:921–930 10.1038/ni1495 [DOI] [PubMed] [Google Scholar]
- Hume D.A. 2008. Macrophages as APC and the dendritic cell myth. J. Immunol. 181:5829–5835 [DOI] [PubMed] [Google Scholar]
- Iannacone M., Moseman E.A., Tonti E., Bosurgi L., Junt T., Henrickson S.E., Whelan S.P., Guidotti L.G., von Andrian U.H. 2010. Subcapsular sinus macrophages prevent CNS invasion on peripheral infection with a neurotropic virus. Nature. 465:1079–1083 10.1038/nature09118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyoda T., Shimoyama S., Liu K., Omatsu Y., Akiyama Y., Maeda Y., Takahara K., Steinman R.M., Inaba K. 2002. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J. Exp. Med. 195:1289–1302 10.1084/jem.20020161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Unutmaz D., Wong P., Sano G., De los Santos K., Sparwasser T., Wu S., Vuthoori S., Ko K., Zavala F., et al. 2002. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 17:211–220 10.1016/S1074-7613(02)00365-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junt T., Moseman E.A., Iannacone M., Massberg S., Lang P.A., Boes M., Fink K., Henrickson S.E., Shayakhmetov D.M., Di Paolo N.C., et al. 2007. Subcapsular sinus macrophages in lymph nodes clear lymph-borne viruses and present them to antiviral B cells. Nature. 450:110–114 10.1038/nature06287 [DOI] [PubMed] [Google Scholar]
- Kassim S.H., Rajasagi N.K., Zhao X., Chervenak R., Jennings S.R. 2006. In vivo ablation of CD11c-positive dendritic cells increases susceptibility to herpes simplex virus type 1 infection and diminishes NK and T-cell responses. J. Virol. 80:3985–3993 10.1128/JVI.80.8.3985-3993.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakai T., Hara T., Lee J.H., Gonda H., Sugai M., Shimizu A. 2004. A novel reticular stromal structure in lymph node cortex: an immuno-platform for interactions among dendritic cells, T cells and B cells. Int. Immunol. 16:1133–1142 10.1093/intimm/dxh113 [DOI] [PubMed] [Google Scholar]
- Lev A., Takeda K., Zanker D., Maynard J.C., Dimberu P., Waffarn E., Gibbs J., Netzer N., Princiotta M.F., Neckers L., et al. 2008. The exception that reinforces the rule: crosspriming by cytosolic peptides that escape degradation. Immunity. 28:787–798 10.1016/j.immuni.2008.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev A., Princiotta M.F., Zanker D., Takeda K., Gibbs J.S., Kumagai C., Waffarn E., Dolan B.P., Burgevin A., Van Endert P., et al. 2010. Compartmentalized MHC class I antigen processing enhances immunosurveillance by circumventing the law of mass action. Proc. Natl. Acad. Sci. USA. 107:6964–6969 10.1073/pnas.0910997107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist R.L., Shakhar G., Dudziak D., Wardemann H., Eisenreich T., Dustin M.L., Nussenzweig M.C. 2004. Visualizing dendritic cell networks in vivo. Nat. Immunol. 5:1243–1250 10.1038/ni1139 [DOI] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397 10.1126/science.1171243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mempel T.R., Henrickson S.E., Von Andrian U.H. 2004. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature. 427:154–159 10.1038/nature02238 [DOI] [PubMed] [Google Scholar]
- Norbury C.C., Malide D., Gibbs J.S., Bennink J.R., Yewdell J.W. 2002. Visualizing priming of virus-specific CD8+ T cells by infected dendritic cells in vivo. Nat. Immunol. 3:265–271 10.1038/ni762 [DOI] [PubMed] [Google Scholar]
- Norbury C.C., Basta S., Donohue K.B., Tscharke D.C., Princiotta M.F., Berglund P., Gibbs J., Bennink J.R., Yewdell J.W. 2004. CD8+ T cell cross-priming via transfer of proteasome substrates. Science. 304:1318–1321 10.1126/science.1096378 [DOI] [PubMed] [Google Scholar]
- Phan T.G., Grigorova I., Okada T., Cyster J.G. 2007. Subcapsular encounter and complement-dependent transport of immune complexes by lymph node B cells. Nat. Immunol. 8:992–1000 10.1038/ni1494 [DOI] [PubMed] [Google Scholar]
- Phan T.G., Green J.A., Gray E.E., Xu Y., Cyster J.G. 2009. Immune complex relay by subcapsular sinus macrophages and noncognate B cells drives antibody affinity maturation. Nat. Immunol. 10:786–793 10.1038/ni.1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst H.C., Tschannen K., Odermatt B., Schwendener R., Zinkernagel R.M., Van Den Broek M. 2005. Histological analysis of CD11c-DTR/GFP mice after in vivo depletion of dendritic cells. Clin. Exp. Immunol. 141:398–404 10.1111/j.1365-2249.2005.02868.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph G.J., Ochando J., Partida-Sánchez S. 2008. Migration of dendritic cell subsets and their precursors. Annu. Rev. Immunol. 26:293–316 10.1146/annurev.immunol.26.021607.090254 [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Salazar-Mather T.P., Biron C.A., Kuziel W.A., Pamer E.G. 2003. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 19:59–70 10.1016/S1074-7613(03)00171-7 [DOI] [PubMed] [Google Scholar]
- Serbina N.V., Jia T., Hohl T.M., Pamer E.G. 2008. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 26:421–452 10.1146/annurev.immunol.26.021607.090326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortman K., Liu Y.J. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2:151–161 10.1038/nri746 [DOI] [PubMed] [Google Scholar]
- Swiecki M., Colonna M. 2010. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol. Rev. 234:142–162 10.1111/j.0105-2896.2009.00881.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.R., Martinez-Pomares L., Stacey M., Lin H.H., Brown G.D., Gordon S. 2005. Macrophage receptors and immune recognition. Annu. Rev. Immunol. 23:901–944 10.1146/annurev.immunol.23.021704.115816 [DOI] [PubMed] [Google Scholar]
- Vremec D., Shortman K. 1997. Dendritic cell subtypes in mouse lymphoid organs: cross-correlation of surface markers, changes with incubation, and differences among thymus, spleen, and lymph nodes. J. Immunol. 159:565–573 [PubMed] [Google Scholar]
- Vremec D., Pooley J., Hochrein H., Wu L., Shortman K. 2000. CD4 and CD8 expression by dendritic cell subtypes in mouse thymus and spleen. J. Immunol. 164:2978–2986 [DOI] [PubMed] [Google Scholar]
- Wu C., Rauch U., Korpos E., Song J., Loser K., Crocker P.R., Sorokin L.M. 2009. Sialoadhesin-positive macrophages bind regulatory T cells, negatively controlling their expansion and autoimmune disease progression. J. Immunol. 182:6508–6516 10.4049/jimmunol.0804247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler-Heitbrock L., Ancuta P., Crowe S., Dalod M., Grau V., Hart D.N., Leenen P.J., Liu Y.J., MacPherson G., Randolph G.J., et al. 2010. Nomenclature of monocytes and dendritic cells in blood. Blood. 116:e74–e80 10.1182/blood-2010-02-258558 [DOI] [PubMed] [Google Scholar]